Introduction
Psoriasis is a T cell-mediated, chronic,
inflammatory and hyperproliferative cutaneous disorder that affects
~2% of the general population. Activated T cells are found in
psoriatic plaques and in the circulation, and have been shown to
set off a series of cellular and molecular reactions resulting in
the creation of psoriatic lesions (1–3).
Reagents that inhibit T-cell activation or function, and that have
been shown to have efficacy in the treatment of psoriasis include
specific immunosuppressive agents, immunomodulatory drugs, fusion
proteins that block T-cell activation or the anergizing of T cells,
cytokines and biologics that inhibit T-cell migration (4–8).
However, recurrence or exacerbation often occurs following disease
resolution when treatment with these agents is suddenly halted,
which indicates that T-cell activation is not effectively blocked.
It is believed that pathogenic T cells are activated in the
periphery by a variety of infectious and non-infectious exogenous
and endogenous factors (1).
However, there is evidence that intrinsic factors play more
important roles than extrinsic factors in the pathogenesis of
psoriasis (9,10). These intrinsic factors may be
involved in spontaneous T-cell activation or proliferation, the
regulation of cytokine production pathways, hematopoietic cell
development, and T-cell development in the thymus (11).
In humans, bone marrow-derived T-cell progenitors
express cluster of differentiation (CD)34 and CD45 (12). These hematopoietic progenitor cells
from the bone marrow travel through the blood to seed the thymus,
attracted by chemokines, including chemokine (C-C motif) ligand 25
(13). The CD4/CD8-double-negative
(DN) population contains the early thymic progenitor cells in the
thymus, and the DN1 population is home to the earliest thymocyte
progenitors at the corticomedullary junction. The outward migration
of these cells to the cortex in response to specific chemokines,
including chemokine (C-X-C motif) ligand 12, is accompanied by
their differentiation into DN2 and DN3 cells (14). Differentiation into DN4 cells and
subsequently, into CD4/CD8-double-positive (DP) cells, occurs along
with the inward migration of the cells from the cortex to the
medulla, where selection and maturation into CD4 or CD8
single-positive (SP) T cells takes place. Having escaped negative
selection, the positively-selected thymocytes mature into naive T
cells and leave the thymus to enter the peripheral circulation
(15).
Several observations indicate a pathogenic role for
bone marrow hematopoietic stem cells (BMHSCs) in psoriasis.
Firstly, Zhang et al reported that in
vitro-differentiated T cells from bone marrow CD34+
progenitor cells of patients with psoriasis are functionally
similar to the circulating T cells of these patients. The
differentiated T cells from the studied patients with psoriasis
showed higher proliferation and a marked capacity to secret Th1
cytokines in response to streptococcal superantigen, and to induce
the overexpression of C-myc and Ki67, but not B-cell lymphoma-extra
large (Bcl-XL), in keratinocytes (16). Secondly, a study of
high-proliferative potential colony-forming cells (HPP-CFCs)
isolated from patients with psoriasis revealed that the bone marrow
of patients with psoriasis is pathogenic by its very nature, and is
deficient for bone marrow hematopoietic cells (17). Thirdly, runt-related transcription
factor 1 (RUNX1), with a restricted expression pattern, is
essential for hematopoietic cell development (18). Furthermore, the genetic defect that
lies between the sodium-hydrogen antiporter 3 regulator 1 and
N-acetyltransferase 9 genes on chromosome 17q25 and which results
in the loss of a RUNX1 binding site may be involved in
psoriasis (19). Finally,
allogeneic bone marrow transplantation (BMT) provides evidence to
support the hematopoietic basis of psoriasis susceptibility.
Reconstituting T-cell populations by allogeneic BMT into psoriatic
individuals results in long-term remission or amelioration of the
disease (20,21). By contrast, BMT from psoriatic
donors into patients with no history of psoriasis results in
development of psoriasis (22).
Therefore, bone marrow hematopoietic precursor cells with abnormal
expression of specific genes are believed to be responsible for the
T cell-related immune dysregulation in psoriasis.
To test this hypothesis, suppression subtractive
hybridization (SSH) was performed on the BMHSCs of a patient with
psoriasis and a healthy control. In the current study, 17
differentially-expressed sequence tags (ESTs), including the CD45
gene were obtained from the forward- and reverse-SSH libraries.
Since CD45 is known to be an important regulator of signal
transduction in the process of T-cell development and activation
(23), the CD45 expression levels
in the bone marrow-derived hematopoietic progenitors and peripheral
blood cells (PBCs) of 20 patients with psoriasis and 10 healthy
subjects were analyzed by quantitative polymerase chain reaction
(qPCR) and flow cytometry.
Materials and methods
Participants
A total of 20 patients with psoriasis vulgaris (16
males and 4 females; median age, 36.8 years; range, 20–58 years)
participated in the study. Eligible patients were those diagnosed
with plaque psoriasis lasting for at least 6 months, a family
history of psoriasis, at least 10% of the total body surface area
affected, and a mean psoriasis area and severity index (PASI) score
of 16.87±1.26 [mean ± standard deviation (SD)]. The control group
consisted of 10 age- and sex-matched healthy individuals. None of
the patients or control subjects had received topical or oral
medications in the 6 months prior to the study. Among the subjects,
the bone marrow samples of a female patient with psoriasis and a
healthy control were used to construct a subtractive cDNA library.
Bone marrow and peripheral blood samples were collected from all
participants and used to check the CD45 expression levels in the
BMHSCs and PBCs. This study was approved by the Ethics Committee of
the First Affiliated Hospital of Dalian Medical University. All
participants were fully aware of the purpose of the study and
provided written informed consent.
Isolation of CD34+ cells from
bone marrow
From each subject, 10 ml of bone marrow aspirate was
drawn into tubes containing sodium heparin. Bone marrow mononuclear
cells (BMMNCs) were isolated from bone marrow aspirates by density
gradient centrifugation in Ficoll1077 (Sigma Chemical Co., St.
Louis, MO, USA) followed by lysis of the remaining erythrocytes
with 0.15 M Tris-ammonium chloride. CD34+ cells were
isolated from BMMNCs with magnetic beads conjugated to anti-human
CD34 antibodies and anti-idiotype antibodies (Dynal Biotech,
Milwaukee, Wisconsin, USA) according to the manufacturer’s
instructions. The purity of the CD34 cells and the percentage of
remaining CD1a CD3 cells were evaluated by flow cytometry with
fluorescein isothiocyanate (FITC)-labeled anti-CD34 antibody,
phycoerythrin (PE)-conjugated anti-CD1a antibody and
FITC-conjugated anti-CD3 antibody (BD Pharmingen, Tokyo,
Japan).
Subtractive cDNA library
construction
Total RNA was extracted with the TRIzol Reagent kit
(Invitrogen Life Technologies, Carlsbad, CA, USA), and mRNA was
purified with the Dynabeads mRNA Purification kit (Dynal Biotech,
Oslo, Norway), according to the manufacturers’ instructions. cDNA
was synthesized and amplified using the SMART PCR cDNA synthesis
kit (BD Biosciences-Clontech, Palo Alto, CA, USA). In the current
study, 2 mg total RNA was used to generate each cDNA population for
use in the subtraction procedure. The manufacturer’s
recommendations were used throughout the cDNA synthesis procedure.
Differentially-expressed genes were identified using the Clontech
PCR-Select cDNA Subtraction kit (BD Biosciences-Clontech). In
brief, the SMART cDNA of the patient and healthy control were
subjected to RsaI digestion, adaptor ligation, two rounds of
subtractive hybridization and subsequent PCR amplifications. The
forward-subtracted cDNA, i.e., cDNA from the patient as the tester
and cDNA from the healthy control as the driver, and the
reverse-subtracted cDNA, i.e., cDNA from the healthy control as the
tester and cDNA from yeast as the driver, were inserted into the
pMD-T18 vector (Takara, Tokyo, Japan) and transformed into
Escherichia coli DH5a. The forward- and reverse-subtracted
cDNA plasmid libraries were constructed and individual colonies
that showed the presence of inserted DNA were randomly picked and
analyzed.
Screening for differentially-expressed
genes by cDNA array dot blot screening
From the two subtracted cDNA libraries, 600
recombinant bacterial clones were picked by blue-white selection on
plates. The white colonies were incubated at 37°C for 12 h in
Eppendorf tubes in 1 ml Luria-Bertani (LB) broth that contained 50
μg/ml ampicillin. Each bacterium in the LB culture was amplified in
a 50 μl PCR system using the nested PCR primers: 1,
5′-TCGAGCGGCCGCCCGGGCAGGT-3′; and 2R, 5′-AGCGTGGTCGCGGCCGAGGT-3′.
PCR was performed as follows: 95°C for 2 min, 35 cycles of 95°C for
30 sec, 62°C for 45 sec and 72°C for 1 min, and 72°C for 6 min. The
PCR product was confirmed on a 2% agarose gel and the remainder was
used for differential screening.
In total, 571 clones with cDNA inserts from two
subtractive libraries were selected randomly for cDNA array dot
blot screening. Briefly, the PCR product (5 μl) was denatured with
an equal volume of 0.5 M NaOH, and 1.5 μl of this mixture was
spotted onto two identical nitrocellulose membranes (Roche, Basel,
Switzerland). The two identical blots were UV cross-linked and
hybridized with digoxigenin (DIG)-labeled forward- and
reverse-subtracted cDNA probes, which were prepared using DIG High
Prime DNA Labeling and Detection Starter kit II (Roche). Hybrids
were detected using an alkaline phosphatase-conjugated anti-DIG
antibody (1:1,000 dilution). The chemiluminescent signals were
generated by treating the membranes with 1% CSPD, and recorded on
an X-ray film (Fuji Biomax MR film; Fujifilm, Minato, Tokyo,
Japan).
Sequencing and sequence analysis
In total, 144 clones identified by differential
screening of the two SSH libraries were sequenced with the
universal M13 sequencing primer using an automatic DNA sequencer
(ABI Applied Biosystems Model 3730). All inserted sequences were
queried for similarity in the National Center for Biotechnology
Information (NCBI) database using the BLASTX program (http://www.ncbi.nlm.nih.gov/BLAST).
Relative quantification of CD45 mRNA
expression in BMHSCs by qPCR
The CD45 EST was selected for further analysis by
qPCR. The primers and TaqMan probe for the target gene were
designed using Primer Select in DNASTAR software (Lasergene,
Madison, WI, USA) and are listed in Table I. The levels of RNA (2 μg of each)
were assayed in the BMHSCs of 20 patients and 10 healthy controls
using the SYBR ExScript RT-PCR kit (Takara). The amplification
conditions were optimized for the ABI PRISM-7500 instrument
(Applied Biosystems). The cycling conditions using TaqMan probe
detection were 95°C for 2 min and 40 cycles of 95°C for 10 sec,
61°C for 10 sec and 72°C for 40 sec. The β-actin gene was selected
as the endogenous control. Relative quantification of target gene
expression was evaluated using the comparative cycle threshold (CT)
method, as previously described (24). The ΔCT value was determined by
subtracting the target CT of each sample from its respective
β-actin CT value. Calculation of ΔΔCT involved using the healthy
control sample ΔCT value as an arbitrary constant to subtract from
the ΔCT values of the patient sample. Differences in the expression
of target genes were determined by calculating
2−ΔΔCT.
 | Table IPrimers and Taqman probe. |
Table I
Primers and Taqman probe.
| Name | Primers and Taqman
probe | Primers for qPCR
(5′→3′) | Length, bp |
|---|
| CD45 | F |
ctgccagctgaggtgcaa | 68 |
| R |
tgcactgtcggtttccctact | |
| Taqman probe |
Fam-aaaccagtcgagtcccaaaacctcaa-Tamra | |
Flow cytometric analysis of CD45
expression in PBCs
For immunofluorescence staining, 1×106
cells were incubated with PerCP-CD45 monoclonal antibodies for 20
min at 4°C. Following two washes with phosphate-buffered saline
that contained 5% fetal calf serum, the labeled cells were analyzed
by FACSCalibur using the CELLQuest software (all BD
Biosciences-Clontech).
Statistical analysis
Data are expressed as the mean ±SD, unless indicated
otherwise. Comparisons of patients with psoriasis and healthy
control subjects were performed with an unmatched t-test.
Spearman’s non-parameter correlation analysis was used to examine
the correlation between disease severity and CD45 expression levels
in the BMHSCs and PBCs. P<0.05 was considered to indicate a
statistically significant difference.
Results
Generation and identification of
differentially-expressed cDNA fragments
Using nested PCR primers 1 and 2R, 300 recombinant
bacteria clones were amplified from the forward-SSH library, and
300 clones were amplified from the reverse-SSH library. Only clones
that had cDNA inserts that carried adaptor 1 and adaptor 2R on the
two ends were amplified. The inserts ranged in size between 200 and
1,000 bp, as assessed by agarose gel electrophoresis. Overall,
284/300 clones from the forward-SSH library and 287/300 clones from
the reverse-SSH library contained inserted fragments, and these
were subjected to further analysis. The remaining clones were
false-positives. From the two SSH libraries, 571 clones with cDNA
insert sizes that ranged in size between 200 and 1,000 bp were
selected to be undergo cDNA array dot blot screening. Of these
clones, 74 positive clones from the forward-SSH library showed
marked signals using the forward-subtracted probe compared with
using the reverse-subtracted probe, and 70 clones from the
reverse-SSH library showed marked signals with the
reverse-subtracted probe compared with the forward-subtracted
probe. These 144 positive clones were selected for further
sequencing.
Sequence analysis of
differentially-expressed genes
A total of 144 clones were sequenced and compared
with NCBI proteins using BLASTX. Significant similarities to known
genes were identified for 9/74 clones (Table II) in the forward-SSH library and
8/70 clones (Table III) in the
reverse-SSH library; the remaining clones showed low-level
similarity to known genes or represented novel genes. The clones
that showed marked similarity to known proteins were linked to: i)
Metabolism; ii) cell wall biogenesis and remodeling; iii) signal
transduction; and iv) stress. Other clones had weaker similarity
with known proteins or represented hypothetical proteins; their
roles were not confirmed.
 | Table IIGene fragments preferentially
expressed in the HSCs of a patient with psoriasis. |
Table II
Gene fragments preferentially
expressed in the HSCs of a patient with psoriasis.
| Clone number | Fragment sizes,
bp | Homologous
gene | Genotype ID | Homology, % | Gene activity |
|---|
| 6046-0259 | 384 | MMP-1 | 4312 | 47 | Matrix
metalloproteinase, degrade all components of extracellular
matrix |
| 6046-0292 | 410 | APOH | 350 | 42 | Apolipoprotein |
| 6046-0288 | 404 | IFNγ | 3458 | 100 | IFN, a soluble
cytokine with antiviral, immunoregulatory and anti-tumor properties
and is a potent activator of macrophages |
| 1067-0526 | 367 | CD45 | 5788 | 100 | Protein tyrosine
phosphatase, an essential regulator of T- and B-cell antigen
receptor signaling |
| 1067-0519 | 400 | SAE1 | 10055 | 100 | As a
SUMO-activating enzyme for the sumoylation of proteins |
| 1067-0533 | 439 | CA3 | 761 | 87 | Carbonic anhydrase
isozymes, catalyze the reversible hydration of carbon dioxide |
| 1067-0469 | 383 | Ribonuclease
T2 | 8635 | 78 | Associated with
human malignancies and chromosomal rearrangement |
| 1067-0529 | 485 | CD99 | 83692 | 55 | An adhesion
molecule during leukocyte extravasation |
| 1067-0535 | 457 | MT-ND4 | 4538 | 94 | Oxidation-reduction
enzyme, involved in oxidative phosphorylation |
 | Table IIIGene fragments preferentially
expressed in HSCs of a healthy control. |
Table III
Gene fragments preferentially
expressed in HSCs of a healthy control.
| Clone number | Fragment sizes,
bp | Homologous
gene | Genotype ID | Homology, % | Gene activity |
|---|
| 6045-0252 | 430 | CYP3A43 | 64816 | 72 | Mono oxygenases
which catalyze many reactions involved in drug metabolism and
synthesis of cholesterol, steroids and other lipids |
| 6045-0264 | 440 | ATP synthase | NULL | 92 | NULL |
| 1066-0441 | 436 | TMSB10 | 9168 | 100 | NULL |
| 1066-0547 | 446 | RABGAP1L | 9910 | 91 | G-protein
modulator |
| 1066-0459 | 500 | ART3 | 419 | 73 | ADP-ribosylation is
a reversible posttranslational modification used to regulate
protein function |
| 1066-0450 | 356 | Squalene
epoxidase | 29230 | 72 | Enzyme that
catalyzes sterol biosyntesis |
| 1066-0453 | 421 | RAPH1 | 65059 | 63 | Regulator of
transmembrane protein, involve-d in signal transduction |
| 1066-0432 | 358 | NDUFB5 | 4711 | 52 | This protein has
NADH dehydrogenase activity and oxidoreductase activity |
Marked expression of CD45 in the BMHSCs
of a patient with psoriasis
To confirm the reliability of SHH and dot blot
screening, the expression levels of CD45 were examined in the
BMHSCs of the 20 patients with psoriasis and the 10 healthy control
subjects using relative qPCR. Following amplification, the CT,
ΔΔCT, ΔΔCT and 2−ΔΔCT values were calculated
(Table IV). Notably, the
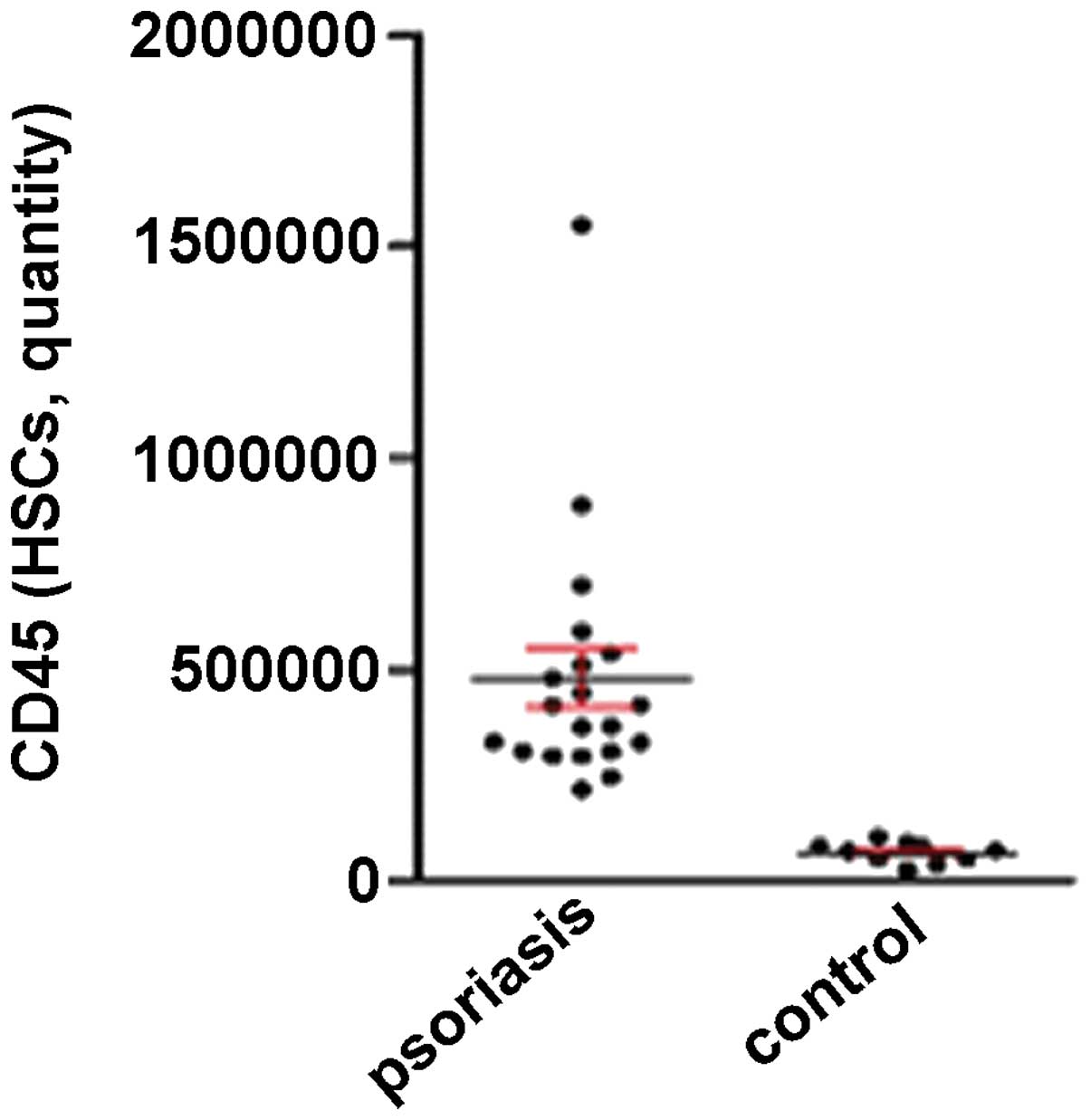
expression level of CD45 was 11.55-fold higher in the BMHSCs of the
patients with psoriasis compared with the healthy controls
(P<0.001; Fig. 1).
 | Table IVRelative abundance of CD45 gene in
HSCs, as determined by qPCR. |
Table IV
Relative abundance of CD45 gene in
HSCs, as determined by qPCR.
| | | CD45 |
|---|
| | |
|
|---|
| Group | CD45, CT | β-actin, CT | ΔCT | ΔΔCT | 2-ΔΔCT |
|---|
| Patients | 19.06±0.58 | 21.40±0.66 | −2.28±1.13 | −3.53±1.31 | 11.55a |
| Normal | 21.42±0.53 | 20.17±0.20 | 0.25±0.53 | 0.00±0.53 | 1 |
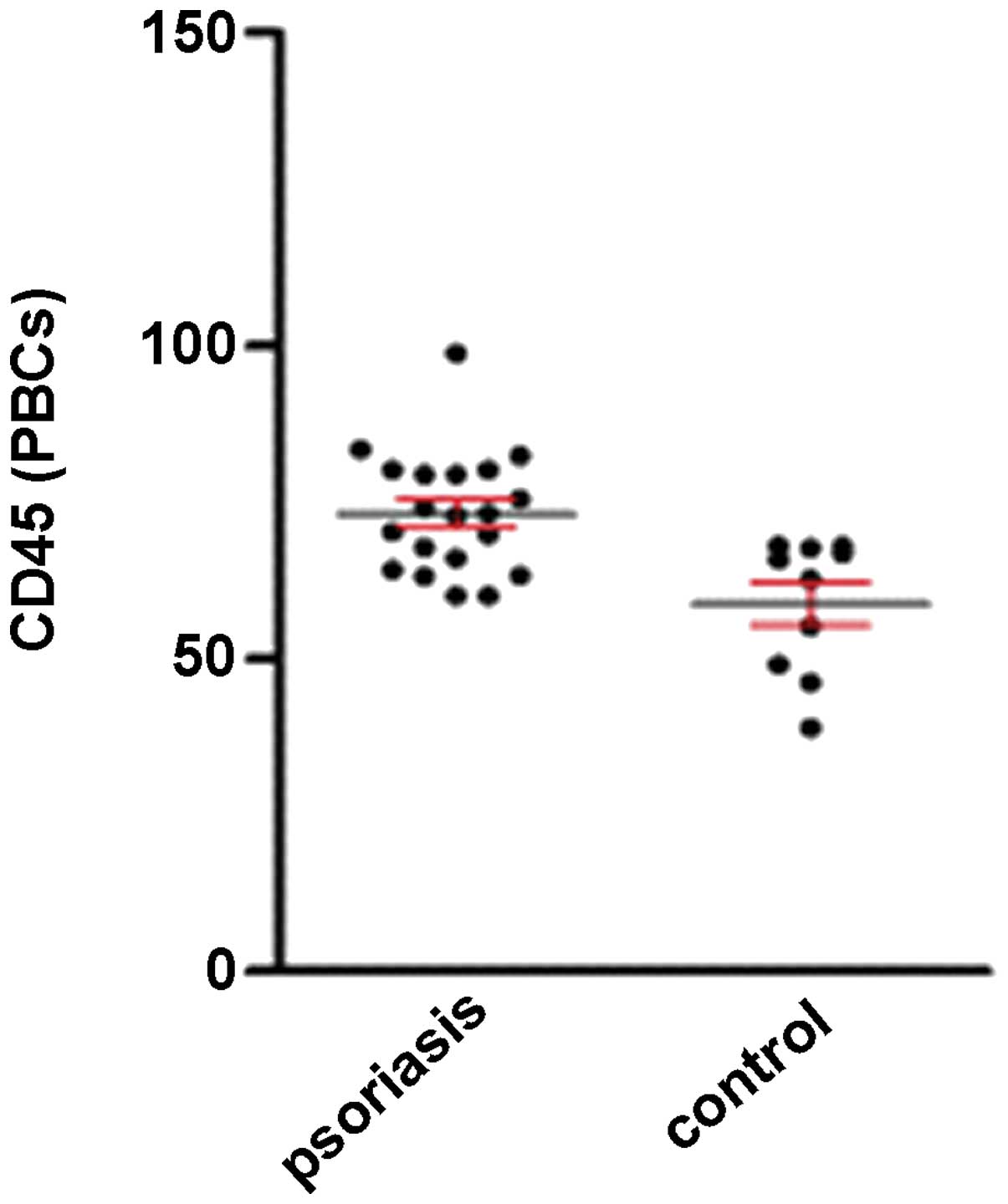
High expression level of CD45 in the PBCs
of patients with psoriasis
As shown in Fig. 2,
the expression level of CD45 in the PBCs was markedly higher in the
patients with psoriasis (73.17±2.14%) compared with the healthy
control subjects (58.81±3.40%) (P<0.05).
Correlation between disease severity and
CD45 expression levels in BMHSCs and PBCs
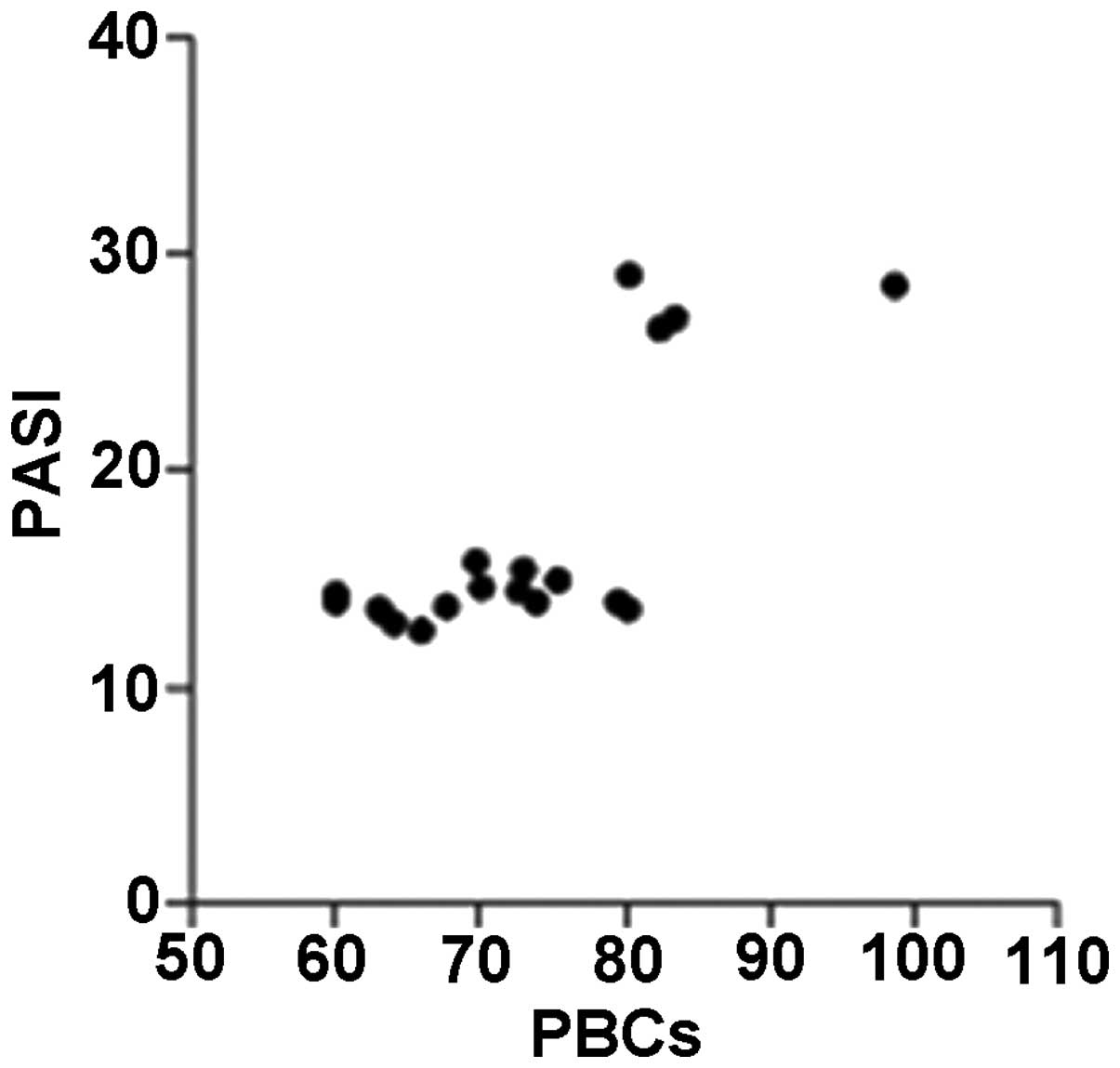
The CD45 expression levels in the PBCs of the
patients with psoriasis were significantly correlated with their
PASI scores (r=0.615; P=0.004), whereas the CD45 expression levels
in the BMHSCs were not correlated with the PASI scores (r=0.321;
P=0.168) (Figs. 3 and 4).
Discussion
Investigations of differential gene expression in
the BMHSCs of patients with psoriasis and healthy controls may lead
to the discovery of candidate genes for the pathogenicity of
psoriasis. In the present study, 17 differentially-expressed genes
were identified by suppression subtractive hybridization. These
genes were sorted into the broad functional categories of hormone
signaling, RNA catabolism, protein ADP DNA base melting,
transcriptional regulation, cell cycle regulation and metabolism.
This diversity of genes indicates that a complex series of
molecular mechanisms is involved in the pathogenesis of
psoriasis.
Of the 17 differentially-expressed genes detected in
the BMHSCs of the patient with psoriasis and a healthy control in
the present investigation, clone 1067-0526, which includes the CD45
EST, was focused upon. CD45, a leukocyte-common antigen, is a
transmembrane protein tyrosine phosphatase that is specifically
expressed on all nucleated hematopoietic cells, from stem cells to
memory cells. In humans, bone marrow-derived T-cell progenitors
have been shown to express CD34 and CD45 (12), and CD45 is an important regulator
of signal transduction at multiple stages of T-cell development
from the early precursors (23).
The expression of CD45 in hematopoietic cells is essential for
early thymocyte development, and mice and humans lacking CD45
expression are severely immunodeficient (25). In CD45-deficient mice, there is
partial blockage of DN1 and DN3 progression and an almost complete
blockage of DP development to SP cells, attributed to an increased
threshold of T-cell receptor (TCR)β, pre-TCR and TCR signaling,
respectively. A major reduction in the numbers of mature SP T cells
is consequently found in the periphery of CD45-deficient mice
(23,26). In the present study, CD45 showed
the highest repeat frequency among the forward-SSH sequences, and
was expressed 11.55-fold higher in the BMHSCs of the patients with
psoriasis compared with those of the controls. Previous studies
have indicated that the BMHSCs from patients with chronic
plaque-type psoriasis differentiate into mature T cells with normal
percentages of SP cells, albeit with increased proliferative
activity (16). Therefore, the
overexpressed CD45 in psoriatic BMHSCs is hypothesized to act as a
positive regulator of signals delivered via TCR expression during
early T-cell development, so that the proliferative ability, but
not the absolute number of mature SP T cells is markedly increased,
further involving the pathogenesis of psoriasis.
CD45 expression on mature hemocytes is critical for
the regulation of immune function, as it functions positively to
regulate T-lymphocyte activation (27). An early study demonstrated that a
CD45-deficient CD4+ T-cell clone lost reactivity to its
specific antigen and the ability to proliferate in response to a
mitogenic signal and anti-CD3 cross-linking, while it retained the
ability to proliferate in response to interleukin 2 (28). This implied that CD45 was important
in transducing the signal initiated by antigen binding to the TCR.
Subsequent studies confirmed this observation. A
CD8+/CD45− deficient T-cell clone that lost
the abilities to cytolyze its appropriate target cells and to
produce cytokines in response to the antigen was observed (29). However, transfection with CD45 cDNA
restored the activation response to TCR ligation (30). In addition, CD45 may regulate
cytokine production and responses to cytokines in other
hematopoietic cells. Previous studies have shown that CD45 is
required for interferon (IFN)-α and IFN-γ production by dendritic
cells and NK cells following stimulation via the immunoglobulin Fc
or major histocompatibility complex-binding receptors (31,32).
Compared with the healthy controls, the expression
levels of CD45 in the PBCs of the patients with psoriasis were
markedly higher in the current study. The CD45 levels in the PBCs
were observed to be closely correlated with the PASI, which is
indicative of psoriasis severity. Dawes et al produced
transgenic mice that expressed an altered level of CD45 and found
that the total level of CD45 expressed was crucial for normal TCR
signaling, lymphocyte proliferation and the production of
cytokines, including IFN-γ and tumor necrosis factor (TNF)-α
(33). Therefore, it is likely
that increased CD45 levels in PBCs are responsible for the abnormal
immune state of patients with psoriasis, which includes increased
lymphocyte proliferation activity and high levels of IFN-γ and
TNF-α production, which further exacerbate psoriasis.
Taken together, the current data indicate that the
abnormalities of hematopoietic progenitor cells that result in the
overexpression of CD45 may lead to similar abnormalities in the
PBCs via hematopoiesis. The increased level of CD45 in PBCs,
particularly on T lymphocytes, may account for the psoriatic
features, including the increased proliferative activity and
elevated secretion of Th1 cytokines.
In the present study, increased levels of IFN-γ were
observed in the BMHSCs of patients with psoriasis, which is in
accordance with the changes in IFN-γ previously observed in the
sera of patients with psoriasis (34). It is generally hypothesized that
serum IFN-γ plays an important role in the pathogenesis of
psoriasis (35). Serum cytokine
concentrations are altered by several processes, including the
production, deposition, degradation and elimination of these
molecules. Furthermore, tissue sources of cytokine production other
than circulating T cells may exist. The origin of the IFN-γ in the
sera of patients with psoriasis is not clear (36). In future studies of psoriasis,
further investigation is required to determine whether the
increased level of IFN-γ in BMHSCs contributes to the increased
serum IFN-γ level, and whether this elevation is conferred upon
peripheral T cells via hematopoiesis.
Other genes that encode proteins that affect hormone
signaling, RNA catabolism, protein ADP DNA base melting,
transcriptional regulation, cell cycle regulation and metabolism
were identified in the present study. However, as there are few
relevant studies on these genes in BMHSCs or in relation to
psoriasis, it is difficult to assign specific roles to these genes
in the development of psoriasis. Future studies that investigate
these gene expression changes may be beneficial in characterizing
the dysfunctional T cells derived from psoriatic BMHSCs.
While several genes, including p15, p21, RUNX1,
HLA-C, and PRKCB were previously found to be
overexpressed in the HSCs of patients with psoriasis (37,38),
these genes were not identified in the present study. The reasons
for this could be that earlier investigations extracted mRNA from
different samples than those employed in the present investigation.
Given the different geographic origins of each sample, there are
likely to be innate differences in the genetic structures of the
two subjects, which may be reflected in their protein profiles. In
addition, these differences may reflect a bias in choosing gene
clones of interest or technical limitations associated with the
methodologies employed.
The present data indicate that the abnormal
expression of genes, such as the augmented expression of CD45 in
the BMHSCs, may induce the production of pathogenic T lymphocytes
that play a role in the progression of psoriasis. The present
findings may have substantial implications for our understanding of
the pathophysiology of psoriasis and the development of novel
treatment strategies.
Acknowledgements
The study was supported by a grant from the National
Natural Science Foundation of China (grant nos. 30872271 and
81171507).
References
|
1
|
Nickoloff BJ: The immunologic and genetic
basis of psoriasis. Arch Dermatol. 135:1104–1110. 2000.PubMed/NCBI
|
|
2
|
Sabat R, Philipp S, Höflich C, et al:
Immunopathogenesis of psoriasis. Exp Dermatol. 16:779–798. 2007.
View Article : Google Scholar
|
|
3
|
van Lingen RG, van de Kerkhof PC, de Jong
EM, et al: Reduced CD26 bright expression of peripheral blood CD8+
T-cell subsets in psoriatic patients. Exp Dermatol. 17:343–348.
2008.PubMed/NCBI
|
|
4
|
Berth-Jones J: The use of ciclosporin in
psoriasis. J Dermatolog Treat. 16:258–277. 2005. View Article : Google Scholar
|
|
5
|
Krueger JG, Wolfe JT, Nabeya RT, et al:
Successful ultraviolet B treatment of psoriasis is accompanied by a
reversal of keratinocyte pathology and by selective depletion of
intraepidermal T cells. J Exp Med. 182:2057–2068. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gottlieb SL, Gilleaudeau P, Johnson R, et
al: Response of psoriasis to a lymphocyte-selective toxin
(DAB389IL-2) suggests a primary immune, but not keratinocyte,
pathogenic basis. Nat Med. 1:442–447. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abrams JR, Kelley SL, Hayes E, et al:
Blockade of T lymphocyte costimulation with cytotoxic T
lymphocyte-associated antigen 4-immunoglobulin (CTLA4Ig) reverses
the cellular pathology of psoriatic plaques, including the
activation of keratinocytes, dendritic cells, and endothelial
cells. J Exp Med. 192:681–694. 2000. View Article : Google Scholar
|
|
8
|
Gordon KB, Papp KA, Hamilton TK, et al:
Efalizumab for patients with moderate to severe plaque psoriasis: a
randomized controlled trial. JAMA. 290:3073–3080. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomfohrde J, Silverman A, Barnes R, et al:
Gene for familial psoriasis susceptibility mapped to the distal end
of human chromosome 17q. Science. 264:1141–1145. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hüffmeier U, Steffens M, Burkhardt H, et
al: Evidence for susceptibility determinant(s) to psoriasis
vulgaris in or near PTPN22 in German patients. J Med Genet.
43:517–522. 2006.PubMed/NCBI
|
|
11
|
Bowcock AM: The genetics of psoriasis and
autoimmunity. Annu Rev Genomics Hum Genet. 6:93–122. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Galić Z, Kitchen SG, Subramanian A, et al:
Generation of T lineage cells from human embryonic stem cells in a
feeder free System. Stem Cells. 27:100–107. 2009.PubMed/NCBI
|
|
13
|
Kondo M, Weissman IL and Akashi K:
Identification of clonogenic common lymphoid progenitors in mouse
bone marrow. Cell. 91:661–672. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Petrie HT and Zúñiga-Pflücker JC: Zoned
out: functional mapping of stromal signaling microenvironments in
the thymus. Annu Rev Immunol. 25:649–679. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Petrie HT: Cell migration and the control
of post-natal T-cell lymphopoiesis in the thymus. Nat Rev Immunol.
3:859–866. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang K, Li X, Yin G, et al: Functional
characterization of T cells differentiated in vitro from bone
marrow-derived CD34 cells of psoriatic patients with family
history. Exp Dermatol. 19:e128–e135. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang K, Zhang R, Li X, et al: The mRNA
expression and promoter methylation status of the p16 gene in
colony-forming cells with high proliferative potential in patients
with psoriasis. Clin Exp Dermatol. 32:702–708. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lacaud G, Gore L, Kennedy M, et al: Runx1
is essential for hematopoietic commitment at the hemangioblast
stage of development in vitro. Blood. 100:458–466. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Helms C, Cao L, Krueger JG, et al: A
putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is
associated with susceptibility to psoriasis. Nat Genet. 35:349–356.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanamori H, Tanaka M, Kawaguchi H, et al:
Resolution of psoriasis following allogeneic bone marrow
transplantation for chronic myelogenous leukemia: case report and
review of the literature. Am J Hematol. 71:41–44. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Woods AC and Mant MJ: Amelioration of
severe psoriasis with psoriatic arthritis for 20 years after
allogeneic haematopoietic stem cell transplantation. Ann Rheum Dis.
65:6972006.PubMed/NCBI
|
|
22
|
Snowden JA and Heaton DC: Development of
psoriasis after syngeneic bone marrow transplant from psoriatic
donor: further evidence for adoptive autoimmunity. Br J Dermatol.
137:130–132. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Byth KF, Conroy LA, Howlett S, et al:
CD45-null transgenic mice reveal a positive regulatory role for
CD45 in early thymocyte development, in the selection of
CD4+CD8+thymocytes, and B cell maturation. J Exp Med.
183:1707–1718. 1996.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)). Method. 25:402–408. 2001.
|
|
25
|
Ward V, Hennig BJ, Hirai K, et al:
Geographical distribution and disease associations of the CD45 exon
6 138G variant. Immunogenetics. 58:235–239. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kishihara K, Penninger J, Wallace VA, et
al: Normal B lymphocyte development but impaired T cell maturation
in CD45-exon6 protein tyrosine phosphatase-deficient mice. Cell.
74:143–156. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hermiston ML, Xu Z and Weiss A: CD45: a
critical regulator of signaling thresholds in immune cells. Annu
Rev Immunol. 21:107–137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pingel JT and Thomas ML: Evidence that the
leukocyte-common antigen is required for antigen-induced T
lymphocyte proliferation. Cell. 58:1055–1065. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weaver CT, Pingel JT, Nelson JO, et al:
CD8+ T-cell clones dificient in the expression of the CD45 protein
tyrosine phosphatase have impaired responses to T-cell receptor
stimuli. Mol Cell Biol. 11:4415–4422. 1991.
|
|
30
|
Koretzky GA, Kohmetscher MA, Kadleck T, et
al: Restoration of T cell receptor-mediated signal transduction by
transfection of CD45 cDNA into a CD45-deficient variant of the
Jurkat T cell line. J Immunol. 149:1138–1142. 1992.PubMed/NCBI
|
|
31
|
Montoya M, Dawes R, Reid D, Lee LN, Piercy
J, Borrow P, Tchilian EZ and Beverley PC: CD45 is required for type
I IFN production by dendritic cells. Eur J Immunol. 36:2150–2158.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shen F, Xu XL, Graf LH, et al:
CD45-cross-linking stimulates IFN-gamma production in NK cells. J
Immunol. 154:644–652. 1995.PubMed/NCBI
|
|
33
|
Dawes R, Petrova S, Liu Z, et al:
Combinations of CD45 isoforms are crucial for immune function and
disease. J Immunol. 176:3417–3425. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takahashi H, Tsuji H, Hashimoto Y,
Ishida-Yamamoto A and Iizuka H: Serum cytokines and growth factor
levels in Japanese patients with psoriasis. Clin Exp Dermatol.
35:645–649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Szegedi A, Aleksza M, Gonda A, et al:
Elevated rate of Thelper1 (T(H)1) lymphocytes and serum IFN-gamma
levels in psoriatic patients. Immunol Lett. 86:277–280. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Arican Ozer, Aral Murat, Sasmaz Sezai, et
al: Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17,
and IL-18 in patients with active psoriasis and correlation with
disease severity. Mediators Inflamm. 24:273–279. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang K, Zhang R, Li X, et al: Promoter
methylation status of p15 and p21 genes in HPP-CFCs of bone marrow
of patients with psoriasis. Eur J Dermatol. 9:141–146.
2009.PubMed/NCBI
|
|
38
|
Yin G, Li J, Wan Y, et al: Abnormality of
RUNX1 signal transduction in psoriatic CD34+ bone marrow cells. Br
J Dermatol. 164:1043–1051. 2011.PubMed/NCBI
|