Introduction
Renal cell carcinoma (RCC) is the most common
malignant tumor of the adult kidney. It accounts for ~3% of adult
malignancies and ~90% of all renal neoplasms (1). RCC is a highly vascularized tumor
that originates in the renal cortex, with a steadily increasing
annual incidence rate of 2.6% (2).
Approximately 30% of patients have metastasis when initially
diagnosed with RCC and up to 30% of patients with clinically
localized disease develop cancer recurrence following surgery
(3,4). Although novel therapeutic strategies
have improved the treatment of RCC, the prognoses of patients with
RCC remain unfavorable, particularly for those with advanced tumors
(5,6).
According to the World Health Organization
International Histological Classification of Kidney Tumors, RCC was
subdivided into clear cell renal cell carcinoma (ccRCC), papillary
RCC, chromophobe RCC, collecting duct carcinoma and unclassified
renal carcinoma, based on histological and genetic differences
(2,7). Approximately 80% of RCCs are
classified as clear cell RCC (ccRCC), which is the most aggressive
form of RCC with the highest rate of metastasis and poorest
survival among common renal malignancies (8,9).
Although great progress has been made, the underlying molecular
mechanisms of tumorigenesis and progression of ccRCC are still not
entirely clear (10–13).
Molecular markers are able to provide information on
the occurrence and aggressiveness of RCC, allowing for the
development of more targeted and effective strategies for early
detection and treatment of RCC (14,15).
Current markers are inadequate to substantially alter existing
diagnostic, therapeutic and prognostic strategies of RCC.
Therefore, it is essential to identify novel renal
cancer-associated genes. By means of bioinformatic approaches, a
novel renal cancer-associated gene newly named Renal Cancer
Differentiation Gene 1 (RCDG1) in the present study [previously
known as chromosome 4 open reading frame 46 (C4orf46); gene ID:
201725], was identified. The expression of RCDG1 in RCC, ccRCC and
normal kidney tissues was examined using western blot and
immunohistochemical analyses. Statistical analysis was then
performed to identify any correlation between RCDG1 levels and
clinicopathological characteristics of the patients.
Materials and methods
Patients and tissue specimens
The tissue specimens used in the present study,
including the paraffin sections of 124 RCC and 92 paired adjacent
normal tissues (located 2.0 cm outside of visible RCC lesions) and
fresh RCC and adjacent normal tissues, were collected from patients
who underwent radical nephrectomy at the Department of Urology,
Peking University Shenzhen Hospital (Shenzhen, China). Clinical and
pathological characteristics of these 124 RCC patients are listed
in Table I. The fresh RCC tissues
and adjacent normal tissues, including eight ccRCCs and four
papillary carcinomas, were stored at −80°C following dissection.
All of these tissue specimens were clinically and pathologically
confirmed to be RCC-positive or normal tissues, by experienced
pathologists of the Pathology Department, Peking University
Shenzhen Hospital (Shenzhen, China). All tissue samples were
classified according to the American Joint Committee for Cancer
(AJCC) and Fuhrman nuclear grading (16). The study was approved by the Ethics
Committee of Peking University Shenzhen Hospital (Shenzhen, China).
Written informed consent was obtained from all patients and the
study was reviewed and approved by the Hospital Ethics
Committees.
 | Table IClinicopathologic characteristics of
124 patients with RCC. |
Table I
Clinicopathologic characteristics of
124 patients with RCC.
| Characteristics | Cases, n (%) |
|---|
| Age (years) |
| <60 | 62 (50.0) |
| ≥60 | 62 (50.0) |
| Gender |
| Male | 88 (71.0) |
| Female | 36 (29.0) |
| Histological
type |
| Clear cell RCC | 60 (48.4) |
| Papillary RCC | 34 (27.4) |
| Chromophobe RCC | 28 (22.6) |
| Collecting duct
carcinoma | 2 (1.6) |
| Fuhrman grade |
| G1–2 | 54 (43.5) |
| G3 | 43 (34.7) |
| G4 | 27 (21.8) |
| AJCC clinical
stage |
| T1 | 78 (62.9) |
| T2 | 33 (26.6) |
| T3–4 | 13 (10.5) |
Perl programming to screen candidate
genes in silico
To screen for novel renal cancer-associated genes
in silico, the following steps were performed as previously
described (17). A secondary
classification database for expressed sequence tag (EST) libraries
was generated based on the Cancer Genome Anatomy Project (CGAP)
information of EST libraries (18). The CGAP EST libraries were
classified into two classes: Libraries from nonfetal, nongerminal
and nonplacental normal tissues (NT), and libraries from renal
cancer. Furthermore, Unigene clusters with <20 ESTs from NT
libraries and >2 ESTs from renal cancer libraries were screened.
The frequency of the best serial analysis of gene expression (SAGE)
tag in NT for each candidate gene was counted based on CGAP SAGE
data and Unigene clusters with <20 SAGE tags from NT were
retained for further analysis. Finally, the candidate genes were
analyzed manually using an Affymetrix HG-U133AB microarray data of
normal tissues downloaded from the University of California at Los
Angeles public core.
Cell lines and culture condition
Human renal cancer cell lines, 786-O, ACHN, 769-P,
Caki-2 and human kidney HEK293T cells were obtained from the Key
Laboratory of Male Reproductive Medicine and Genetics (Guangdong,
China) were used for western blot analysis of RCDG1 expression in
this study. These cells were cultured in Dulbecco’s Modified
Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA, USA)
supplemented with 10% fetal bovine serum, at 37°C in a humidified
incubator containing 5% CO2.
Quantitative polymerase chaine reaction
(qPCR) evaluation for the mRNA of RCDG1
Total RNA from 12 paired renal caner tissues and
adjacent normal tissues was extracted using TRIzol solution
(Invitrogen, Carlsbad, CA, USA), treated with Revert Aid First
Strand cDNA Synthesis kit (MBI Fermentas Inc., Burlington, ON,
Canada) to obtain the cDNA templates according to the
manufacturer’s instructions. The cDNA was then subjected to qPCR
for evaluation of the relative mRNA levels of RCDG1 and GAPDH (as
an internal control) with the corresponding primer pairs: Sense:
5′-GGAGACGCAGCCTTTTCATTA-3′ and antisense:
5′-GTCCCGCCACGTTTTAAGGA-3′ for RCDG1; and sense:
5′-CACCAGGGCTGCTTTTAACTC-3′ and antisense:
5′-GAAGATGGTGATGGGATTTC-3′ for GAPDH. The reaction mixture was set
up in a total volume of 20 μl, consisting of 1 μl cDNA template
synthesized previously, 10 μl SYBR Green master mix (Invitrogen), 1
μl of each primer (sense and antisense primer) and RNase-free
water. Cycling parameters were set as follows: 95°C for 2 min,
followed by 40 cycles of 95°C for 15 sec, 55°C for 30 sec and 72°C
for 40 sec. The expression levels were calculated using the ΔΔCt
method.
Western blot analysis of RCDG1
expression
Collected cells of cell lines used in this study and
the frozen fresh samples were homogenised on ice in three volumes
of lysis buffer [150 mM NaCl, 20 mM Tris-HCl (pH 7.4), 0.1% SDS, 1%
sodium deoxycholate, 1% Triton X-100, 5 mgml aprotinin, and 1 mgml
leupeptin; Thermo Fisher Scientific, Waltham, MA, USA]. Protein
concentration was quantified using the Pierce Bicinchoninic Acid
Protein Assay kit (Thermo Fisher Scientific) and protein samples
(100 μg) were separated by 10% SDS-PAGE and transferred onto
nitrocellulose membranes. After blocking the membranes with 10%
fat-free milk at room temperature for 2 h, the membranes were
incubated in primary antibodies overnight at 4°C. RCDG1 (previously
known as C4orf46) and β-actin proteins were identified with the
primary antibodies rabbit polyclonal anti-C4orf46 (1:1,000, Sigma,
St. Louis, MO, USA) to detect RCDG1 and sc-47778 (1:400, Santa Cruz
Biotechnology, Santa Cruz, CA, USA), respectively. The membranes
were washed three times with Tris-buffered saline containing
Tween-20 and incubated for 2 h with secondary antibody goat
anti-rabbit immunoglobulin (Ig)G-horse radish peroxidase (HRP)
(sc-2004) or goat anti-mouse IgG-HRP (sc-2005). The protein bands
were detected with the Immun-Star™ HRP Chemiluminescence kit
peroxide buffer and luminolenhancer (Bio-Rad Laboratories,
Hercules, CA, USA). Each assay was repeated at least three
times.
Immunohistochemistry (IHC)
IHC analysis of RCDG1 was performed according to
standard procedures. Briefly, paraffin-embedded samples were cut
into 5 μm sections, dewaxed in xylene and rehydrated in a
descending ethanol series, followed by incubation in 3% hydrogen
peroxide solution for 20 min. Antigen retrieval was performed by
boiling the sections in a microwave oven for 2×15 min in 0.01 M
citrate buffer (pH 6.0). The sections were washed with
phosphate-buffered saline (PBS) three times for 5 min, and the
sections were then treated with 10% bovine serum albumin for 30 min
at 37°C to block non-specific protein binding. For the
immunostaining of RCDG1, the specimens were treated with rabbit
polyclonal antibody anti-C4orf46 (1:800, Sigma, USA) to detect
RCDG1 overnight at 4°C. The samples were then rinsed with PBS three
times and treated with the anti-rabbit IHC kit (Maixin Bio; Fujian,
China) at 37°C for 30 min. Subsequently, the slides were stained
with 3′3-diaminobenzidine tetrahydrochloride (Maixin Bio, Fujian,
China) for 3 min, counterstained with hematoxylin (Maixin Bio),
dehydrated, and mounted. Negative controls were prepared with
omission of the primary antibodies.
Evaluation of the staining was carried out by two
independent pathologists who were blinded to the clinicopathologic
variables with a two-score system of immunointensity (II) and
immunopositivity (IP) (19,20).
II was graded as follows: 0, no staining; 1, weakly stained; 2,
moderately stained; 3, highly stained. The percentage of cells with
IP was graded as follows: 0, ≤1; 1, 2–25; 2, 26–50; 3, 51–75 and 4,
≥75%. All of the paraffin-embedded sections were given final scores
based on the multiplications of the II and IP score. A final score
of 0–12 was graded as negative (I, 0–1), weak (II, 2–4), moderate
(III, 5–8), and strong (IV, 9–12). In case of any discrepancy, the
specimens were evaluated by the two observers together until a
final score was agreed.
Statistical analysis
Statistical analyses were performed using SPSS 17.0
(IBM, Armonc, NY, USA). The χ2 test was used to make
comparisons between RCC tissues and adjacent normal tissues, as
well as the comparison between ccRCC and non-ccRCC tissues.
Relationships between the expression of RCDG1 and clinicopathologic
variables were calculated using the Kruskal-Wallis and Mann-Whitney
rank sum tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
Results of in silico screening and qPCR
evaluation
A total of 32 candidate clusters were first screened
out by Perl programming based on EST data. Following secondary
analysis using SAGE and microarray data, the data was narrowed to
nine clusters, with reconfirmed high expression in normal tissues.
The nine clusters were ranked according to the number of EST from
renal cancer and qPCR was performed to evaluate the renal cancer
specificity. In the first five genes evaluated, C4orf46 (chromosome
4 open reading frame 46) was highly specific for renal cancer and
it was therefore temporarily termed RCDG1 (Fig. 1).
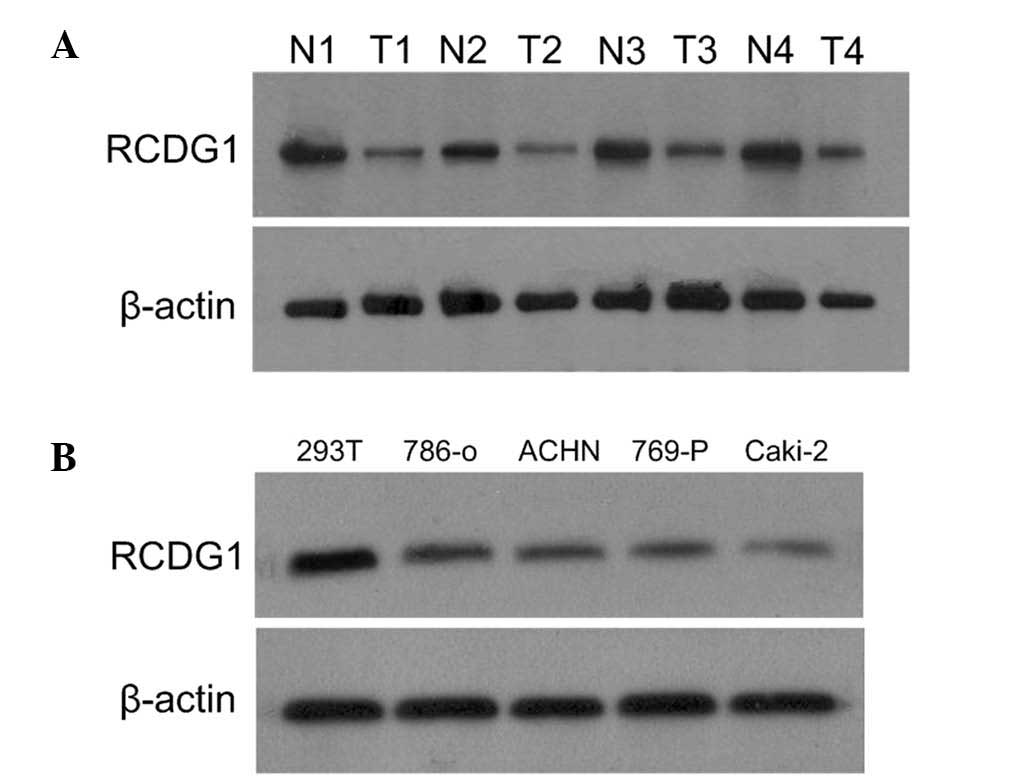
Western blot analysis of RCDG1 protein
levels in RCC tissues and cell lines
Western blotting was performed to determine the
expression levels of RCDG1 in RCC and adjacent normal tissues, as
well as in RCC cell lines and HEK-293T cells. As shown in Fig. 2A, RCDG1 protein was expressed in
both RCC tissues and adjacent normal tissues. The expression levels
of RCDG1 in RCC tumor tissues (T) were significantly lower as
compared with those of adjacent normal tissues (N). This difference
in expression was consistently observed in RCC lines (786-O, ACHN,
769-P and Caki-2) as compared with normal HEK293T cells (Fig. 2B).
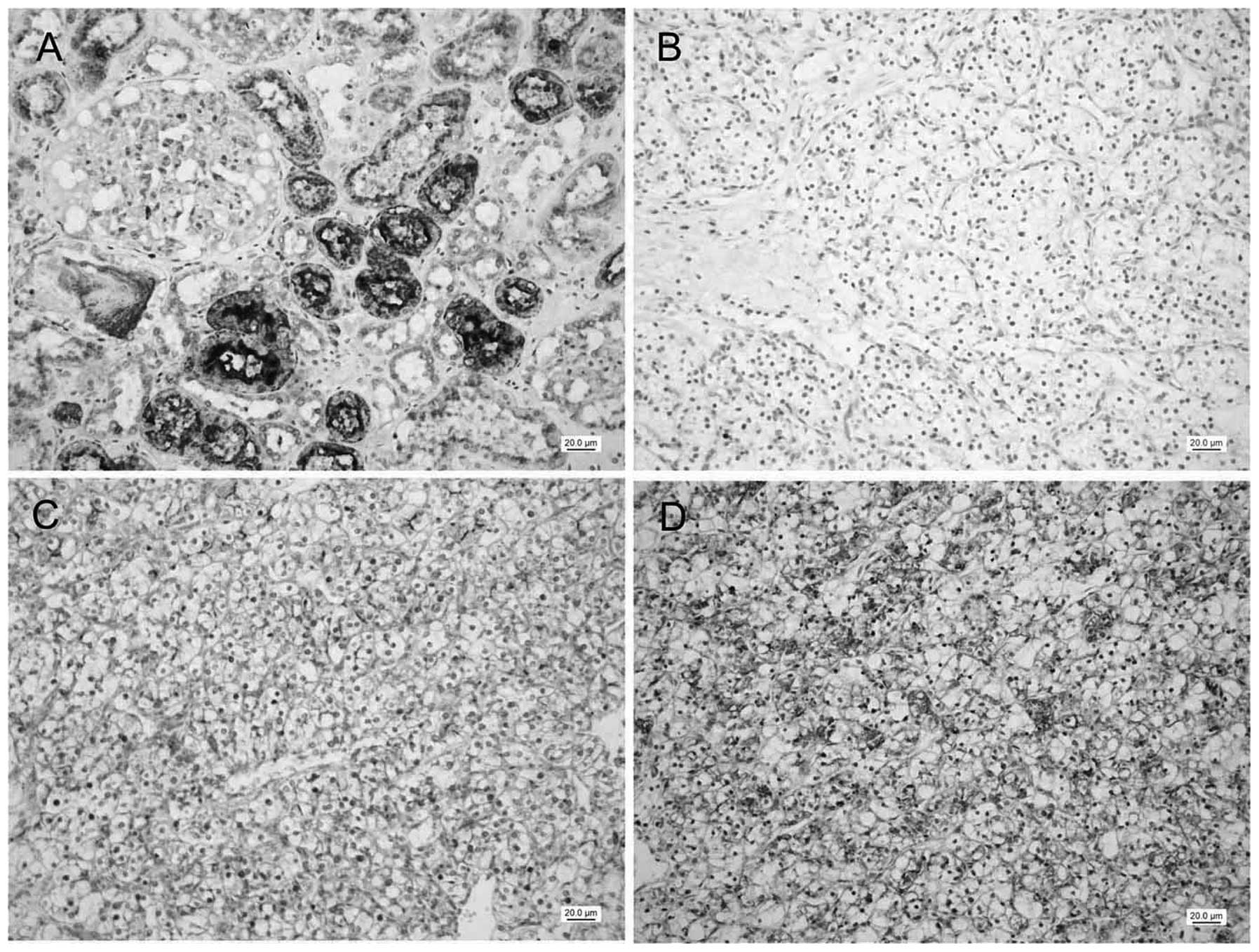
IHC analysis of the expression of RCDG1
in RCC tissues and adjacent normal tissues
In total, 124 RCC tissues and 92 cases of adjacent
normal tissues were used for detection of RCDG1 protein expression
by IHC. In normal renal tissues, 90 (97.8%) cases showed positive
immunostaining (score ≥2) with a total average score of 9.6±0.3 and
85 (92.4%) cases showed strong staining (score ≥9) of RCDG1. As
shown in Fig. 3, epithelial cells
in renal tubules, including the proximal tubules and distal
convoluted tubules, showed strong cytoplasmic staining of RCDG1. By
contrast, 66 (53.2%) cases showed positive staining and none of the
tissue samples were found to be strongly stained. Statistic
analysis demonstrated RCDG1 protein expression levels in RCC
tissues were significantly lower than those in normal tissues
(P<0.001 by χ2 test; Table II).
 | Table IIExpression of RDCG1 in renal cell
carcinoma and normal tissues (χ2 test). |
Table II
Expression of RDCG1 in renal cell
carcinoma and normal tissues (χ2 test).
| | RDCG1 expression, n
(%) | |
|---|
| |
| |
|---|
| Histology | Positive cases, n
(%) | I | II | III | IV | P-value |
|---|
| Normal (n=92) | 90 (97.8) | 2 (2.2) | 1 (1.1) | 4 (4.3) | 85 (92.4) | <0.001 |
| RCC (n=124) | 66 (53.2) | 58 (46.8) | 45 (36.3) | 21 (16.9) | 0 | |
| ccRCC (n=60) | 23 (38.3) | 37 (61.7) | 15 (25.0) | 8 (13.3) | 0 | 0.005 |
| Non-ccRCC (n=64) | 43 (67.2) | 21 (32.8) | 30 (46.9) | 13 (20.3) | 0 | |
Twenty-three (38.3%) cases of ccRCC tissues showed
positive staining of RCDG1 (Table
II). Respectively, 21 cases of papillary RCC (61.8%) showed
positive staining and 21 cases of chromophobe RCC tissues (75.0%)
showed positive immunostaining (Table III). Of the two cases of
collecting duct carcinoma, one of the tissue samples was moderately
stained, whereas the other was negative. The results revealed RCDG1
expression levels in ccRCC tissues were significantly lower than
those in non-ccRCC tissues (P=0.005, χ2 test, Table II).
 | Table IIICorrelation between RDCG1 expression
and clinicopathologic characteristics of patients with non-clear
cell renal cell carcinoma. |
Table III
Correlation between RDCG1 expression
and clinicopathologic characteristics of patients with non-clear
cell renal cell carcinoma.
| RDCG1 expression, n
(%) | |
|---|
|
| |
|---|
|
Characteristics | I | II | III | P-value |
|---|
| Age (years) |
| <60 (n=30) | 12 (40.0) | 14 (46.7) | 4 (13.3) | 0.145 |
| ≥60 (n=34) | 9 (26.5) | 16 (47.0) | 9 (26.5) | |
| Gender |
| Male (n=49) | 18 (36.7) | 22 (44.9) | 9 (18.4) | 0.233 |
| Female (n=15) | 3 (20.0) | 8 (53.3) | 4 (26.7) | |
| Histological
type |
| Papillary RCC
(n=34) | 13 (38.2) | 13 (38.2) | 8 (23.6) | 0.938 |
| Chromophobe RCC
(n=28) | 7 (25.0) | 17 (60.7) | 4 (14.3) | |
| Collecting duct
carcinoma (n=2) | 1 (50.0) | 0 | 1 (50.0) | |
| Fuhrman grade |
| G1–2 (n=29) | 10 (34.5) | 15 (51.7) | 4 (13.8) | 0.310 |
| G3 (n=24) | 9 (37.5) | 10 (41.7) | 5 (20.8) | |
| G4 (n=11) | 2 (18.2) | 5 (45.4) | 4 (36.4) | |
| Tumor stage |
| T1 (n=35) | 15 (42.9) | 14 (40.0) | 6 (17.1) | 0.239 |
| T2 (n=21) | 4 (19.0) | 13 (62.0) | 4 (19.0) | |
| T3–T4 (n=8) | 2 (25.0) | 3 (37.5) | 3 (37.5) | |
Correlation between RCDG1 expression and
clinicopathological characteristics in ccRCC and in non-ccRCC
samples
It was next investigated whether the expression of
RCDG1 correlated to the patients’ clinicopathological
characteristics in ccRCC and non-ccRCC tissues. In ccRCC tissues,
as shown in Table IV, RCDG1
expression was negatively correlated with the Fuhrman grade and
cases with lower RCDG1 expression showed a significantly higher
Fuhrman grade (P=0.008, Kruskal-Wallis test), while no correlation
was found with age, gender and tumor state. No significant
correlation was observed between RCDG1 expression and any of the
characteristics measured in non-ccRCC tissues (Table III).
 | Table IVCorrelation between RDCG1 expression
and clinicopathologic characteristics of patients with clear cell
renal cell carcinoma. |
Table IV
Correlation between RDCG1 expression
and clinicopathologic characteristics of patients with clear cell
renal cell carcinoma.
| RDCG1 expression, n
(%) | |
|---|
|
| |
|---|
|
Characteristics | I | II | III | P-value |
|---|
| Age (years) |
| <60 (n=32) | 18 (56.3) | 9 (28.1) | 5 (15.6) | 0.359 |
| ≥60 (n=28) | 19 (67.9) | 6 (21.4) | 3 (10.7) | |
| Gender |
| Male (n=39) | 22 (56.4) | 11 (28.2) | 6 (15.4) | 0.263 |
| Female (n=21) | 15 (71.4) | 4 (19.1) | 2 (9.5) | |
| Fuhrman grade |
| G1–2 (n=25) | 10 (40.0) | 11 (44.0) | 4 (16.0) | 0.008 |
| G3 (n=19) | 12 (63.2) | 4 (21.0) | 3 (15.8) | |
| G4 (n=16) | 15 (93.7) | 0 | 1 (6.3) | |
| Tumor stage |
| T1 (n=43) | 24 (55.8) | 13 (30.2) | 6 (14.0) | 0.376 |
| T2 (n=12) | 10 (83.3) | 0 | 2 (16.7) | |
| T3–T4 (n=5) | 3 (60.0) | 2 (40.0) | 0 | |
Discussion
Although numerous environmental and genetic factors
have been associated with RCC, the definitive mechanisms involved
in the initiation and progression of RCC have remained elusive
(2,21). Recent identification and potential
application of molecular tumor markers is expected to reform the
clinical staging of RCC and to have an important role in the early
diagnosis, individualized treatment and prognostic prediction of
RCC patients (22). Until
recently, there has been limited use of these molecular markers for
RCC (13). Numerous tumor markers
have been found to be associated with tumor progression and
prognoses of RCC patients. A study by Chuang et al (23) showed that tumor necrosis factor-α
(TNF-α) was able to promote invasion and epithelial-mesenchymal
transition of kidney cancer (23).
Mutations of the Von Hippel-Lindau (VHL) gene were considered
critical for the initiation of ccRCC and loss-of-function mutations
have been shown to be correlated with a poor prognosis for patients
with ccRCC (24,25). High expression of carbonic
anhydrase IX, which is regulated by the Von Hippel-Lindau (VHL)
protein (pVHL), was suggested to be correlated with a favorable
prognosis and a greater likelihood of response to systemic
treatment for metastatic disease (26,27).
However, there is still a need to discover renal cancer-associated
genes, which promote the mechanisms of pathogenesis, invasion and
metastasis of RCC.
A renal cancer-associated gene was newly identified
in the present study through bioinformatic, western blot and
immunohistochemical analyses. This gene was preliminarily named
RCDG1 in this study, however, it is registered as C4orf46
(chromosome 4 open reading frame 46). RCDG1 is located on 4q32.1
with an mRNA of 3,545 bp which encodes a small, conserved and
uncharacterized protein C4orf46 (PRO_0000335689).
Immunohistochemical staining showed that the protein was
predominantly located in the cytoplasm of epithelial cells in the
proximal tubules as well as the distal convoluted tubules.
In the present study, western blotting was performed
to evaluate the expression of the RCDG1 protein in RCC and normal
kidney tissues, RCC cell lines and a normal kidney cell line. The
results demonstrated that RCDG1 was significantly downregulated in
RCC tissues and renal cancer cell lines, as compared with normal
tissues and cell lines. An IHC assay of RCDG1 in paraffin sections
of paired RCC and adjacent normal tissues showed comparable results
to the western blot analysis. Furthermore, statistical analysis
revealed that RCDG1 had a diverse expression pattern across
different types of RCC and the downregulation was more marked in
ccRCC tissues as compared with other types of RCC (non-ccRCC)
tissues. Further analysis showed RCDG1 expression was statistically
correlated with the Fuhrman grade in ccRCC cases but not in other
types of RCC tissues. This suggested that reduced expression of
RCDG1 may be involved in the occurrence and differentiation of
ccRCC.
The number of samples used in the present study was
moderate but sufficient to reveal the statistically significant
differences. The functions of RCDG1 in epithelial cells of renal
tubules, involvement in cellular pathways, transcriptional control
and mechanisms of downregulation in RCC, however, remain to be
elucidated. Recent advances in experimental techniques using
knockdown and transgenic overexpression of target genes have
facilitated further understanding of the pathogenesis, behavior and
molecular biology of cancers (28,29).
Functional experiments on renal cancer cell lines through RNA
interfere and overexpression of RCDG1 may provide further
information for understanding the roles of RCDG1 in RCC.
Furthermore, comprehensive analysis of the transcriptional
regulation of RCDG1 may help identify the mechanisms of the
tumorigenesis and progression of RCC, offering a new target for the
emerging targeted therapies.
In conclusion, the present study newly identified a
renal cancer-associated gene, preliminarily named RCDG1. RCDG1 was
shown to be significantly downregulated in RCC tissues, most
markedly in ccRCC tissues. RCDG1 expression was shown to be
negatively correlated with the Fuhrman grade in ccRCC, suggesting
that the downregulation of RCDG1 may be involved in the
tumorigenesis of RCC and the differentiation of ccRCC. Further
functional analysis of RCDG1 will offer additional information
regarding the role of this gene in RCC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 81101922), Medical
Scientific Research Foundation of Guangdong Province of China (nos.
A2012584 and A2013606) and Science and Technology Development Fund
Project of Shenzhen (no. JCYJ20130402114702124).
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2
|
Patel C, Ahmed A and Ellsworth P: Renal
cell carcinoma: a reappraisal. Urol Nurs. 32:182–190.
2012.PubMed/NCBI
|
|
3
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar
|
|
4
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang Z, Chu PG, Woda BA, et al: Analysis
of RNA-binding protein IMP3 to predict metastasis and prognosis of
renal-cell carcinoma: a retrospective study. Lancet Oncol.
7:556–564. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Suh JH, Oak T, Ro JY, Truong LD, Ayala AG
and Shen SS: Clinicopathologic features of renal cell carcinoma in
young adults: a comparison study with renal cell carcinoma in older
patients. Int J Clin Exp Pathol. 2:489–493. 2009.PubMed/NCBI
|
|
7
|
Lopez-Beltran A, Scarpelli M, Montironi R
and Kirkali Z: 2004 WHO classification of the renal tumors of the
adults. Eur Urol. 49:798–805. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheville JC, Lohse CM, Zincke H, Weaver AL
and Blute ML: Comparisons of outcome and prognostic features among
histologic subtypes of renal cell carcinoma. Am J Surg Pathol.
27:612–624. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Choi YD, Kim KS, Ryu S, et al: Claudin-7
is highly expressed in chromophobe renal cell carcinoma and renal
oncocytoma. J Korean Med Sci. 22:305–310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu L, Wang G, Shevchuk MM, Nanus DM and
Gudas LJ: Activation of HIF2α in kidney proximal tubule cells
causes abnormal glycogen deposition but not tumorigenesis. Cancer
Res. 73:2916–2925. 2013.
|
|
11
|
Metcalf JL, Bradshaw PS, Komosa M, Greer
SN, Stephen Meyn M and Ohh M: K63-ubiquitylation of VHL by SOCS1
mediates DNA double-strand break repair. Oncogene. 20:1055–1065.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang N, Wu P, Wu L, et al: The
differential expression of vascular endothelial growth inhibitor
isoforms, VEGI251, VEGI174 and VEGI192 in human clear-cell renal
cell carcinoma. Cancer Genomics Proteomics. 10:47–53. 2013.
|
|
13
|
Wood CG: Molecular markers of prognosis in
renal cell carcinoma: Insight into tumor biology helps define risk
and provides targets for therapy. J Surg Oncol. 94:264–265. 2006.
View Article : Google Scholar
|
|
14
|
Fritzsche FR, Riener MO, Dietel M, Moch H,
Jung K and Kristiansen G: GOLPH2 expression in renal cell cancer.
BMC Urol. 8:152008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsimafeyeu I, Demidov L, Stepanova E, Wynn
N and Ta H: Overexpression of fibroblast growth factor receptors
FGFR1 and FGFR2 in renal cell carcinoma. Scand J Urol Nephrol.
45:190–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cairns P: Renal cell carcinoma. Cancer
Biomark. 9:461–473. 2010.
|
|
17
|
Lai Y, Yu Z, Wang Y and Ye J:
Identification of PCAG1 as a novel prostate cancer-associated gene.
Mol Med Rep. 7:755–760. 2013.PubMed/NCBI
|
|
18
|
Krizman DB, Wagner L, Lash A, Strausberg
RL and Emmert-Buck MR: The Cancer Genome Anatomy Project: EST
sequencing and the genetics of cancer progression. Neoplasia.
1:101–106. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun S, Du R, Gao J, et al: Expression and
clinical significance of Notch receptors in human renal cell
carcinoma. Pathology. 41:335–341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maaser K, Daubler P, Barthel B, et al:
Oesophageal squamous cell neoplasia in head and neck cancer
patients: upregulation of COX-2 during carcinogenesis. Br J Cancer.
88:1217–1222. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Janzen NK, Kim HL, Figlin RA and
Belldegrun AS: Surveillance after radical or partial nephrectomy
for localized renal cell carcinoma and management of recurrent
disease. Urol Clin North Am. 30:843–852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lam JS, Pantuck AJ, Belldegrun AS and
Figlin RA: Protein expression profiles in renal cell carcinoma:
staging, prognosis, and patient selection for clinical trials. Clin
Cancer Res. 13:703s–708s. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chuang MJ, Sun KH, Tang SJ, et al:
Tumor-derived tumor necrosis factor-alpha promotes progression and
epithelial-mesenchymal transition in renal cell carcinoma cells.
Cancer Sci. 99:905–913. 2008. View Article : Google Scholar
|
|
24
|
Linehan WM, Lerman MI and Zbar B:
Identification of the von Hippel-Lindau (VHL) gene. Its role in
renal cancer. JAMA. 273:564–570. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schraml P, Struckmann K, Hatz F, et al:
VHL mutations and their correlation with tumour cell proliferation,
microvessel density, and patient prognosis in clear cell renal cell
carcinoma. J Pathol. 196:186–193. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bui MH, Seligson D, Han KR, et al:
Carbonic anhydrase IX is an independent predictor of survival in
advanced renal clear cell carcinoma: implications for prognosis and
therapy. Clin Cancer Res. 9:802–811. 2003.PubMed/NCBI
|
|
27
|
Stillebroer AB, Mulders PF, Boerman OC,
Oyen WJ and Oosterwijk E: Carbonic anhydrase IX in renal cell
carcinoma: implications for prognosis, diagnosis, and therapy. Eur
Urol. 58:75–83. 2010. View Article : Google Scholar
|
|
28
|
Di Cello F, Shin J, Harbom K and Brayton
C: Knockdown of HMGA1 inhibits human breast cancer cell growth and
metastasis in immunodeficient mice. Biochem Biophys Res Commun.
2013.PubMed/NCBI
|
|
29
|
Loyd CM, Diaconu D, Fu W, et al:
Transgenic overexpression of keratinocyte-specific VEGF and Ang1 in
combination promotes wound healing under nondiabetic but not
diabetic conditions. Int J Clin Exp Pathol. 5:1–11. 2012.
|