Introduction
Previous studies have indicated that numerous
proteins are associated with the development of hepatocellular
carcinoma (HCC) (1–8). Hepatitis B virus (HBV) encodes three
envelope proteins in the pre-S/S open reading frame that are termed
large, middle and small (major) surface proteins (9). Following the integration of viral
sequences into the genome, the translation of C-terminally
truncated surface proteins of HBV frequently occurs. It has been
suggested that the expression of certain genes, activated by these
translated viral sequences, may contribute to hepatocarcinogenesis
(10).
In a previous study (10), the transactivating potential of
middle hepatitis B surface protein C-terminally truncated at amino
acid 167 (MHBst167) on the HBV regulatory element was
investigated. The data indicated that MHBst167 is a
pleiotropic, non-liver-specific transactivator which can modulate
ubiquitous transcription factors that are associated with
proliferation and inflammation (10). In order to further reveal the
biological roles of MHBst167, the present study
investigated cellular proteins interacting with the carboxyl
terminus of MHBst167, an important functional region. To
identify these proteins, a GAL4-based yeast two-hybrid system
(Clontech Laboratories, Inc., Mountain View, CA, USA), using the
MHBst167 cDNA as bait, was utilized to screen a human
fetal liver cDNA library. The aim of the study was to provide novel
insights into the biological functions of MHBst167.
Materials and methods
Yeast strains and plasmids
The Matchmaker® GAL4-based two-hybrid
system 3 and vector pACT2 containing a human fetal liver cDNA
library were obtained from Clontech Laboratories, Inc. The yeast
strain AH109 (MATα, trp1–901, leu2–3, 112, ura3–52, his3–200,
gal4Δ, gal80Δ, LYS2:GAL1UAS-GAL1TATA-HIS3,
GAL2UAS-GAL2TATA-ADE2,
URA3:MEL1UAS-MEL1TATA-LacZ) contained the
pGBKT7–53 plasmid, encoding the DNA-binding domain (DNA-BD) mouse
p53 fusion protein. The yeast strain Y187 (MATα, ura3–52, his3–200,
ade2–101, trp1–901, leu2–3, 112, gal4, gal80, met-,
URA3::GAL1UAS-GAL1TATA-lacZ, MEL1) contained
the pTD1-1 plasmid, in which vector pACT2 encoded the AD/simian
virus 40 large T antigen fusing protein. AH109 was used for cloning
bait plasmids and Y187 was used for cloning library plasmids. The
yeast-Escherichia coli shuttle plasmids pGBKT7 DNA-BD
cloning plasmid, pGADT7 DNA-activation domain (DNA-AD) cloning
plasmid, pGBKT7–53 control plasmid, pGADT7, pGBKT7-Lam control
plasmid and pCL1 were obtained from Clontech Laboratories, Inc.
Chemical agents and culture media
The pGEM-T vector and Taq DNA polymerase were
purchased from Promega Corp. (Madison, WI, USA). T4 DNA ligase,
EcoRI and BamHI restriction endonucleases were
purchased from Takara (Shiga, Japan), and c-Myc monoclonal antibody
and goat anti-mouse immunoglobulin G antibody conjugated with
horseradish peroxidase were obtained from Zhongshan Company
(Zhongshan, China). Tryptone and yeast extracts were obtained from
Oxoid (Thermo Fisher Scientific, Inc., Waltham, MA, USA). X-α-Gal
and yeast peptone dextrose adenine, synthetic defined
(SD)/-tryptophan (Trp), SD/-leucine (Leu), SD/-Trp/-Leu,
SD/-Trp/-Leu/-histidine (His) and SD/-Trp/-Leu/-His/-adenine (Ade)
media were purchased from Clontech Laboratories, Inc. Glass beads
were purchased from Sigma (St. Louis, MO, USA), while lysis buffer
[20 g/l Triton X-100, 10 g/l SDS, 10 mmol/l NaCl, 10 mmol/l
Tris-HCl (pH 8.0) and 1 mmol/l Na2EDTA] and phenol,
chloroform and isoamyl alcohol (volume fraction 25:24:1) were also
obtained from Clontech Laboratories, Inc.
Construction of bait plasmid and
expression of MHBst167
The extracted MHBst167 DNA was amplified
by polymerase chain reaction (PCR) from the A7 plasmid (HBV strain
subtype adr). The primer sequences, which contained EcoRI
and BamHI sites, respectively, for cloning were as follows:
Sense, 5′-gaattcatggtcaccttgaggtgg-3′; antisense,
5′-ggatcccaaacaggagatgaaggtcct-3′. The PCR conditions were as
follows: 94°C for 4 min, denaturation at 94°C for 50 sec, annealing
at 58°C for 50 sec, extension at 72°C for 1 min, 35 cycles. The PCR
product was cloned into the pGEM-T vector and the primary structure
of the insert was confirmed by direct sequencing. The
auto-sequencing assay was performed by the Shanghai Hua Nuo
Biological Corporation (Shanghai, China). The fragment encoding the
MHBst167 was digested from the
pGEM-T-MHBst167 using EcoRI and BamHI
restriction enzymes, and ligated into the pGBKT7 vector. The pGBKT7
vector expressed proteins fused to amino acids 1–147 of the GAL4
DNA-BD and the pGADT7 vector expressed proteins fused to amino
acids 768–881 of the GAL4 DNA-AD. The MHBst167 gene was
inserted into the pGBKT7 multiple cloning site. The resulting
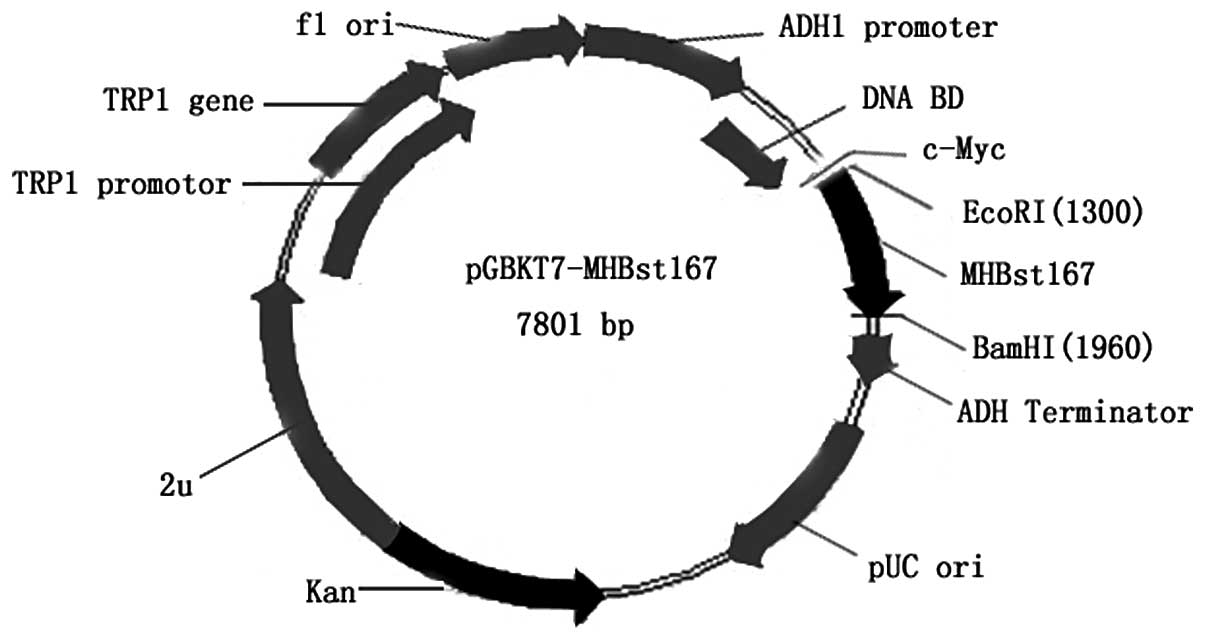
plasmid, pGBKT7-MHBst167 (Fig. 1), containing the full-length
MHBst167 gene, could direct expression of the DNA-BD,
c-Myc and MHBst167 fusion protein. The plasmid was
transformed into yeast strain AH109 using a lithium acetate method
as previously described (11).
Yeast two-hybrid screen
The yeast two-hybrid screening procedure used was a
modification of the method described by Gietz et al
(11). The pACT2-cDNA plasmid
genome was isolated following the standard protocol. AH109 yeast
cells containing pGBKT7-MHBst167 were transformed into
the Y187 yeast strain containing the pACT2-cDNA liver library
(Clontech Laboratories, Inc.) by the lithium acetate method, in
accordance with the manufacturer’s instructions. Cells were plated
and selected for on quadruple dropout (QDO) media lacking leucine,
tryptophan, histidine and adenine. After 6–18 days of growth at
30°C, the yeast colonies were transferred onto plates containing
X-α-Gal to check for the expression of the MEL1 reporter gene.
Positive interactions were detected by the appearance of
blue-colored colonies. Segregation analysis and mating experiments
were performed to exclude the false positives, ensuring that only
true positive colonies were selected. Following the sequencing of
the positive colonies, the sequences were BLASTed with GenBank
(blast.ncbi.nlm.nih.gov/Blast.cgi) to
analyze the function of the genes.
Results
Identification of the plasmid
The 501-bp fragment of MHBst167 was
generated by PCR amplification of the HBV plasmid (subtype adr),
sequenced and analyzed using Vector NTI® 6 (Life
Technologies, Carlsbad, CA, USA) and BLAST database homology. The
PCR product was cloned with the pGEM-T vector. Following digestion
with EcoRI/BamHI restriction enzymes, the fragments
were ligated in-frame into the pGBKT7 and pGADT7
EcoRI/BamHI sites. Restriction enzyme analysis of
pGBKT7-MHBst167 and pGADT7-MHBst167,
respectively, with EcoRI/BamHI yielded two products:
7,300 bp empty pGBKT7 and 501 bp MHBst167, and 7,900 bp
empty pGADT7 and 501 bp MHBst167. Analysis of the PCR
products by agarose gel electrophoresis (Fig. 2) showed the products with the
expected size (501 bp, MHBst167). The
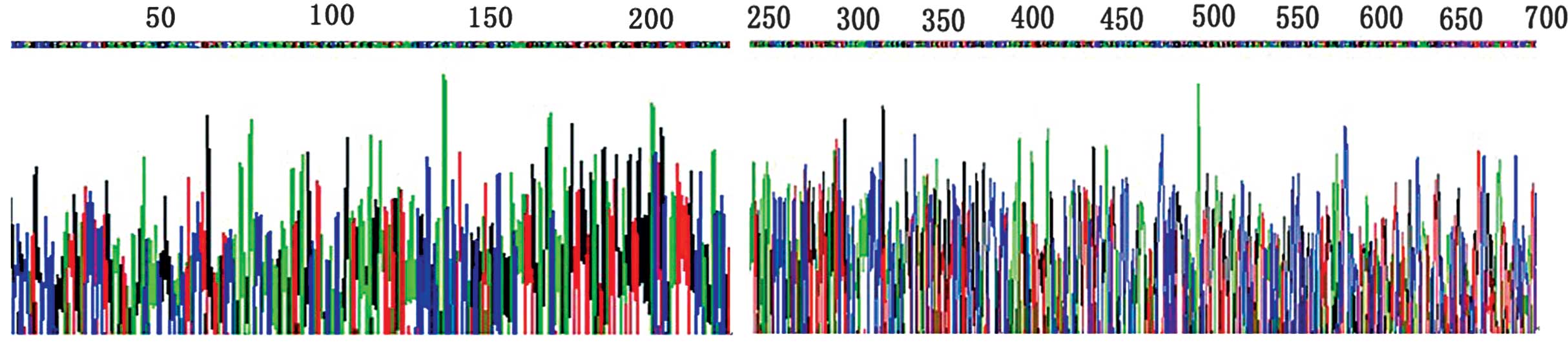
pGEM-T-MHBst167 sequence was confirmed by direct
sequencing (Fig. 3).
Screening of the fetal liver cell cDNA
library
Plasmids from the blue-colored colonies containing
only pGBKT7-MHBst167, as the bait for screening the
human fetal liver cell cDNA library, were isolated. Sixty-six
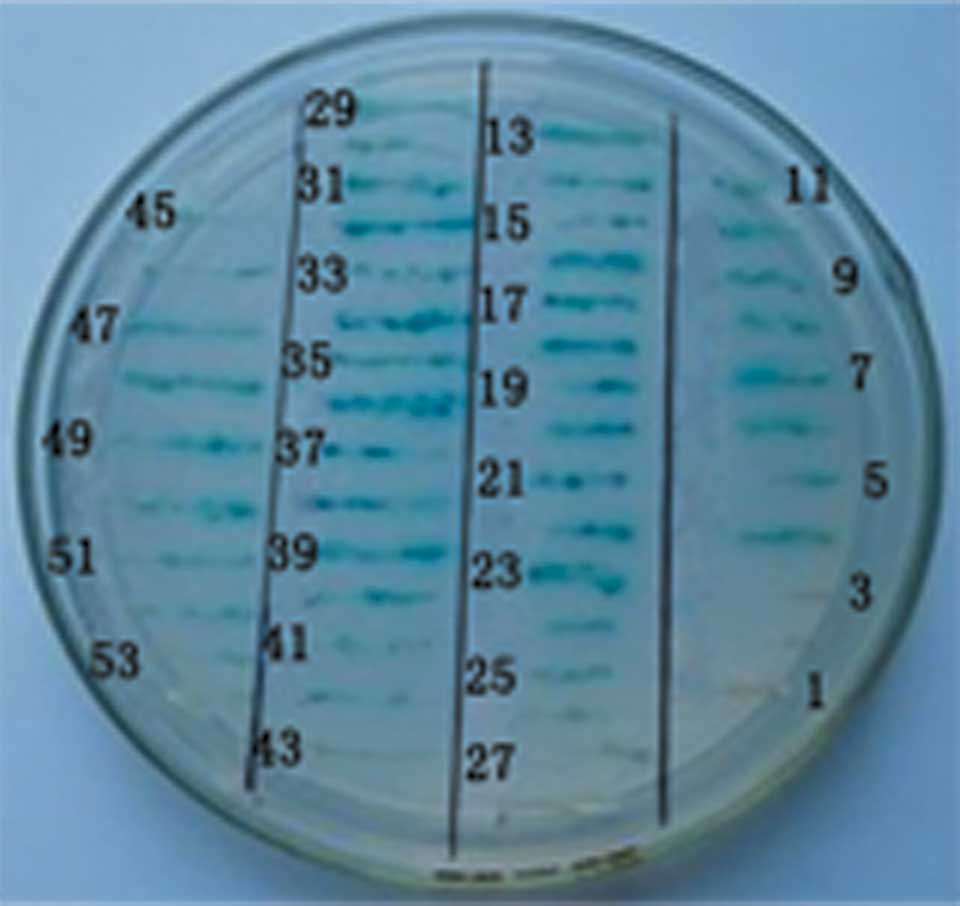
clones grew on the QDO media. The clones were further selected for
by X-α-Gal assay and, again, blue colonies were picked. Following
elimination, 52 positive clones were further tested by the
specificity of the selective SD/-Trp-Leu-His-Ade/X-α-Gal media and
expression (Fig. 4). Since
pACT2-cDNA plasmids contained two restriction endonuclease sites
for recognition by BglII either side of the multiple cloning
site, the gene fragments of the fetal liver cell cDNA library
(pACT2-cDNA) screened were released by BglII digestion
(Fig. 5).
Analysis of cDNA sequencing and
homology
Seven positive colonies grew on the selective
SD/-Trp-Leu-His-Ade/X-α-Gal media. These colonies were prescreened
by BglII digestion to ensure that only colonies with
different inserts were subjected to sequencing. The seven colonies
from the cDNA library were sequenced and analyzed using BLAST
software provided by the National Center for Biotechnology
Information. The seven sequences had high similarity to known
genes. A summary of the identified genes is shown in Table I.
 | Table IComparison between positive clones and
similar sequences in GenBank. |
Table I
Comparison between positive clones and
similar sequences in GenBank.
| High similarity to
known genes | No. of similar
clones | Homology (%) |
|---|
| Homo sapiens
ADP-ribosylation factor 1, mRNA (cDNA clone IMAGE: 4024984) | 2 | 98 |
| Full-length cDNA
clone CS0DM004YC15 of fetal liver of Homo sapiens | 1 | 100 |
| Homo sapiens
ALDOB gene, virtual transcript | 1 | 100 |
| Homo sapiens
complement component 3, mRNA | 1 | 99 |
| Homo sapiens
BAC clone GS1–306C12 from 7 | 1 | 100 |
| Homo sapiens
vitronectin (serum spreading factor, somatomedin B, complement
S-protein), mRNA | 1 | 98 |
Discussion
Genetic alterations associated with human HCC have
been reported for numerous genes; however, at present these are not
sufficient to distinguish between HCC and precancerous liver
diseases, including hepatitis, hepatic fibrosis and cirrhosis
(12). It has previously been
suggested that the expression of certain genes, activated by
translated HBV sequences, may contribute to hepatocarcinogenesis
(10). Large HBV surface protein
and MHBst belong to the HBV PreS2 activator protein family. PreS2
activators have been suggested to act in a similar manner to
promoters of tumorigenesis, modulating the control of proliferation
through the activation of certain key enzymes (13). The ability of PreS2 activator
proteins to activate transcription is associated with the
cytoplasmic orientation of the PreS2 domain. While MHBst represents
a standard for PreS2 activator proteins, the full-length MHBs
exhibits no transcriptional activator activity, since the PreS2
domain is oriented into the lumen of the endoplasmic reticulum. For
functional MHBst to be generated from the full-length MHBs, the 3′
end of the preS2/S gene, which encodes the last 70 amino acids in
the sequence, must be deleted during the integration process. This
sequence of amino acids corresponds to the third hydrophobic region
of the S domain of the protein (14).
The potential of MHBst167 to activate
transcription is mediated via various ubiquitous transcription
factors, which are involved in the activation of proto-oncogenes or
genes associated with inflammation. The overexpression of
proto-oncogenes induced by MHBst167 may be causative of
over-proliferation or may maintain mitogenesis; as such, it has
been proposed that an association exists between transactivation by
integrated HBV sequences and the development of HCC (10).
The present study used a yeast two-hybrid system as
an approach for detecting protein-protein interactions. By this
method, weak protein interactions may be identified which may not
be observed using other in vitro assays, such as
immunoprecipitation. The yeast two-hybrid system 3 utilized in this
study was commercially available from Clontech Laboratories, Inc.
(15–17). This system was selected since the
promoters modulating HIS3, ADE2 and MEL1 reporter gene expression
in the AH109 yeast strain result in significantly fewer false
positives. Furthermore, the simple mating protocol is time- and
labor-efficient. As such, the system facilitates the identification
of rare protein-protein interactions and produces more reproducible
results (18,19).
Following the screening of a fetal liver cDNA
library, this study identified seven putative clones associated
with MHBst167. The proteins identified were as follows:
i) and ii) Homo sapiens ADP-ribosylation factor 1; iii)
full-length cDNA clone CS0DM004YC15 of the fetal liver of Homo
sapiens; iv) Homo sapiens ALDOB gene; v) Homo
sapiens complement component 3 (C3); vi) Homo sapiens
bacterial artificial chromosome (BAC) clone GS1–306C12 from
chromosome 7; and vii) serum spreading factor (SF).
The first interacting protein identified in this
study, ADP-ribosylation factor 1, is a GTPase required for the
exocytosis of cytotoxic T-lymphocyte antigen 4 (CTLA-4), a process
that is also dependent on phospholipase D activity. It has been
suggested that ADP-ribosylation factor 1 and phospholipase D act to
stimulate the rapid translocation of specialized compartments
containing CTLA-4 in regulatory T cells to the plasma membrane
(20). Poly(ADP-ribose) polymerase
(PARP) is an enzyme predominantly localized in the nucleus and is
responsible for catalyzing poly-ADP-ribosylation. The activity of
PARP is associated with numerous biological processes, including
DNA repair, cell proliferation and malignant transformation. It has
been shown that the expression of PARP in HCC is increased in
patients with liver cirrhosis, with higher expression levels in
less-differentiated tumors (21).
The present study showed that MHBst167
also interacted with the full-length cDNA clone CS0DM004YC15 of the
fetal liver of Homo sapiens and the Homo sapiens
ALDOB gene. ALDOB is an enzyme involved in glycolysis and
gluconeogenesis and is the only isoenzyme of aldolase expressed in
differentiated hepatocytes. The impaired function of ALDOB has been
associated with hereditary fructose intolerance, a recessively
inherited disorder of carbohydrate metabolism. To date, 29
mutations have been identified in the ALDOB gene that impair the
functioning of the enzyme (22).
ALDOB has additionally been associated with HCC. In a study by
Kinoshita et al (12),
ALDOB was shown to be downregulated in primary HCC tissues in 90%
of a cohort of 20 patients, as compared with healthy controls.
Therefore, the underexpression of key ALDOB gene products may be
important in the development and/or progression of HCC.
C3 is a key protein involved in the complement
immune system. In a previous study of patients with chronic liver
disease caused by HBV infection, C3 expression levels were revealed
to be lower in the serum of patients than those in the serum of
healthy controls. Furthermore, the levels of circulating immune
complexes (CICs) in the patients were increased (23). It was suggested that the decreased
C3 levels may have been a result of decreased synthesis and/or an
increased consumption by the CICs (23). Results from a study by Takezaki
et al (24) indicated that
C3 levels may exhibit diagnostic power in the detection of HCC in
patients with liver cirrhosis.
The present study showed that MHBst167
also interacted with the Homo sapiens BAC clone GS1–306C12
from chromosome 7.
Another important protein interacting with
MHBst167 from the fetal liver cDNA library was serum SF
(Homo sapiens vitronectin). Human serum SF is a secreted
glycoprotein that is involved in the promotion of cell adhesion and
spreading. SF has been shown to be synthesized and secreted into
culture by HepG2 human HCC cells (25). A study by Inuzuka et al
(26) analyzed plasma vitronectin
levels in patients with liver disease and compared these levels
with various parameters of liver function and the severity of liver
cirrhosis, graded according to Child’s criteria. The plasma
vitronectin level was low in all liver disease groups as compared
with that in the healthy controls, although the difference was only
significant between the controls and patients with HCC and
decompensated cirrhosis. Furthermore, the vitronectin level was
inversely correlated with the severity of cirrhosis. These results
suggested that the plasma vitronectin level may be used as an
indicator of the synthetic function of the liver in patients with
liver diseases and that it may also be a marker of the severity of
cirrhosis. Immunoelectron microscopy in the same study revealed the
presence of vitronectin in the rough endoplasmic reticulum of
hepatocytes, as well as around certain cells close to sites of
necrosis and on collagen fibers (26).
These interacting proteins, identified by the
yeast-two hybrid system, are closely associated with carbohydrate
metabolism, tumor development/progression and immunoregulation.
These data may provide novel insights into the functions of
MHBst167, the pathogenesis of HBV-related diseases and
malignant transformation. Further studies are required to
investigate how the interactions between MHBst167 and
the proteins identified in this study affect the occurrence and
development of chronic hepatitis B, hepatic fibrosis and HCC.
Acknowledgements
This study was previously published as a poster: Zhi
Qun Li, Song Zhang, Yang Zhu and Jun Cheng: Screening of hepatocyte
proteins binding with C-terminally truncated surface antigen middle
protein of hepatitis B virus (MHBst167) by yeast two-hybrid system.
International Journal of Infectious Diseases 2010.07: 98. The
authors wish to thank Chinese PLA 302 Hospital for the supply of
financial and technological assistance in carrying out this
research.
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
MHBst167
|
middle hepatitis B surface protein
C-terminally truncated at amino acid position 167
|
|
PCR
|
polymerase chain reaction
|
|
DNA-BD
|
DNA binding domain
|
|
DNA-AD
|
DNA activation domain
|
|
HCC
|
hepatocellular carcinoma
|
|
QDO
|
quadruple dropout medium lacking
leucine, tryptophan, histidine and adenine
|
|
CIC
|
circulating immune complex
|
|
BAC
|
bacterial artificial chromosome
|
|
SF
|
spreading factor
|
References
|
1
|
Kong XB, Yang ZK, Liang LJ, Huang JF and
Lin HL: Overexpression of P-glycoprotein in hepatocellular
carcinoma and its clinical implication. World J Gastroenterol.
6:134–135. 2000.PubMed/NCBI
|
|
2
|
Sun JJ, Zhou XD, Liu YK and Zhou G: Phase
tissue intercellular adhesion molecule-1 expression in nude mice
human liver cancer metastasis model. World J Gastroenterol.
4:314–316. 1998.PubMed/NCBI
|
|
3
|
Xiao CZ, Dai YM, Yu HY, Wang JJ and Ni CR:
Relationship between expression of CD44v6 and nm23-H1 and tumor
invasion and metastasis in hepatocellular carcinoma. World J
Gastroenterol. 4:412–414. 1998.PubMed/NCBI
|
|
4
|
Feng T, Zeng ZC, Zhou L, Chen WY and Zuo
YP: Detection of human liver-specific F antigen in serum and its
preliminary application. World J Gastroenterol. 5:175–176.
1999.PubMed/NCBI
|
|
5
|
Lin GY, Chen ZL, Lu CM, Li Y, Ping XJ and
Huang R: Immunohistochemical study on p53, H-rasp21, c-erbB-2
protein and PCNA expression in HCC tissues of Han and minority
ethnic patients. World J Gastroenterol. 6:234–238. 2000.PubMed/NCBI
|
|
6
|
Huang XF, Wang CM, Dai XW, et al:
Expressions of chromogranin A and cathepsin D in human primary
hepatocellular carcinoma. World J Gastroenterol. 6:693–698.
2000.PubMed/NCBI
|
|
7
|
Mei MH, Xu J, Shi QF, Yang JH, Chen Q and
Qin LL: Clinical significance of serum intercellular adhesion
molecule-1 detection in patients with hepatocellular carcinoma.
World J Gastroenterol. 6:408–410. 2000.PubMed/NCBI
|
|
8
|
Zhao Cai Yan, Li Yue Lin, Liu Su Xia and
Feng Zhong Jun: Changes of IL-6 and relevant cytokines in patients
with hepatocellular carcinoma and their clinical significance.
World J Gastroenterol. 4(Suppl 2): 332000.
|
|
9
|
Wang HC, Wu HC, Chen CF, et al: Different
types of ground glass hepatocytes in chronic hepatitis B virus
infection contain specific pre-S mutants that may induce
endoplasmic reticulum stress. Am J Pathol. 163:2441–2449. 2003.
View Article : Google Scholar
|
|
10
|
Caselmann WH, Renner M, Schlüter V,
Hofschneider PH, Koshy R and Meyer M: The hepatitis B virus
MHBst167protein is a pleiotropic transactivator
mediating its effect via ubiquitous cellular transcription factors.
J Gen Virol. 78:1487–1495. 1997.
|
|
11
|
Gietz RD, Triggs-Raine B, Robbins A,
Graham KC and Woods RA: Identification of proteins that interact
with a protein of interest: applications of the yeast two-hybrid
system. Mol Cell Biochem. 172:67–69. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kinoshita M and Miyata M: Underexpression
of mRNA in human hepatocellular carcinoma focusing on eight loci.
Hepatology. 36:433–438. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hildt E, Munz B, Saher G, Reifenberg K and
Hofschneider PH: The PreS2 activator MHBs(t) of hepatitis B virus
activates c-raf-1/Erk2 signaling in transgenic mice. EMBO J.
21:525–535. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lauer U, Weiss L, Hofschneider PH and
Kekulé AS: The hepatitis B virus pre-S/S(t) transactivator is
generated by 3′ truncations within a defined region of the S gene.
J Virol. 66:5284–5289. 1992.PubMed/NCBI
|
|
15
|
Fields S and Song O: A novel genetic
system to detect protein-protein interactions. Nature. 340:245–246.
1989. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evangelista C, Lockshon D and Fields S:
The yeast two-hybrid system: prospects for protein linkage maps.
Trends Cell Biol. 6:196–199. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kharel Y, Takahashi S, Yamashita S and
Koyama T: In vivo interaction between the human dehydrodolichyl
diphosphate synthase and the Niemann-Pick C2 protein revealed by a
yeast two-hybrid system. Biochem Biophys Res Commun. 318:198–203.
2004. View Article : Google Scholar
|
|
18
|
Vidalain PO, Boxem M, Ge H, Li S and Vidal
M: Increasing specificity in high-throughput yeast two-hybrid
experiments. Methods. 32:363–370. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
von Mering C, Krause R, Snel B, et al:
Comparative assessment of large-scale data sets of protein-protein
interactions. Nature. 417:399–403. 2002.PubMed/NCBI
|
|
20
|
Mead KI, Zheng Y, Manzotti CN, et al:
Exocytosis of CTLA-4 is dependent on phospholipase D and ADP
ribosylation factor-1 and stimulated during activation of
regulatory T cells. J Immunol. 174:4803–4811. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shimizu S, Nomura F, Tomonaga T, et al:
Expression of poly(ADP-ribose) polymerase in human hepatocellular
carcinoma and analysis of biopsy specimens obtained under
sonographic guidance. Oncol Rep. 12:821–825. 2004.
|
|
22
|
Esposito G, Santamaria R, Vitagliano L, et
al: Six novel alleles identified in Italian hereditary fructose
intolerance patients enlarge the mutation spectrum of the aldolase
B gene. Hum Mutat. 24:5342004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Joshi N, Ayesha Q and Habibullah CM:
Immunological studies in HBV-related chronic liver diseases. Indian
J Pathol Microbiol. 33:351–354. 1990.PubMed/NCBI
|
|
24
|
Takezaki E, Murakami S, Nishibayashi H,
Kagawa K and Ohmori H: A clinical study of complements as a marker
of a hepatocellular carcinoma. Gan No Rinsho. 36:2119–2122.
1990.(In Japanese).
|
|
25
|
Barnes DW and Reing J: Human spreading
factor: synthesis and response by HepG2 hepatoma cells in culture.
J Cell Physiol. 125:207–214. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Inuzuka S, Ueno T, Torimura T, et al:
Vitronectin in liver disorders: biochemical and immunohistochemical
studies. Hepatology. 15:629–636. 1992. View Article : Google Scholar : PubMed/NCBI
|