Introduction
Hepatocellular carcinoma (HCC) is the third leading
cause of cancer-related mortality worldwide, with up to 75,0000
novel cases reported annually (1).
Despite the fact that the major risk factors, including viral
hepatitis (B and C) and/or alcohol abuse, have been identified, the
therapeutic options remain limited due to incomplete understanding
of the cellular and molecular mechanisms involved in the
pathogenesis of HCC (2).
Therefore, further investigation of the mechanisms involved in the
genesis and development of HCC is important for identifying novel
therapeutic approaches to improve the clinical outcomes for
patients with HCC.
Notch signaling is a classical signaling pathway,
which has an important role in regulating cell fate decisions,
proliferation and apoptosis of multiple cell lineages (3). Notch signaling comprises several
transmembrane receptors and their ligands, transcription factors,
as well as negative and positive modifiers. Notch receptors (Notch
1–4) and two groups of ligands, Jagged (Jagged1 and Jagged2) and
Delta-like (DLL1, DLL3, DLL4), have been identified in mammals
(4). Notch signaling has been
identified to be associated with numerous types of tumor, including
human HCC, as evidenced by its contribution to the formation of
liver tumors in mice (5). One
Notch signaling ligands, Jagged2, is expressed widely in human
organs, including the heart, skeletal muscle and pancreas (6). It has been demonstrated that a
missense mutation in the Jagged2 gene leads to mouse
syndactylism with digit malformation (7). Mice with null mutations of
Jagged2 also exhibit a number of defects, including cleft
palate, syndactyly and thymic abnormalities (6). Additionally, Jagged2 also appears to
have an important role in various diseases, including malignant
tumors. Jagged2 was reported to mediate lung adenocarcinoma
epithelial-mesenchymal transition (EMT) and metastasis in mice
(8). In hypoxic conditions,
Jagged2 promoted breast cancer metastasis and self-renewal of
cancer stem-like cells (9). Notch3
and Jagged2 contribute to gastric cancer development and glandular
differentiation (10). Notably,
the overexpression of Jagged2 has been identified in >90% of
pancreatic cancer cell lines (11). However, the role of Jagged2 in the
genesis and development of HCC remains elusive.
Sonic hedgehog (Shh) signaling has a key role in the
regulation of embryogenesis, adult tissue homeostasis and
carcinogenesis (12,13). It is activated by Shh ligand
binding to the Patched-1 receptor, which relieves the repression of
transducer protein Smoothened and subsequently triggers activation
of the Gli family of transcription factors, including Gli1. The
activated Hedgehog signaling regulates cell proliferation,
survival, angiogenesis and EMT in HCC (14). A transcriptome analysis identified
Jagged2 as a sonic hedgehog-regulated factor, where inhibition of
Shh signaling by a dominant-negative version of Gli3
(Gli3Rep) resulted in the loss of Jagged2 expression.
Conversely, constitutive activation of the Shh pathway, using
Gli3Act resulted in the upregulation of Jagged2
(13,15). Nevertheless, the correlation
between Gli1 and Jagged2 in human HCC remains poorly
understood.
In the present study, the Jagged2 expression and its
relation with Gli1 protein in human HCC was investigated, and then
the correlation between Jagged2 expression and the
clinicopathological features of HCC were analyzed, detailing its
potential as a prognostic marker for HCC patients.
Materials and methods
Patients and tissue specimens
A total of 58 patients were recruited, including 15
females and 43 males (mean age, 51.7 years; range, 24–78). The
patients had not been treated with preoperative chemotherapy or
interventional embolization. All of the patients received curative
liver resection in the Department of Hepatobiliary Surgery, First
Affiliated Hospital of Medical College of Xi’an Jiaotong
University, (Xi’an, China) between December 2009 and June 2010. All
of the procedures were approved by the Xi’an Jiaotong University
Ethics Committee and informed consent forms were signed by each
patient. A total of 58 HCC tissues and matched normal
tumor-adjacent tissues (>2 cm distance from the margin of the
resection) were collected with an area of 0.5×0.5 cm and stored
immediately in paraformaldehyde for immunohistochemistry. All of
the patients were followed-up, either via visits or telephone
contact, with a median follow-up time of 23 months (range, 3–36
months) after liver resection. The data of clinical features was
obtained from the medical records of each patient. The maximum
diameter of the tumor, intrahepatic metastasis, histological
Edmonson classification and tumor-node-metastasis (TNM) stage were
gathered from the pathological records and confirmed by two
experienced pathologists who were blinded to the clinical data and
the results of immunohistochemical staining.
Immunohistochemical analysis
Horseradish peroxidase staining was applied for
immunohistochemical analysis. All of the paraformaldehyde-fixed
paraffin sections were incubated at 60°C for ≥4 h. Next, the
sections were dewaxed in dimethylbenzene and rehydrated in alcohol
of diminishing concentrations. All of the sections were then placed
into citrate buffer and boiled for 15 min for antigen retrieval. A
total of 3.0% hydrogen peroxide was utilized for eliminating the
endogenous peroxidase. The sections were then blocked by 10% goat
serum at 37°C for 30 min and incubated at 4°C overnight with
primary antibody directed against Jagged2 (1:100; Santa Cruz
Biotechnology Inc., Santa Cruz, CA, USA; sc-5604) or Gli1 (1:100;
Santa Cruz Biotechnology Inc.; sc-20687). The biotinylated goat
anti-rabbit secondary antibody (Beijing Zhongshan Goldenbridge
Biotechnology Co., Ltd., Beijing, China) was used to detect the
primary antibody at 37°C for 45 min. Next, the horseradish
peroxidase-streptavidin conjugate was used to react with
biotinylated secondary antibody at 37°C for 30 min. The sections
were reacted with diaminobenzidine and counterstained with
hematoxylin. Finally, the sections were dehydrated in increasing
concentrations of alcohol, transparentized by dimethylbenzene and
coverslipped onto glass slides.
All of the stained sections were observed by two
independent experienced pathologists in a blinded manner. Each
stained section obtained a final score based on the intensity and
percentage of positive cells following semi-quantitative
assessment. The staining intensity was grouped into four grades: 0,
negative staining; 1, weakly positive staining; 2, moderately
positive staining; and 3, strongly positive staining. The stained
sections with different percentages of positive cells were scored
appropriately: 0 (<5%), 1 (5–25%), 2 (26–50%), 3 (51–75%) and 4
(>75%). These two scores were the mean of ten different high
magnification (x400) fields and were multiplied to calculate the
final score of each stained section. The sections with a total
score of >1 were defined as exhibiting positive staining for the
above proteins (16).
Statistical analysis
Each quantitative value was expressed as the mean ±
standard deviation or the median. Comparison of Jagged2 or Gli1
protein expression between HCC tissue and normal tumor-adjacent
tissue was calculated by a paired-samples t-test. Spearman’s rank
correlation coefficient test was used to analyze the correlation
between Jagged2 and Gli1 protein expression. The differences in
Jagged2 or Gli1 expression in the HCC tissues with different
clinical features were compared by the Mann-Whitney U-test. The
Kaplan-Meier method was used to obtain the survival curves of
different clinical characteristics and the differences between
these survival curves were analyzed using a log-rank test. The Cox
proportional hazard regression model was applied to determine the
independent prognostic factors in multivariate survival analysis.
SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) was used in this
statistical analysis and P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of Jagged2 and Gli1 in HCC and
matched normal tumor-adjacent tissues
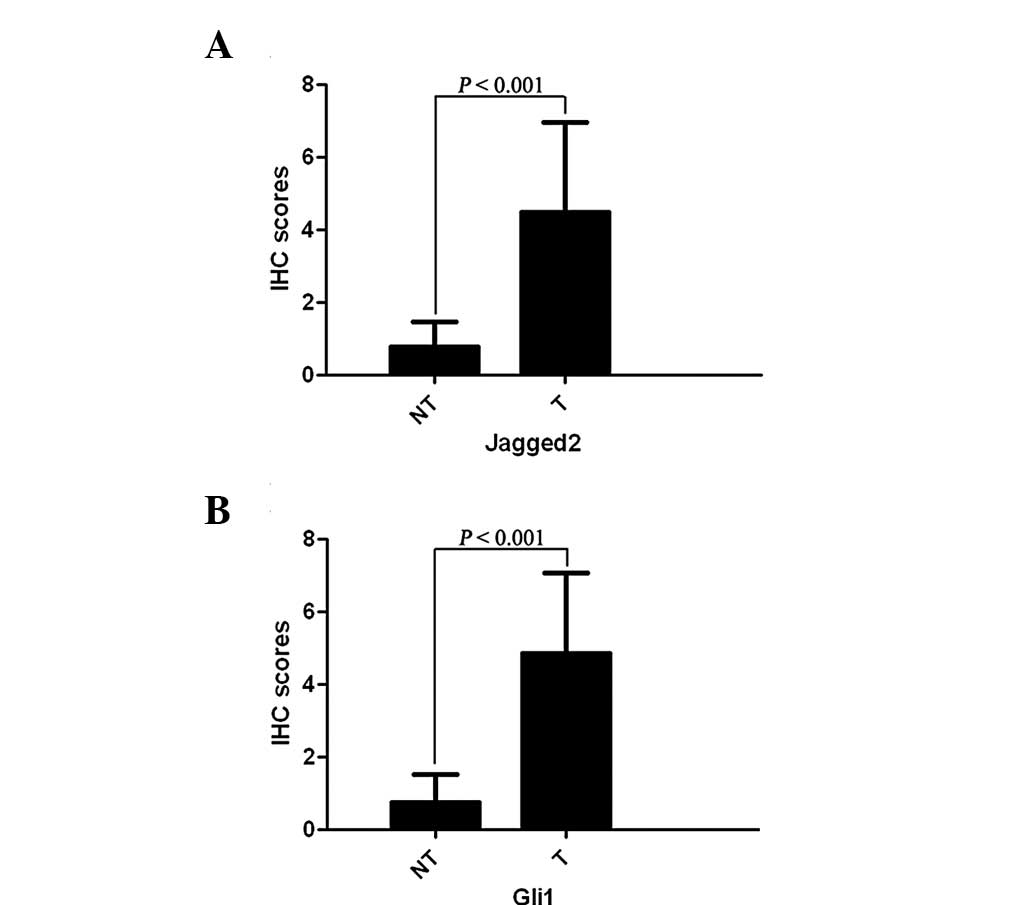
Immunohistochemical staining demonstrated that
Jagged2 protein expression in HCC tissues was found to be
significantly higher than that in the matched normal tumor-adjacent
tissues (4.47±2.48 vs. 0.78±0.68; P<0.001; Fig. 1A). The same was observed for Gli1
protein expression (4.84±2.24 vs. 0.74±0.79; P<0.001; Fig. 1B). The positive staining of Jagged2
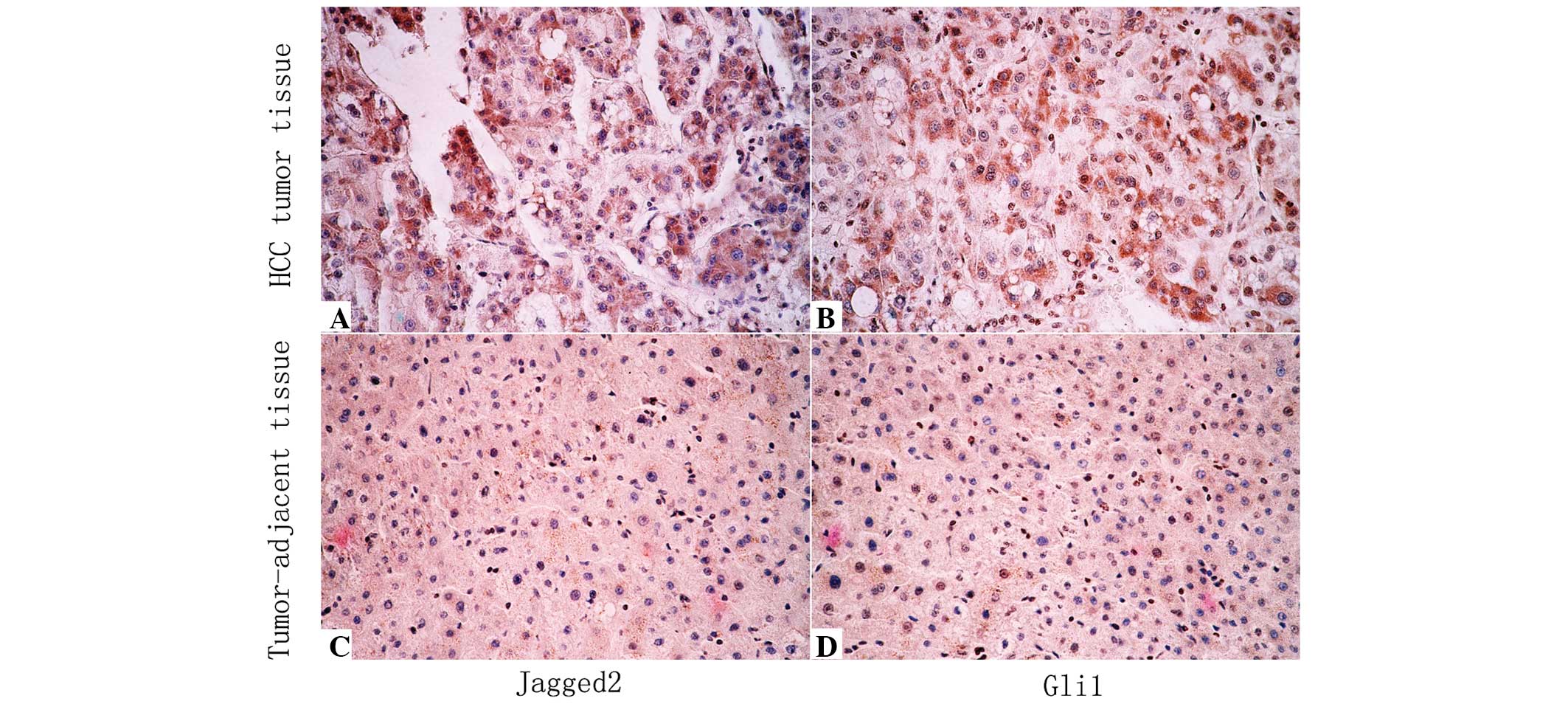
was observed mainly in the cytoplasm, while Gli1 was localized in
the cytoplasm and nuclei in HCC cells (Fig. 2A–B).
Correlation between Jagged2 and Gli1
A previous study indicated that there is a
correlation between Jagged2 and Gli1 expression in various
malignancies; however, this association has not been observed in
HCC (15). Using the Spearman’s
rank correlation coefficient test, Jagged2 was demonstrated to be
positively correlated with Gli1 protein expression (r=0.643,
P<0.001; Fig. 3)
Correlation between Jagged2 or Gli1
protein expression in HCC tissues and clinicopathological
features
The correlation between Jagged2 or Gli1 protein
expression and clinicopathological parameters in 58 HCC patients
was analyzed by the Mann-Whitney U-test, and the results are shown
in Table I. Jagged2 protein
expression was significantly correlated with intrahepatic
metastasis (P=0.007), advanced TNM stage (P=0.001) and high
Edmonson pathological classification (P=0.004). Gli1 protein
expression was also closely associated with intrahepatic metastasis
(P=0.024), advanced TNM stage (P=0.007) and high Edmonson
pathological classification (P=0.034).
 | Table ICorrelation between the
clinicopathological characteristics and protein expression of
Jagged2 and Gli1 in the 58 hepatocellular carcinoma patients. |
Table I
Correlation between the
clinicopathological characteristics and protein expression of
Jagged2 and Gli1 in the 58 hepatocellular carcinoma patients.
| Characteristic | U-value
(Jagged2) | P-value
(Jagged2) | U-value (Gli1) | P-value (Gli1) |
|---|
| Age (<50/≥50) | 341.0 | 0.214 | 393.0 | 0.679 |
| Sex
(male/female) | 306.5 | 0.770 | 302.0 | 0.705 |
| AFP (<400/≥400
ug/dl) | 222.0 | 0.457 | 194.0 | 0.183 |
| HBsAg
(positive/negative) | 247.5 | 0.389 | 256.0 | 0.479 |
| Cirrhosis
(yes/no) | 246.0 | 0.554 | 260.0 | 0.749 |
| Tumor size (<5/≥5
cm) | 350.5 | 0.263 | 382.5 | 0.583 |
| Intrahepatic
metastasis (yes/no) | 126.0 | 0.007 | 149.5 | 0.024 |
| Hepatic capsule
invasion (yes/no) | 298.5 | 0.503 | 273.5 | 0.255 |
| Portal vein tumor
thrombus (yes/no) | 185.5 | 0.748 | 177.0 | 0.618 |
| TNM stage
(I–II/III–IV) | 220.5 | 0.001 | 230.0 | 0.007 |
| Edmonson
classification (I–II/III–IV) | 209.0 | 0.004 | 256.5 | 0.034 |
Analysis of risk factors for patient
survival
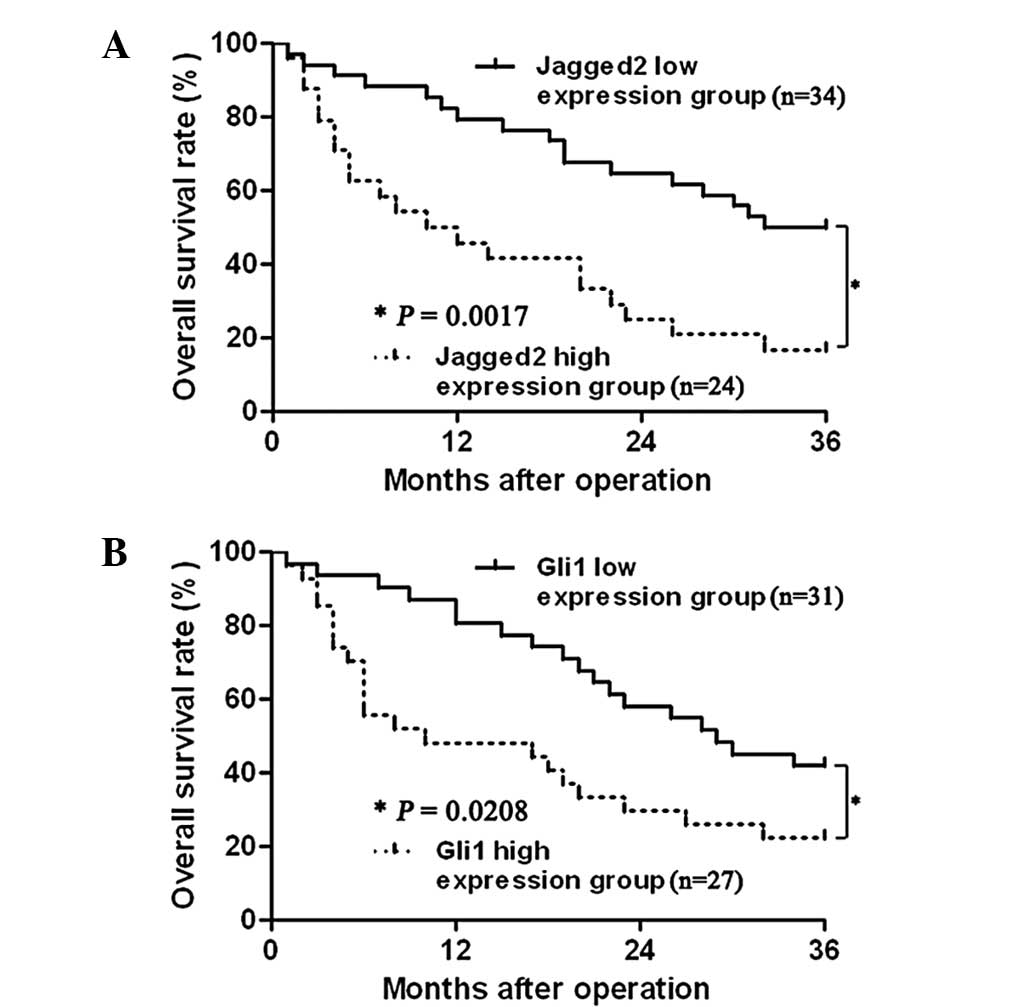
The Kaplan-Meier method was used in univariate
analysis. Median values of Jagged2 and Gli1 were used as cut-off
points to divide the patients into two groups: The low expression
group and the high expression group. As demonstrated in Table II, Jagged2, Gli1, α-fetoprotein
(AFP level), maximum tumor diameter, intrahepatic metastasis, TNM
stage and Edmonson pathological classification were associated with
patient 3-year overall survival (P<0.05, respectively). The
survival curves of variables are demonstrated in Fig. 4. With further investigation, Cox
proportional hazard regression analysis was conducted with the
seven variables that were previously determined, to select the
potential risk factors for determining prognosis. The results
demonstrated that TNM stage, Edmonson pathological classification
and Jagged2 expression were potential risk factors, which affected
the prognosis of HCC patients (P<0.05, respectively; Table III).
 | Table IIUnivariate analysis of Kaplan-Meier
method. |
Table II
Univariate analysis of Kaplan-Meier
method.
| Variable |
χ2-value | P-value |
|---|
| Jagged2 expression
(low/high) | 17.728 | 0.000 |
| Gli1 expression
(low/high) | 4.612 | 0.032 |
| AFP (<400/≥400
ug/dl) | 4.585 | 0.032 |
| Tumor size (<5/≥5
cm) | 4.491 | 0.034 |
| Intrahepatic
metastasis (yes/no) | 13.837 | 0.000 |
| TNM stage
(I–II/III–IV) | 20.419 | 0.000 |
| Edmonson
classification (I–II/III–IV) | 12.014 | 0.001 |
 | Table IIICox proportional hazard regression
mode. |
Table III
Cox proportional hazard regression
mode.
| Variable | B | SE | Wald | Exp (B) | P-value | 95% CI |
|---|
| Gli1 expression
(low/high) | −0.711 | 0.428 | 2.755 | 0.491 | 0.097 | 0.212–1.137 |
| Jagged2 expression
(low/high) | 1.309 | 0.417 | 9.866 | 3.701 | 0.002 | 1.636–8.375 |
| AFP (<400/≥400
ug/dl) | 0.459 | 0.452 | 1.030 | 1.582 | 0.310 | 0.653–3.834 |
| Tumor size (<5/≥5
cm) | −0.329 | 0.455 | 0.525 | 0.719 | 0.469 | 0.295–1.753 |
| Intrahepatic
metastasis (yes/no) | 0.873 | 0.460 | 3.593 | 3.749 | 0.058 | 0.971–5.901 |
| TNM stage
(I–II/III–IV) | 1.979 | 0.582 | 11.567 | 7.234 | 0.001 | 2.313–22.626 |
| Edmonson
classification (I–II/III–IV) | 1.321 | 0.411 | 10.352 | 2.393 | 0.001 | 1.676–8.385 |
Discussion
As one of the ligands of Notch signaling, Jagged2
expression was found to be associated with a variety of tumor
types. However, there is a controversy regarding the effect of
Jagged2 on tumorigenesis and tumor development. One study reported
that Jagged2 regulated the expression of cytokines, which promote
antitumor immunity (17). However,
another study rejected this and demonstrated that Jagged2 enhances
cell growth, invasion and migration in two uveal melanoma cell
lines (Mel285 and Mel290) (18).
In the cutaneous melanoma cell line, Jagged2 was the most
markedly overexpressed gene in the highly invasive clone, with an
RNA level ~15-fold higher than that in the less invasive cells
(19). Furthermore, when Jagged2
was induced at the transcriptional level under hypoxic conditions,
it was significantly correlated with angiogenic processes in breast
cancer, renal cell carcinoma and epithelial tumor cells (20). However, to the best of our
knowledge no study has been conducted regarding the association
between HCC and Jagged2. In the present study, the results
identified that Jagged2 expression in HCC tissues was notably
higher than that in the matched normal tumor-adjacent tissues. In
addition, the clinical pathological parameter analysis demonstrated
that Jagged2 was significantly correlated with intrahepatic
metastasis, advanced TNM stage and high Edmonson pathological
classification, suggesting that Jagged2 may have an important role
in tumorgenesis and tumor development. This observation is
consistent with those of previous studies concerning the oncogenic
effect of Jagged2 in several malignant tumor types (8–11).
It was also identified that patients with low Jagged2 expression
had an improved outcome compared with patients with high Jagged2
expression. Furthermore, Cox proportional hazard regression mode
analysis suggested that Jagged2 was an independent factor for the
prediction of poor long-term HCC survival following radical liver
resection.
Hedgehog/Gli1 signaling pathway is involved in a
variety of human cancer types and aberrant activation of this
signaling has also been identified to be associated with HCC
(21–23). Recent studies identified that the
transcription factor Gli1 was associated with poor prognosis among
the patients with HCC and the expression of the Gli1 gene in
tumor tissues was significantly correlated with disease-free
survival and overall survival rates (24). Using immunohistochemical staining,
the present study demonstrated that Gli1 expression in HCC tissues
was higher than that in the matched tumor-adjacent tissues. Gli1
was significantly correlated with intrahepatic metastasis, advanced
TNM stage and high Edmonson pathological classification, and the
3-year overall survival of the patients with Gli1 high expression
was lower than that in those with low Gli1 expression.
Additionally, the AFP level, maximum tumor diameter, intrahepatic
metastasis, TNM stage and Edmonson pathological classification were
associated with the 3-year overall survival. According to the
clinical research, HCC patients with advanced TNM stage and high
Edmonson pathological classification have a poor long-term
survival.
Hedgehog signaling induced Jagged2 upregulation and
tumor growth factor-β1 secretion to promote the motility and
invasiveness of cancer cells (18). A previous transcriptome analysis
identified Jagged2 as a sonic hedgehog-regulated factor (15). However, the accurate mechanisms
involved in Hedgehog signaling-induced Jagged2 upregulation remain
unclear. In the present study, it was demonstrated that Jagged2
protein expression was positively correlated with Gli1. This result
indicated that Gli1, the key transcriptional factor in Shh
signaling, may regulate Jagged2 expression in HCC.
In conclusion, the present study demonstrated that
Jagged2 and Gli1 were overexpressed in HCC tissues and Jagged2 was
positively correlated with Gli1 protein expression. High-expression
of Jagged2 or Gli1 was associated with a poor 3-year overall
survival in HCC patients and Jagged2 acted as an independent
predictor of an unfavorable prognosis. Elucidating the regulation
between Gli1 and Jagged2 expression, and the mechanisms involved in
promoting HCC invasion and metastasis by Jagged2, requires further
investigation.
Acknowledgements
This study was supported by grants from the National
Natural Scientific Foundation of China (grant nos. 81272645 and
81072052 to Qingguang Liu and 81071897 to Yingmin Yao), the
Research Fund for the doctoral Program of High Education of China
from Ministry of Education (grant no. 20120201120090 to Xin Zheng),
the Fundamental Research Funds for the Central Universities
sponsored by Xian Jiaotong University to Xin Zheng, the Clinical
medicine specialty of seven year clinical research and the
Innovation Fund for students granted by the First Affiliated
Hospital of Xi’an Jiaotong University (13ZD18).
References
|
1
|
Guo C, Liu Q, Zhang L, Yang X, Song T and
Yao Y: Double lethal effects of fusion gene of wild-type p53 and
JunB on hepatocellular carcinoma cells. J Huazhong Univ Sci
Technolog Med Sci. 32:663–668. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tu K, Zheng X, Zhou Z, et al: Recombinant
human adenovirus-p53 injection induced apoptosis in hepatocellular
carcinoma cell lines mediated by p53-Fbxw7 pathway, which controls
c-Myc and cyclin E. PLoS One. 8:e685742013. View Article : Google Scholar
|
|
3
|
Suwanjunee S, Wongchana W and Palaga T:
Inhibition of gamma-secretase affects proliferation of leukemia and
hepatoma cell lines through Notch signaling. Anticancer Drugs.
19:477–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gao J, Dong Y, Zhang B, et al: Notch1
activation contributes to tumor cell growth and proliferation in
human hepatocellular carcinoma HepG2 and SMMC7721 cells. Int J
Oncol. 41:1773–1781. 2012.PubMed/NCBI
|
|
5
|
Villanueva A, Alsinet C, Yanger K, et al:
Notch signaling is activated in human hepatocellular carcinoma and
induces tumor formation in mice. Gastroenterology. 143:1660–1669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deng Y, Madan A, Banta AB, Friedman C,
Trask BJ, Hood L and Li L: Characterization, chromosomal
localization, and the complete 30-kb DNA sequence of the human
Jagged2 (JAG2) gene. Genomics. 63:133–138. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang R, Lan Y, Chapman HD, et al: Defects
in limb, craniofacial, and thymic development in Jagged2 mutant
mice. Genes Dev. 12:1046–1057. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Y, Ahn YH, Gibbons DL, et al: The
Notch ligand Jagged2 promotes lung adenocarcinoma metastasis
through a miR-200-dependent pathway in mice. J Clin Invest.
121:1373–1385. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xing F, Okuda H, Watabe M, et al:
Hypoxia-induced Jagged2 promotes breast cancer metastasis and
self-renewal of cancer stem-like cells. Oncogene. 30:4075–4086.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang H, An HJ, Song JY, Kim TH, Heo JH,
Ahn DH and Kim G: Notch3 and Jagged2 contribute to gastric cancer
development and to glandular differentiation associated with MUC2
and MUC5AC expression. Histopathology. 61:576–586. 2012.PubMed/NCBI
|
|
11
|
Mullendore ME, Koorstra JB, Li YM, et al:
Ligand-dependent Notch signaling is involved in tumor initiation
and tumor maintenance in pancreatic cancer. Clin Cancer Res.
15:2291–2301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng X, Vittar NB, Gai X, et al: The
transcription factor GLI1 mediates TGFβ1 driven EMT in
hepatocellular carcinoma via a SNAI1-dependent mechanism. PLoS One.
7:e495812012.
|
|
13
|
Katoh Y and Katoh M: Hedgehog signaling
pathway and gastrointestinal stem cell signaling network. Int J Mol
Med. 18:1019–1023. 2006.PubMed/NCBI
|
|
14
|
Robbins DJ, Fei DL and Riobo NA: The
Hedgehog signal transduction network. Sci Signal. 5:re62012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rabadán MA, Cayuso J, Le Dréau G, et al:
Jagged2 controls the generation of motor neuron and oligodendrocyte
progenitors in the ventral spinal cord. Cell Death Differ.
19:209–219. 2012.PubMed/NCBI
|
|
16
|
Tu K, Zheng X, Zan X, Han S, Yao Y and Liu
Q: Evaluation of Fbxw7 expression and its correlation with the
expression of c-Myc, cyclin E and p53 in human hepatocellular
carcinoma. Hepatol Res. 42:904–910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Choi K, Ahn YH, Gibbons DL, et al:
Distinct biological roles for the Notch ligands Jagged-1 and
Jagged-2. J Biol Chem. 284:17766–17774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Asnaghi L, Handa JT, Merbs SL, Harbour JW
and Eberhart CG: A role for Jag2 in promoting uveal melanoma
dissemination and growth. Invest Ophthalmol Vis Sci. 54:295–306.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gütgemann A, Golob M, Müller S, Buettner R
and Bosserhoff AK: Isolation of invasion associated cDNAs in
melanoma. Arch Dermatol Res. 293:283–290. 2001.PubMed/NCBI
|
|
20
|
Pietras A, von Stedingk K, Lindgren D,
Påhlman S and Axelson H: JAG2 induction in hypoxic tumor cells
alters Notch signaling and enhances endothelial cell tube
formation. Mol Cancer Res. 9:626–636. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie J, Murone M, Luoh SM, et al:
Activating Smoothened mutations in sporadic basal-cell carcinoma.
Nature. 391:90–92. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Karhadkar SS, Bova GS, Abdallah N, et al:
Hedgehog signaling in prostate regeneration, neoplasia and
metastasis. Nature. 431:707–712. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Velcheti V and Govindan R: Hedgehog
signaling pathway and lung cancer. J Thorac Oncol. 2:7–10. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Che L, Yuan YH, Jia J and Ren J:
Activation of sonic hedgehog signaling pathway is an independent
potential prognosis predictor in human hepatocellular carcinoma
patients. Chin J Cancer Res. 24:323–331. 2012. View Article : Google Scholar
|