Introduction
Glioma is the most common human brain tumor and is
associated with high mortality and disability. Traditional
therapies, including surgery, radiotherapy and chemotherapy have
not achieved satisfactory outcomes. As with other types of tumor,
genetic mutation is likely to be the cause of glioma. It is
possible to treat glioma through regulating the expression of
certain genes. The human N-myc downstream regulated gene
(hNDRG) family is a family of differentiation-associated
genes, which consists of four members, NDRG1, -2, -3
and -4 (1). NDRG2
was originally cloned from a normal human whole brain cDNA library
through subtractive hybridization in our laboratory (Experimental
Teaching Center of Basic Medicine, The Fourth Military Medical
University, Xi’an, China) in 1999 when analyzing different genes in
gliomas and their associated normal tissue. The NDRG2 gene
is located on chromosome 14q11.2 (2) and it has been reported that
NDRG2 may function as a tumor suppressor gene (3). NDRG2 is highly expressed in
numerous normal tissues, whereas its expression is low or
undetectable in various tumors, including lung and colon cancer.
Furthermore, transfection of NDRG2 into human cancer cell
lines results in growth inhibition (4–6).
Therefore, in the present study, it was hypothesized that
NDRG2 is a tumor suppressor gene, which may be involved in
tumorigenesis and tumor progression. NDRG2 expression was
assessed in human gliomas, and the methylation status of the CpG
islands within the NDRG2 promoter region was analyzed in the
glioma samples and their adjacent tissues. Furthermore, the
correlation between NDRG2 expression and the methylation
status of the NDRG2 promoter was investigated. The findings
of the present study support a role for NDRG2 as a tumor
suppressor gene and provide a basis for further investigation into
the mechanism underlying NDRG2-induced tumor growth
inhibition.
Materials and methods
Tissue samples
The present study was approved by the Ethics
Committee of the Fourth Military Medical University (Xi’an, China).
Fifty-three glioma tissue samples and 26 adjacent normal tissue
samples were collected from patients who underwent surgery at the
Xijing and Tangdu Hospital of the Fourth Military Medical
University (Xi’an, China) between November 2006 and September 2007.
The patients included 31 males and 22 females. The median patient
age was 42 years (range, 13–70 years). None of the patients had
received radiotherapy or chemotherapy prior to surgery. Among the
patients included in the present study, there were 24 cases of
astrocytoma, 19 cases of anaplastic astrocytoma and 10 cases of
glioblastoma multiform astrocytoma. Tumors were graded according to
the pathological classification criteria of glioma established by
the World Health Organization (7).
There were 24, 19 and 10 cases of grade II, III and IV tumors,
respectively. All patients provided informed consent according to
institutional guidelines and remained under continuous medical
supervision and assistance in accordance with the Declaration of
Helsinki. All samples were snap-frozen in liquid nitrogen and
stored at −70°C until analysis, or fixed in 10% formaldehyde and
embedded in paraffin for subsequent analysis.
Cell culture
U251 human glioblastoma cells were obtained from
American Type Culture Collection (Rockville, MD, USA) and were
cultured in Dulbecco’s modified Eagle’s medium (Invitrogen Life
Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum in a
humidified 5% CO2 atmosphere at 37°C. U251 cells served
as a typical glioblastoma sample for the subsequent methylation
analysis.
Quantitative polymerase chain reaction
(qPCR)
Total RNA was extracted from each tissue sample
(50–100 μg) using TRIzol® reagent (Invitrogen Life
Technologies) according to the manufacturer’s instructions. Total
RNA was reverse-transcribed using a ThemoScript™ RT-PCR system
(Invitrogen Life Technologies). Primers were designed from the
conserved region of their respective cDNA sequences and were
synthesized by Beijing Aoke Biotechnology Co., Ltd. (Beijing,
China). The primer sequences were as follows: Forward, 5′-GAG ATA
TGC TCT TAA CCA CCC G-3′ and reverse, 5′-GCT GCC CAA TCC ATC CAA-3′
for NDRG2 (GenBank accession no. AF159092; amplicon size, 90
bp); and forward, 5′-ATC ATG TTT GAG ACC TTC AAC A-3′ and reverse,
5′-CAT CTC TTG CTC GAA GTC CA-3′ for β-actin (GenBank
accession no. NM001101; amplicon size, 318 bp). The ΔΔCT method
(8) was used to calculate the fold
gene expression relative to the lowest tumor grade in the specific
analysis. Human β-actin mRNA served as the reference
transcript.
Western blot analysis
Total protein was extracted from the 100-μg tissue
samples from each of the eight cases of grade II, III and IV tumors
using 1 ml lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.25%
sodium taurodeoxycholate, 50 mM NaF, 5 mM EDTA, 1 mM
phenylmethanesulfonyl fluoride, 1 mM Na3NO4,
20 μg/ml aprotinin, 1 μg/ml leupeptin and 1 μg/ml pepstatin) as
described previously (9). Equal
quantities of protein were resolved using discontinuous SDS-PAGE
(6% lamination gel and 12% separation gel). Proteins were
transferred from the gel onto a pyroxylin membrane (Takara, Dalian,
China) for 2.5 h at a constant voltage of 100 V. The membranes were
blocked with Tris-buffered saline (TBS; pH 7.0) containing 5%
non-fat dried milk for 1 h at room temperature. The membrane was
subsequently washed in TBS containing Tween-20 (TBST) and incubated
overnight at 4°C with anti-NDRG2 (Abnova Co., Ltd., Taipei, Taiwan)
and -tubulin antibodies (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA). Following agitation for 1 h using a Vortex Genie 2
(Scientific Industries, Inc., Bohemia, NY, USA) the membranes
underwent four 5 min washes with TBST. The membranes were incubated
with two horseradish peroxidase-conjugated secondary antibodies
(Beijing Zhongshan Jinqiao Biotechnology Co. Ltd., Beijing, China)
at room temperature and were agitated for 1 h in a shaker and
washed with TBST four times. Immunoreactive bands were visualized
using an enhanced chemiluminescence (ECL) method and ECL reagents
(Pierce Biotechnology, Rockford, IL, USA).
Immunohistochemistry
In order to detect the expression of NDRG2, the
tissue samples were subjected to immunohistochemistry as previously
described (10). In brief, 5-μm
sections of paraffin-embedded, formalin-fixed tissue samples were
treated with 3% H2O2 at room temperature for
10 min to inhibit any endogenous peroxidase activity. Sections were
washed three times with phosphate-buffered saline (PBS) and
pretreated for antigen retrieval by microwaving in 10 mM citrate
buffer (pH 6.0) for 15 min at high power. Sections were
subsequently incubated with primary antibodies against NDRG or
Tubulin (Santa Cruz Biotechnology) overnight at 4°C and underwent
three 5 min washes with PBS containing 0.1% Tween-20. Following
washing, slides were incubated with biotin-labeled secondary
antibodies (Santa Cruz Biotechnology, Inc.) for 10 min at room
temperature and washed again. The staining was visualized using an
avidin-biotin complex and counterstaining was performed using
hematoxylin (11).
Methylation detection using bisulfite
sequencing PCR
DNA was extracted from the tumor tissues and U251
cells using a DNA extraction kit (Tiangen Biotechnology Co., Ltd.,
Beijing, China) according to the manufacturer’s instructions and
was stored at −20°C. Normal lymphocyte DNA was modified using a
SssI methyltransferase kit (New England Biolabs, Ipswich, MA, USA)
according to the manufacturer’s instructions. Genomic DNA was
modified using bisulfite (Sigma-Aldrich, St. Louis, MO, USA)
according to the method described by Herman et al (12). Non-methylated genomic DNA from
normal human peripheral blood lymphocytes served as negative
control. Genomic DNA was obtained from human lymphocytes, whose CpG
sites had been completely methylated, using the SssI
methyltransferase and served as positive control. Deionized water
was the blank control. Two overlapping fragments from the
NDRG2 promoter region, spanning 32 CpG sites between
nucleotides 20,564,110 and 20,562,476 (Genbank accession no.
NC_000014) were PCR-amplified from sodium bisulfite-modified DNA
using the following primers: Forward, 5′-AAG GAG AGT TTA TTT TAG
GGT GTG-3′ and reverse 5′-TAC CCA AAA TCC TAAT ACC TCT C-3′
(9). The amplified sequence was
530 bp and included 32 CpG sites. The recovered PCR products were
ligated into the pMD19-T Vector (Takara Bio Inc., Shiga, Japan) and
transformed into E. coli competent cells (GM109). Extracted
plasmids were digested using the PstI and EcoRI
restriction endonucleases (Takara) according to the manufacturer’s
instructions. Restriction digests were ethanol precipitated prior
to gel analysis.
Statistical analysis
Data are presented as the mean ± standard deviation.
Paired t-tests were performed to assess the differences in NDRG2
mRNA expression between tumor and adjacent normal tissue samples.
Mean mRNA levels were compared between the three tumor grades using
variance analysis and P<0.05 was considered to indicate a
statistically significant difference. Mean protein levels were
compared between the three tumor grades using variance analysis.
The proportion of promoter methylation in normal and tumor tissues
among the three grades was analyzed using Pearson’s χ2
test. The number of methylation sites in each sample was compared
between grades using the Kruskal-Wallis H test.
Results
NDRG2 expression is reduced in
glioma
Immunohistochemistry was used to analyze NDRG2
expression in 26 human glioma samples (grade II, n=9; grade III,
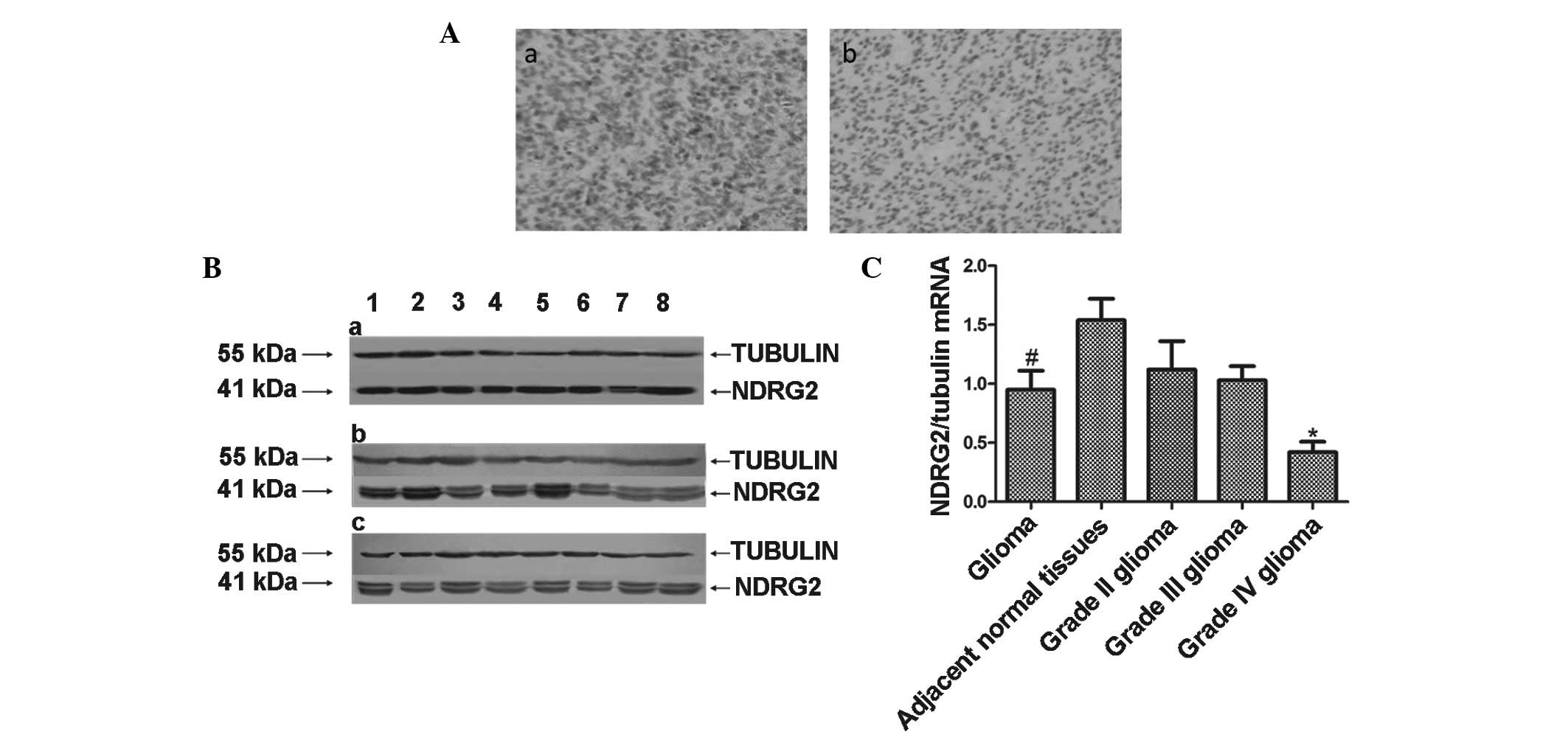
n=9; and grade IV, n=8). As shown in Fig. 1A, the expression of NDRG2 was
observed to be significantly higher in adjacent normal tissues
(Fig. 1A-a) compared with glioma
tissues (Fig. 1A-b) and NDRG2 was
observed to be primarily expressed in the cytoplasm. NDRG2 staining
was also found to decrease with increasing glioma malignancy (data
not shown). Western blot analysis was performed to further assess
the protein expression of NDRG2 in glioma tissues. The protein
expression of NDRG2 in the grade II and III glioma samples was
found to be 1.75- and 1.7-fold of that in the grade IV glioma
samples, respectively. This difference in NDRG2 protein expression
among the glioma grades was considered to be statistically
significant (P<0.05; Fig.
1B).
Furthermore, NDRG2 mRNA expression was assessed and
the findings were in accordance with those that were identified via
immunohistochemistry and western blot analysis. In 26 human brain
normal tissue samples, the expression of NDRG2 mRNA was found to be
1.7-fold that in the glioma samples (P<0.05). Furthermore, NDRG2
mRNA expression in the grade II and III glioma samples was observed
to be 2.7- and 2.5-fold of that in grade IV glioma samples.
Moreover, a significant negative correlation was identified between
NDRG2 mRNA expression and the glioma grade (P<0.05; Fig. 1C).
NDRG2 promoter methylation detection in
glioma
To further investigate the mechanisms underlying the
decrease in NDRG2 expression observed in the glioma tissue samples,
NDRG2 promoter methylation was detected in glioma tissue samples
using bisulfite sequencing PCR. In bisulfite sequencing PCR,
cytosine (C) residues in methylated CpG sites of the NDRG2 promoter
are not deaminated into uracil (U); therefore, remain as a C or
guanine in the PCR product. C residues of non-methylated CpG sites
are deaminated into U residues and are transformed into thymine or
adenine in the PCR product. Sequencing revealed that none of the
CpG sites in the NDRG2 promoter region in the genomic DNA from
normal human lymphocytes were methylated; however, following
treatment with the SssI methyltransferase, all of the CpG sites
were methylated, thus, validating the experimental procedures.
Methylated CpG sites were detected in 19 out of 41 of the glioma
tissue samples (46.3%), compared with four out of the 22 adjacent
normal tissues (18.2%), which was considered to be a significant
difference (P<0.05; Table I).
In addition, 66.7% of the 15 grade IV glioma samples, 33.3% of the
12 grade III glioma samples and 35.7% of the 14 grade II glioma
samples were observed to be methylated.
 | Table IN-myc downstream regulated gene 2
methylation in glioma and adjacent normal tissue samples. |
Table I
N-myc downstream regulated gene 2
methylation in glioma and adjacent normal tissue samples.
| Group | Samples (n) | Methylated (n) | Methylation rate
(%) |
|---|
| Glioma tissue | 41 | 19 | 46.3a |
| Adjacent normal
tissue | 22 | 4 | 18.2 |
| Grade IV | 15 | 10 | 66.7 |
| Grade III | 12 | 4 | 33.3 |
| Grade II | 14 | 5 | 35.7 |
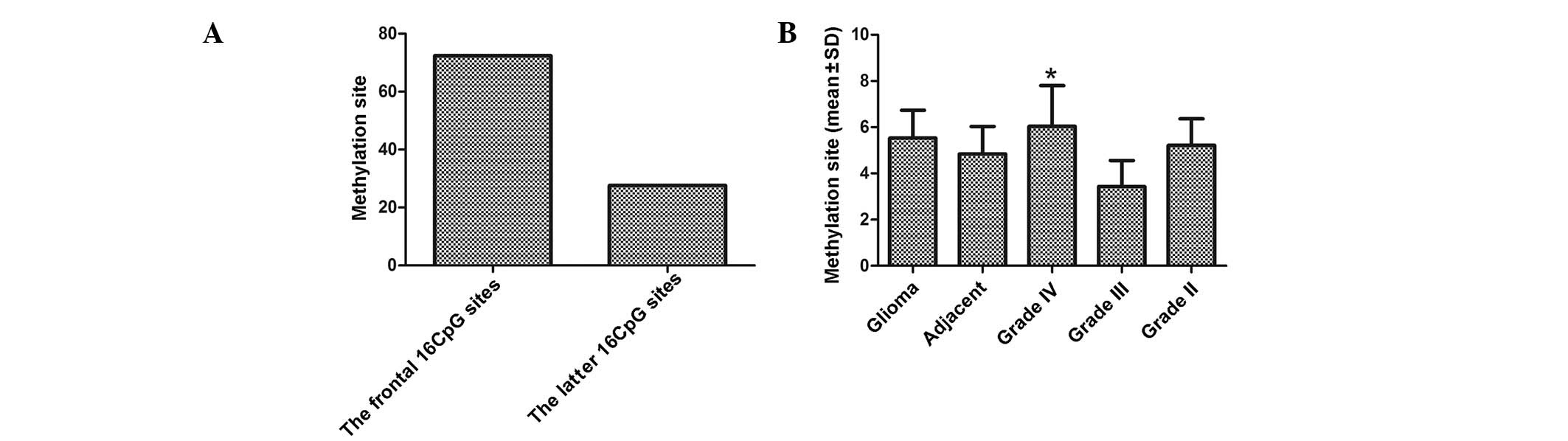
Distribution of methylation sites
Further assays were performed in order to analyze
the distribution of methylated CpG sites in the NDRG2 promoter
region. As shown in Fig. 2A,
methylation was predominantly observed in the frontal 16 CpG sites,
which were 63 of 87 (72.4%) as compared with 24 of 87 (27.6%) in
the latter 16 CpG sites (P<0.05). The average number of
methylation sites in 19 methylation-positive glioma tissues was
5.53±1.21, compared with 4.85±1.18 in the four adjacent normal
tissue samples. No significant difference was observed in the
average number of methylation sites in glioma tissue compared with
that in the normal tissue (P>0.05). The average number of
methylation sites in the 10 methylated grade IV, four methylated
grade III and five methylated grade II samples was 6.04±1.76,
3.44±1.12 and 5.22±1.14, respectively (Fig. 2B). The average number of
methylation sites in the grade IV samples compared with that in the
grade III and II samples was significantly different
(P<0.05).
DNA sequencing
To further investigate the methylation of the NDRG2
promoter region, DNA sequencing of the NDRG2 promoter was performed
in different tissues and cells. Sequencing revealed that the CpG
sites in the NDRG2 promoter region were methylated in the U251
cells and glioma tissues (Fig.
3).
Discussion
Immunohistochemistry and qPCR analysis revealed that
NDRG2 expression was significantly decreased in glioma tissues
compared with adjacent normal tissues and that the decrease in
NDRG2 expression was associated with increased glioma malignancy.
These findings indicate that decreased NDRG2 expression may have a
role in the occurrence and development of glioma. Furthermore,
bisulfite sequencing PCR revealed that NDRG2 promoter methylation
was responsible for the decrease in NDRG2 expression in glioma.
These findings may have important implications for the use of NDRG2
as a potential target for gene therapy in patients with glioma.
NDRG2 is a novel tumor suppressor candidate
gene. Previous studies have shown that NDRG2 expression is low in
numerous types of tumor tissue and cancer cell lines (11,13–15).
Moreover, induced expression of NDRG2 in cultured cancer cells has
been reported to inhibit the cell cycle at G1-phase,
indicating that NDRG2 expression inhibits cancer cell proliferation
(16). However, the mechanisms
underlying the downregulation of NDRG2 in glioma and NDRG2-induced
inhibition of cell proliferation are yet to be elucidated. Based on
a novel epigenetic theory, in the present study, it was
hypothesized that the downregulated NDRG2 expression observed in
tumors, including glioma, may be due to hypermethylation of the
NDRG2 promoter region, as no NDRG2 coding sequence mutant
has been detected in tumors. The majority of previous
investigations of NDRG2 expression have been performed in tumors
and their adjacent normal tissues. In the present study, NDRG2
expression and methylation were anlalyzed among three glioma grades
(II, III, IV), as well as in adjacent normal tissues.
Research has shown that NDRG2 expression in certain
types of tumor, including glioma, is significantly lower compared
with that in corresponding normal tissue. In the present study, a
negative correlation was observed between NDRG2 mRNA expression and
the tumor grade. Furthermore, a negative correlation was identified
between NDRG2 protein expression and tumor grade (P<0.05;
Fig. 1). In addition, in the
glioblastoma group (grade IV), NDRG2 expression was significantly
decreased, although still detectable, demonstrating that NDRG2 may
have potential as a biomarker to determine prognosis. These data
indicate that NDRG2 is a candidate glioma suppressor gene,
and NDRG2 may be important in the development and progression of
gliomas.
Previous studies have shown a correlation between
CpG island methylation and tumorigenesis. CpG island methylation
results in tumorigenesis via transcriptional silencing and loss of
function through inducing changes in chromatin structure (17–19).
In the present study the methylation rate in the NDRG2 promoter
region was observed to be higher in glioma tissues compared with
that in adjacent normal tissues. Furthermore, the methylation rate
of grade IV glioma was markedly higher when compared with grade III
and II glioma (Table I). One grade
II sample exhibited a high number of methylation sites, however,
the reason for this was unknown. No significant difference was
observed between the number of methylation sites in gliomas
compared with those in adjacent normal tissues. This may be due to
the small sample size; however, a greater percentage of methylation
was observed in the grade IV samples than in the grade III samples.
These findings indicate that methylation of the NDRG2 promoter
region is important in the development and progression of
glioma.
In the present study, methylation was detected at a
higher frequency in the frontal 16 CpG sites compared with the
latter 16 CpG sites. This indicates that the sequence including the
frontal 16 CpG sites of the NDRG2 promoter may have a significant
effect in the inhibition of gene transcription. Methylation sites
in the NDRG2 promoter region were also sequenced in a glioblastoma
sample, adjacent normal tissue sample and U251 human glioblastoma
cells. It was hypothesized that hypermethylation of the NDRG2
promoter may lead to repressed NDRG2 protein expression and
aberrant cell proliferation, which may contribute to the
pathogenesis of malignant glioma. Although it is statistically
higher than that of normal tissue, the methylation rate of glioma
samples is <50%, indicating that other mechanisms, for example
the regulation of the histone structure, may contribute to NDRG2
downregulation in glioma.
In conclusion, the present study demonstrated that
NDRG2 may be a tumor suppressor gene and methylation of the
NDRG2 promoter region may be one of the mechanisms by which NDRG2
expression is downregulated in gliomas. This mechanism of glioma
tumorigenesis may provide an important insight for glioma
treatment. Furthermore, it has been reported that demethylation
using 5-Aza-2′-deoxycytidine or S-adenosyl-methionine is capable of
restoring the expression of certain genes (13). Therefore, demethylating agents may
have potential for use in glioma treatment.
Acknowledgements
The authors would like to thank Dr Zhang Jian and Dr
Zhang Jing from the Department of Biochemistry and Molecular
Biology, and Dr Lin Wei from the Department of Neurosurgery of the
Xijing Institute of Clinical Neuroscience (Fourth Military Medical
University, Xi’an, China), as well as Dr Peter Roerig (Department
of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany)
for their technological support. The present study was supported by
a grant from the National Natural Science Foundation of China
(grant no. 30672168).
References
|
1
|
Qu X, Zhai Y, Wei H, et al:
Characterization and expression of three novel
differentiation-related genes belong to the human NDRG gene family.
Mol Cell Biochem. 229:35–44. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chun DY, Bo YL, Ping LX, et al: Exploring
a new gene containing ACP like domain in human brain and expression
it in E. coli. Prog Biochem Biophys. 28:72–76. 2001.(In
Chinese).
|
|
3
|
Lusis EA, Watson MA, Chicoine MR, et al:
Integrative genomic analysis identifies NDRG2 as a candidate tumor
suppressor gene frequently inactivated in clinically aggressive
meningioma. Cancer Res. 65:7121–7126. 2005. View Article : Google Scholar
|
|
4
|
Jian L, Ping LX, Xin LS, et al: Expression
pattern of ndr2 gene, a candidate tumor-suppressor, in different
human tissues and tumors. Prog Biochem Biophys. 29:223–227.
2002.(In Chinese).
|
|
5
|
Ellen TP, Ke Q, Zhang P and Costa M:
NDRG1, a growth and cancer related gene: regulation of gene
expression and function in normal and disease states.
Carcinogenesis. 29:2–8. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choi SC, Yoon SR, Park YP, et al:
Expression of NDRG2 is related to tumor progression and survival of
gastric cancer patients through Fas-mediated cell death. Exp Mol
Med. 39:705–714. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cavenee W, Furnari F, Nagane M, et al:
Diffusely infiltrating astrocytomas. Pathology and Genetics of
Tumours of the Nervous System. Kleihues P and Cavenee WK:
International Agency for Research on Cancer; Lyon, France: pp.
10–21. 2000
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
9
|
Tamai K, Shiina M, Tanaka N, et al:
Regulation of hepatitis C virus secretion by the Hrs-dependent
exosomal pathway. Virology. 422:377–385. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Franz M, Grün K, Richter P, et al: Extra
cellular matrix remodelling after heterotopic rat heart
transplantation: gene expression profiling and involvement of
ED-A+ fibronectin, alpha-smooth muscle actin and
B+ tenascin-C in chronic cardiac allograft rejection.
Histochem Cell Biol. 134:503–517. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Chu D, Chu X, et al: Decreased
expression of NDRG2 is related to poor overall survival in patients
with glioma. J Clin Neurosci. 18:1534–1537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee DC, Kang YK, Kim WH, et al: Functional
and clinical evidence for NDRG2 as a candidate suppressor of liver
cancer metastasis. Cancer Res. 68:4210–4220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Melotte V, Qu X, Ongenaert M, et al: The
N-myc downstream regulated gene (NDRG) family: diverse functions,
multiple applications. FASEB J. 24:4153–4166. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim YJ, Yoon SY, Kim JT, et al: NDRG2
expression decreases with tumor stages and regulates
TCF/beta-catenin signaling in human colon carcinoma.
Carcinogenesis. 30:598–605. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu XL, Liu XP, Lin SX, et al: NDRG2
expression and mutation in human liver and pancreatic cancers.
World J Gastroenterol. 10:3518–3521. 2004.PubMed/NCBI
|
|
17
|
Kang GH, Shim YH, Jung HY, Kim WH, Ro JY
and Rhyu MG: CpG island methylation in premalignant stages of
gastric carcinoma. Cancer Res. 61:2847–2851. 2001.PubMed/NCBI
|
|
18
|
Szyf M, Pakneshan P and Rabbani SA: DNA
methylation and breast cancer. Biochem Pharmacol. 68:1187–1197.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Toyota M, Suzuki H, Sasaki Y, et al:
Epigenetic silencing of microRNA-34b/c and B-cell translocation
gene 4 is associated with CpG island methylation in colorectal
cancer. Cancer Res. 68:4123–4132. 2008. View Article : Google Scholar : PubMed/NCBI
|