Introduction
Breast cancer is one of the most common types of
cancer among females (1). Although
progress has been made in diagnosis and adjuvant therapies, the
pathogenesis of breast cancer remains unknown. Furthermore, drug
resistance also remains a major clinical obstacle for the
successful treatment of breast cancer. Bone morphogenetic proteins
(BMPs), belonging to the TGF-β superfamily, had been confirmed in
several studies to be important in cell proliferation,
differentiation and apoptosis during tumor progression (2–4). In
breast cancer, it was revealed that BMP2, BMP4, BMP6, BMP7, BMP9
and BMP10 were involved in breast cancer development, progression
and drug resistance (5–10). However, few studies focusing on the
epigenetic regulation of the expression of BMPs have been
conducted. Recently, it was recognized that DNA methylation was a
common event in human cancer, which induced the loss of tumor
suppressor gene expression (11–12).
Previous studies have also demonstrated that the downregulation of
BMP2 was silenced by aberrant DNA methylation in bone formation and
gastric carcinomas (13,14). To date, the epigenetic regulation
of BMP2 in breast cancer and its association with tumor progression
and drug resistance remains to be elucidated.
The present study detected the mRNA expression and
the promoter methylation status of BMP2 in breast cancer tissues
and breast cancer cell lines. In addition, BMP2 expression and
cancer cell chemoresistance was also evaluated in an in
vitro cell model. Our data indicated that the downregulation of
BMP2 caused by promoter hypermethylation may contribute to breast
cancer progression and cancer cell drug resistance.
Materials and methods
Patients and tissue specimens
A total of 32 breast cancer tissues and
patient-matched adjacent normal breast tissues were obtained from
patients who underwent surgery at the Cancer Hospital of Sichuan
Province (Chengdu, China). All tumor tissues were reviewed by an
experienced pathologist using World Health Organization
recommendations on histopathological typing. For total RNA
isolation, the specimens were frozen in liquid nitrogen and stored
at −80°C. There were no patients who received chemotherapy or
radiotherapy prior to surgery. The study protocol conformed to the
local ethical standards of the institutional review board of the
The PLA General Hospital of Chengdu Military Region (Chengdu,
Sichuan, China).
Cell culture
The MCF-7 human breast cancer cell line was
purchased from the Shanghai Institute for Biological Sciences,
Chinese Academy of Sciences (Shanghai, China) and MCF-7/ADR, a
doxorubicin resistant subline of MCF-7, was generated by
continuously culturing the drug-sensitive parental cell line MCF-7
in medium containing incrementally increasing concentrations of
doxorubicin (Sigma-Aldrich, St. Louis, MO, USA). The cells were
cultured in RPMI-1640 (Gibco-BRL, Carlsbad, CA, USA) supplemented
with 10% fetal calf serum, 100 U/ml penicillin and 100 U/ml
streptomycin at 37°C and 5% CO2. To avoid the effects of
the drug, the MCF-7/ADR drug resistant cell line was cultured in
drug-free medium for >2 weeks prior to subsequent
experiments.
Treatment of cells with
5-aza-2′-deoxycytidine (5-aza-dc)
The cells were seeded at a density of
1×105 cells in 6-well plates. Following 24 h, the cells
were treated with 5 μM 5-aza-dc (Sigma-Aldrich). Total cellular RNA
and protein were isolated from the cells 0 and 3 days after the
addition of 5-aza-dc as described above.
RNA isolation and quantitative polymerase
chain reaction (qPCR)
The total RNA of 32 cancer and adjacent normal
breast tissues were extracted using TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. Total RNA was retrotranscribed using the RevertAid™
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham,
MA, USA) according to the manufacturer’s instructions. qPCR was
performed with the SYBR-Green PCR master mix (Takara Bio, Inc.,
Shiga, Japan) using a Mastercycler ep realplex (Eppendorf, Hamburg,
Germany). The levels of mRNA expression were quantified following
normalization with endogenous control GAPDH using the
2−ΔΔCT method (15).
The primer sequences used for PCR were as follows: Forward:
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse:
5′-GTAGAGGCAGGGATGATGTTCT-3′ for GAPDH (product size: 132 bp); and
forward: 5′-CCCAGCGTGAAAAGAGAGAC-3′ and reverse:
5′-GAGACCGCAGTCCGTCTAAG-3′ for bmp2 (product size: 168 bp). The
experiments were performed independently for each sample and at
least three technical replicates were run for each treated sample
and controls.
Western blot analysis
Whole cell extracts were lysed using lysis buffer
pulsed protease inhibitor (Millipore, Billerica, MA, USA).
Equivalent quantities of protein were resolved by SDS-PAGE,
transferred onto polyvinylidene difluoride membranes (GE Healthcare
Biosciences, Piscataway, NJ, USA), inhibited with
phosphate-buffered saline (PBS) containing 0.2% Tween-20 and 5%
non-fat dry milk and incubated with primary antibodies against BMP2
(1:1,000; rabbit polyclonal to BMP2) and actin (1:2,000, mouse
monoclonal to actin) obtained from Abcam (Cambridge, MA, USA).
Antibody binding was revealed by incubation with horseradish
peroxidase-conjugated secondary antibodies (Invitrogen Life
Technologies) and an Enhanced Chemiluminescnce Plus immunoblotting
detection system (GE Healthcare Biosciences). The signals were
quantified using NIH Image J 1.63 software (National Institutes of
Health, Bethesda, MA, USA).
Methylation-specific PCR (MSP) and
bisulfite genomic sequencing
Genomic DNA of MCF-7 or MCF-7/ADR was extracted
using a TIANamp Genomic DNA kit (Tiangen Biotech (Beijing) Co.,
Ltd., Beijing, China). Bisulfite conversion of genomic DNA was
performed using the EZ DNA Methylation-Gold kit (Zymo Research,
Irvine, CA, USA) according to the manufacturer’s instructions. MSP
was performed on bisulfite-modified DNA. The primer sequences of
BMP-2 for the unmethylated reaction were: Forward:
5′-GGGTTAGTGTTGAGTGGATTATT-3′ and reverse:
5′-CACAACACTAAAAATCAACTCC-3′ and for the methylated reaction
forward: 5′-TTAGCGTCGAGTGGATTATC-3′ and reverse:
5′-GACGCTAAAAATCGACTCC-3′. PCR products were analyzed by
electrophoresis in a 2.0% agarose gel containing golden view (SBS
Genetech Co., Ltd., Beijing, China). For bisulfite sequencing of
DNA samples, sodium bisulfite-treated genomic DNA was amplified
using the following primers: Forward: 5′-GTGGTTTTTGTTGTTYGG-3′ and
Reverse: 5′-TCTACCTTACTCCAATACACCC-3′. The PCR reaction products
were gel purified and cloned into the pGEM-T Vector system
according the manufacturer’s instructions (Promega Corporation,
Madison, WI, USA). The colonies were sequenced on a genetic
analyzer (Applied Biosystems, Foster City, CA, USA) to analyze the
methylated cytosine level.
Small interfering RNA (siRNA)
transfection
siRNAs (Guangzhou RiboBio Co., Ltd., Guangzhou,
Guangdong, China) were transfected using Lipofectamine™ 2000 and
OptiMEM (Invitrogen Life Technologies) according to the
manufacturer’s instructions. Following 72 h of siRNA transfection,
the cell lysate was prepared and western blotting was performed as
described above.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT)
assay
MCF-7 cells were seeded in 96-well plates at a
density of 1×104 cells per well and incubated overnight
in 10% fetal bovine serum medium. The cells were transfected with
BMP2 specific siRNA or negative control siRNA. Then, the cells were
treated with different concentrations of adriamycin (0–200 mg/l).
Following incubation for 48 h at 37°C, 20 μl MTT (Sigma-Aldrich)
solution (5 mg/ml in PBS) was added to each well and incubation
continued for a further 4 h at 37°C. The medium was then removed
and the MTT crystals were solubilized using 100 μl
dimethylsulfoxide. Absorbance was measured at 560 nm using a
microplate reader (Thermo Fisher Scientific). Absorbance readings
were subtracted from the value of blank wells. The reduction in
cell growth was calculated as a percentage of control absorbance in
the absence of any drug. Data are presented as the mean ± standard
deviation (SD) of at least three independent experiments.
Statistical analysis
Data are expressed as the mean ± SD. The paired
t-test and independent sample t-test were applied for statistical
analysis using SPSS software version 13.0 (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
DNA methylation suppresses BMP2
expression in breast cancer tissues
The present study performed quantitative analysis of
BMP2 mRNA in 32 breast cancer tissues and matched adjacent normal
tissues by qPCR. Among the 32 pairs of samples, nine samples of
breast cancer tissue demonstrated a higher expression of BMP2
compared with the adjacent normal tissue, and 23 samples of breast
cancer tissue demonstrated a lower expression of BMP2 compared with
the adjacent normal tissue. Compared with the normal tissues, BMP2
mRNA expression was significantly lower in cancer tissues
(P<0.01; Fig. 1A). Furthermore,
data demonstrated that the mRNA expression of BMP2 decreased
between tumor stage 1 and stage 3. There was a significantly higher
level of BMP2 mRNA in stage 1 than in stage 2 and 3 tumors
(Fig. 1B). Subsequently, the BMP2
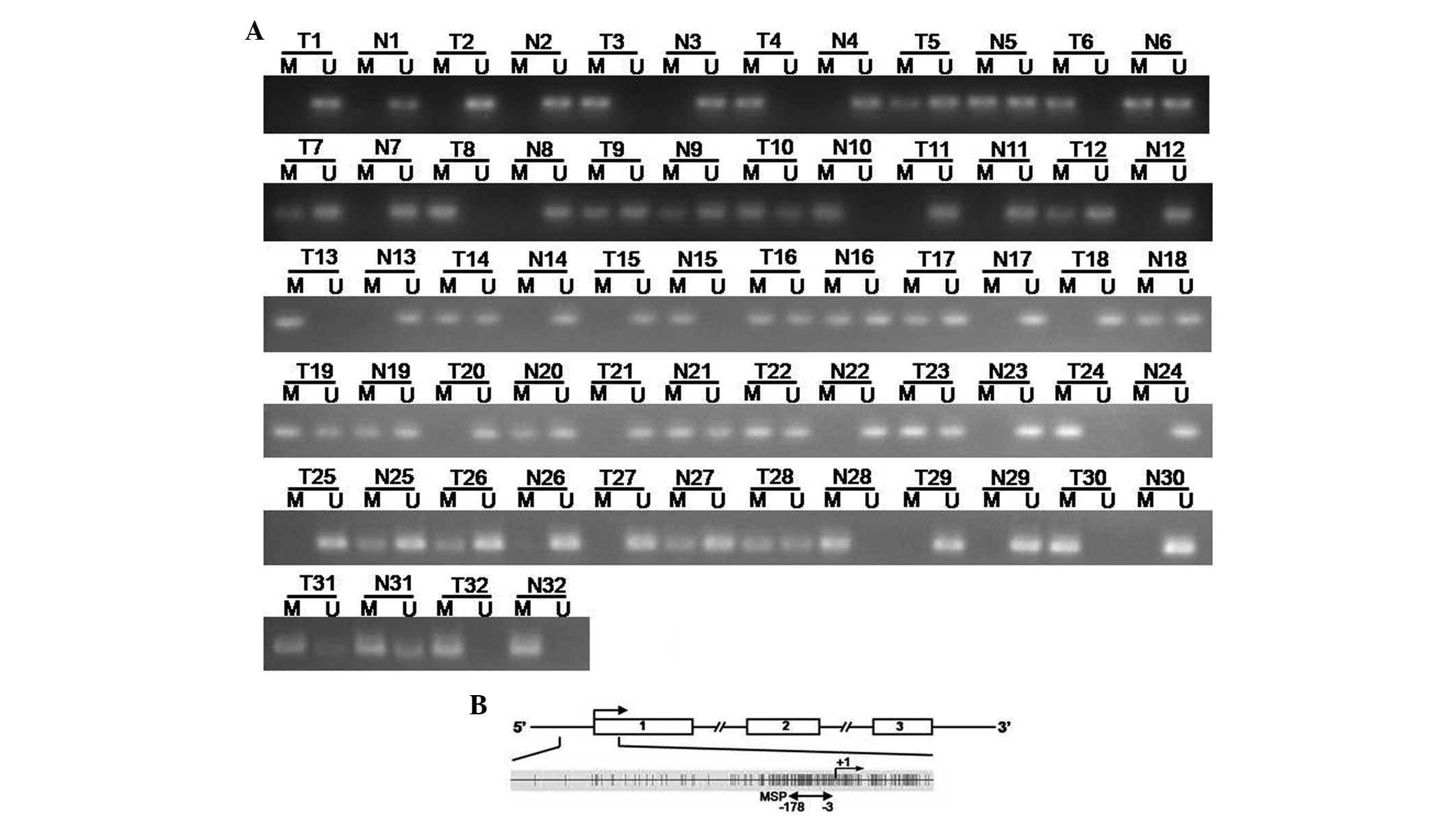
methylation status was detected by MSP. Aberrant methylation was
detected in 22 out of 32 (68.75%) tumors, which was more frequent
than that in the paired adjacent normal tissues [15 out of 32
(46.87%); Fig. 2A].
DNA methylation suppresses the expression
of BMP2 in drug resistant breast cancer cells
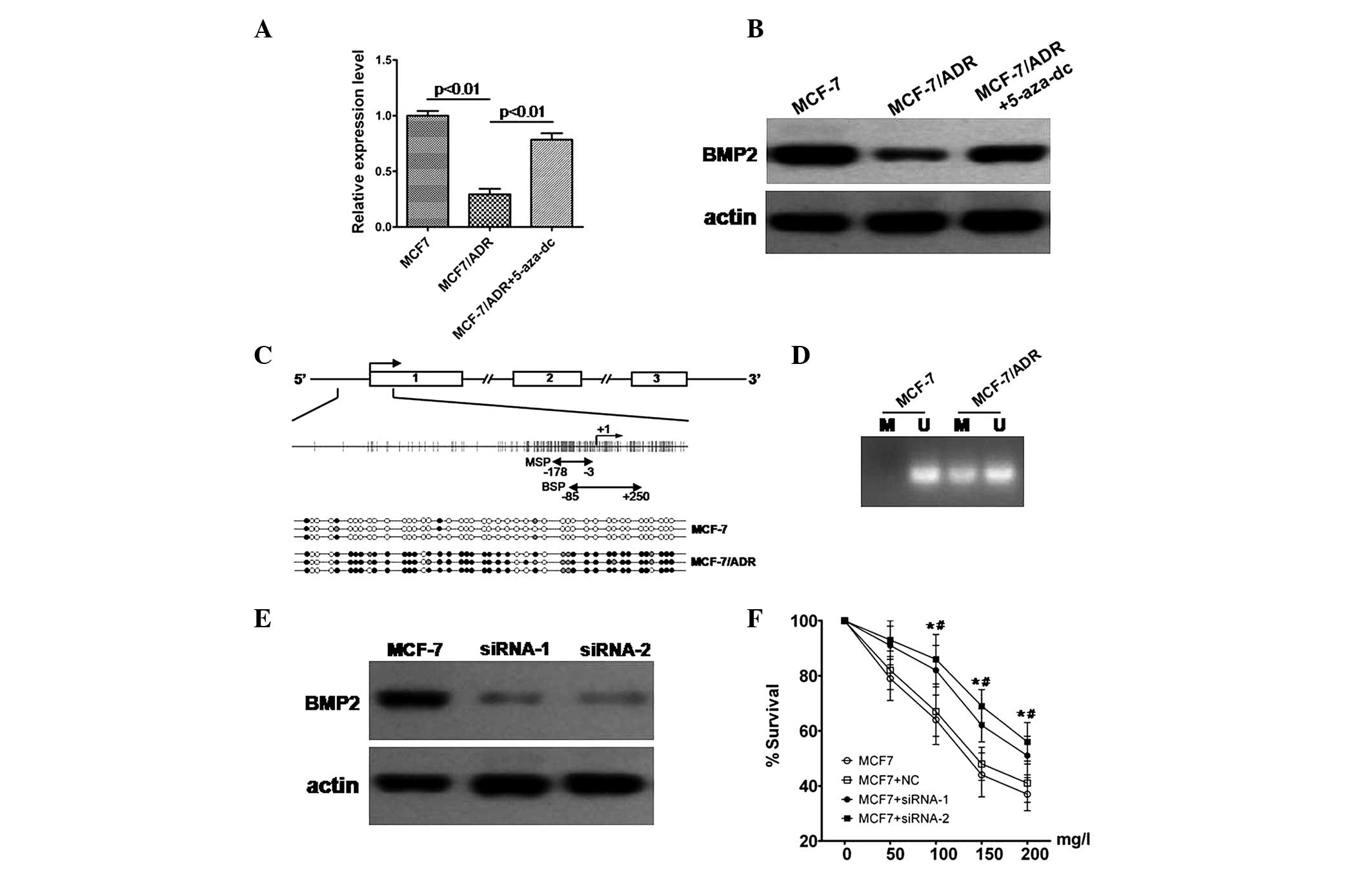
qPCR and western blot analysis demonstrated that
BMP2 was expressed in the breast cancer cell line MCF-7 and was
downregulated in doxorubicin resistant MCF-7/ADR cells (Fig. 3A and B). Following treatment with a
DNA methyltransferase inhibitor 5-aza-dc, the downregulation of
BMP2 was reversed in MCF-7/ADR cells (Fig. 3A and B). This indicated that the
expression of BMP2 may be involved in DNA methylation regulation.
Thus, MSP and bisulfite-sequencing PCR (BSP) were employed to
investigate the promoter methylation status of BMP2. MSP analyses
revealed that CpG islands of the BMP promoter were unmethylated in
MCF-7 cells. However, in MCF-7/ADR cells, they were partially
methylated (Fig. 3D). Furthermore
the MSP results were confirmed by BSP. The results demonstrated
that MCF-7/ADR cells showed dense methylation of the promoter CpG
islands (Fig. 3C).
Knockdown of BMP2 by siRNA increases the
chemoresistance of MCF-7 cells
In order to evaluate the effect of BMP2
downregulation in MCF-7 cells, the negative control siRNA and
BMP2-specific siRNA were transfected into the MCF-7 cells and qPCR
and western blot analysis detected the efficacy of the
downregulation of expression of the BMP2 gene. The results
indicated that siRNA was able to significantly downregulate the
expression of BMP2 (Fig. 3E).
Then, the MTT assay was employed to detect the effects of BMP2
expression downregulation by siRNA transfection on doxorubicin
resistance. The survival ratio of MCF-7 cells, MCF-7 cells
transfected with negative control siRNA and BMP2-specific siRNA-1
and siRNA-2 were analyzed following treatment with a range of
increasing concentrations of doxorubicin for 48 h. The survival
ratio of cells transfected with BMP2-specific siRNA-1 or siRNA-2
was significantly higher than cells transfected with negative
control siRNA or MCF-7 cells (Fig.
3F). No significant difference between the MCF-7 cells and
cells transfected with negative control siRNA was identified.
Discussion
The present study found that BMP2 expression was
decreased in breast cancer tissues and was closely associated with
DNA hypermethylation. Furthermore, the downregulation of BMP2
induced by DNA hypermethylation was able to enhance the
chemoresistance of the MCF-7 breast cancer cell line. All these
results indicated that aberrant methylation regulated BMP2
expression and was important in breast cancer development and drug
resistance.
BMPs are a group of growth factors that belong to
the TGF-β superfamily. Previous studies confirmed that BMPs are
important in cell proliferation, differentiation and apoptosis
during development of the embryo and tumor progression (2–4). In
breast cancer, the role of BMPs has not been well characterized,
although several studies have focused on this subject (16,17).
BMP2 was reported to inhibit breast cancer cell proliferation
through p21 and PTEN (18,19). In addition, BMP2 may be involved in
tumor angiogenesis, invasion and hormone-independent growth of
breast cancer (20,21). The data of the present study also
confirmed that BMP2 was involved in breast cancer development and
progression. However, there are few studies investigating the
association between BMP2 and cancer cell drug resistance, and the
mechanisms underlying the regulation of BMP2 expression. In the
present study, the in vitro cell model revealed that the
downregulation of BMP2 was able to enhance the chemoresistance of
MCF-7 breast cancer cells. All these data indicated that BMP2 may
be important in breast cancer development and drug resistance.
DNA methylation was recognized as one of the most
common forms of gene expression regulation (22). In various types of human cancer,
DNA methylation patterns are associated with cancer development and
progression (23). In breast
cancer, numerous studies have confirmed that DNA methylation is
involved in tumorigenesis by inhibition of tumor suppressor gene
expression, including estrogen receptor, hox5a, twist and
E-cadherin (24–28). The present study revealed that DNA
hypermethylation of BMP2 was closely associated with the
downregulation of BMP2 in clinical breast cancer specimens.
Furthermore, in an in vitro cell model, DNA hypermethylation
led to the downregulation of BMP2 and resulted in enhanced
chemoresistance of MCF-7 cells. These data indicted that DNA
hypermethylation may cause breast cancer cell drug resistance
through silencing BMP2 expression.
In conclusion, the results of the present study
indicated that promoter hypermethylation suppression of BMP2 is a
frequent event in human breast cancer. This aberrant epigenetic
event may be important in the development and drug resistance of
breast cancer, and our data may provide a novel prognostic marker
and therapeutic strategy for breast cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
Slattery ML, John EM, Torres-Mejia G, et
al: Genetic variation in bone morphogenetic proteins and breast
cancer risk in hispanic and non-hispanic white women: The breast
cancer health disparities study. Int J Cancer. 132:2928–2939. 2013.
View Article : Google Scholar
|
|
3
|
Thawani JP, Wang AC, Than KD, et al: Bone
morphogenetic proteins and cancer: review of the literature.
Neurosurgery. 66:233–246. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buijs JT, Petersen M, van der Horst G, et
al: Bone morphogenetic proteins and its receptors; therapeutic
targets in cancer progression and bone metastasis? Curr Pharm.
16:1291–1300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ye S, Park BH, Song KJ, et al: In vivo
inhibition of bone morphogenetic protein-2 on breast cancer cell
growth. Spine (Phila Pa 1976). 38:E143–E150. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo D, Huang J and Gong J: Bone
morphogenetic protein 4 (BMP4) is required for migration and
invasion of breast cancer. Mol Cell Biochem. 363:179–190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lian WJ, Liu G, Liu YJ, et al:
Downregulation of BMP6 enhances cell proliferation and
chemoresistance via activation of the ERK signaling pathway in
breast cancer. Oncol Rep. 30:193–200. 2013.PubMed/NCBI
|
|
8
|
Rodriguez-Martinez A, Alarmo EL, Saarinen
L, et al: Analysis of BMP4 and BMP7 signaling in breast cancer
cells unveils time-dependent transcription patterns and highlights
a common synexpression group of genes. BMC Med Genomics. 4:802011.
View Article : Google Scholar
|
|
9
|
Wang K, Feng H, Ren W, et al: BMP9
inhibits the proliferation and invasiveness of breast cancer cells
MDA-MB-231. J Cancer Res Clin Oncol. 137:1687–1696. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye L, Bokobza S, Li J, et al: Bone
morphogenetic protein-10 (BMP-10) inhibits aggressiveness of breast
cancer cells and correlates with poor prognosis in breast cancer.
Cancer Sci. 101:2137–2144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
You JS and Jones PA: Cancer genetics and
epigenetics: two sides of the same coin? Cancer Cell. 22:9–20.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanwal R and Gupta S: Epigenetic
modifications in cancer. Clin Genet. 81:303–311. 2012. View Article : Google Scholar
|
|
13
|
Wen XZ, Akiyama Y, Baylin SB, et al:
Frequent epigenetic silencing of the bone morphogenetic protein 2
gene through methylation in gastric carcinomas. Oncogene.
25:2666–2673. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu B, Wang H, Wang J, et al: Epigenetic
regulation of BMP2 by 1,25-dihydroxyvitamin D3 through DNA
methylation and histone modification. PLoS One. 8:e614232013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
16
|
Alarmo EL, Kuukasjärvi T, Karhu R, et al:
A comprehensive expression survey of bone morphogenetic proteins in
breast cancer highlights the importance of BMP4 and BMP7. Breast
Cancer Res Treat. 103:239–246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Davies SR, Watkins G, Douglas-Jones A, et
al: Bone morphogenetic proteins 1 to 7 in human breast cancer,
expression pattern and clinical/prognostic relevance. J Exp Ther
Oncol. 7:327–338. 2008.PubMed/NCBI
|
|
18
|
Ghosh-Choudhury N, Ghosh-Choudhury G,
Celeste A, et al: Bone morphogenetic protein-2 induces cyclin
kinase inhibitor p21 and hypophosphorylation of retinoblastoma
protein in estradiol-treated MCF-7 human breast cancer cells.
Biochim Biophys Acta. 1497:186–196. 2000. View Article : Google Scholar
|
|
19
|
Waite KA and Eng C: BMP2 exposure results
in decreased PTEN protein degradation and increased PTEN levels.
Hum Mol Genet. 12:679–684. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Clement JH, Raida M, Sänger J, et al: Bone
morphogenetic protein 2 (BMP-2) induces in vitro invasion and in
vivo hormone independent growth of breast carcinoma cells. Int J
Oncol. 27:401–407. 2005.PubMed/NCBI
|
|
21
|
Raida M, Clement JH, Leek RD, et al: Bone
morphogenetic protein 2 (BMP-2) and induction of tumor
angiogenesis. J Cancer Res Clin Oncol. 131:741–750. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jones PA: Functions of DNA methylation:
islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
De Carvalho DD, Sharma S, You JS, et al:
DNA methylation screening identifies driver epigenetic events of
cancer cell survival. Cancer Cell. 21:655–667. 2012.PubMed/NCBI
|
|
24
|
Hervouet E, Cartron PF, Jouvenot M, et al:
Epigenetic regulation of estrogen signaling in breast cancer.
Epigenetics. 8:237–245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pang JM, Dobrovic A and Fox SB: DNA
methylation in ductal carcinoma in situ of the breast. Breast
Cancer Res. 15:2062013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Connolly R and Stearns V: Epigenetics as a
therapeutic target in breast cancer. J Mammary Gland Biol
Neoplasia. 17:191–204. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dagdemir A, Durif J, Ngollo M, et al:
Breast cancer: mechanisms involved in action of phytoestrogens and
epigenetic changes. In Vivo. 27:1–9. 2013.PubMed/NCBI
|
|
28
|
Huang Y, Nayak S, Jankowitz R, et al:
Epigenetics in breast cancer: what’s new? Breast Cancer Res.
13:2252011.
|