Introduction
Malignant glioma is the most common brain tumor,
with a high ability of invasion, metastasis and recurrence and poor
prognosis (1). Current treatment
options, including surgery, chemotherapy and radiotherapy, have
undesirable outcomes, and therefore, novel therapeutic strategies
are required (2).
Gene therapy has been shown to be an alternative,
efficient therapeutic strategy for glioma treatment. Tumor
suppressor genes, such as p53, and small hairpin (sh)RNA for
oncogenes, can be delivered into glioma cells (3,4).
The WNT/β-catenin pathway has been well documented
to be essential for the proliferation, survival, metastasis and
neoangiogenesis of glioma cells (5). The WNT gene family consists of 19
members, which are soluble ligands for the cellular transmembranal
receptors Frizzled and low density lipoprotein receptor-related
protein (6). For glioma, WNT5a and
WNT7b (particularly WNT5a) are overexpressed and secreted into the
extracellular space in an autocrine or paracrine manner (6). Subsequently, WNT proteins activate
downstream signaling through β-catenin-dependent and -independent
mechanisms. The activation of the WNT/β-catenin pathway ultimately
regulates cell proliferation, motility and tumorigenicity of glioma
cells (7). Decreased expression of
WNT inhibitors is a predominant mechanism causing ectopic
activation of WNT signaling in glioma. Epigenetic silencing of WNT
pathway inhibitor genes by supermethylation of their promoters
accounts for the enhanced activation of WNT signaling in certain
cases of glioma (8). A study using
β-catenin small interfering (si)RNA demonstrated that targeting the
WNT pathway reduces glioma growth (9). Suppression of the WNT pathway
therefore appears to be an effective anti-glioma strategy.
The TIKI gene family of proteases have been
previously identified as a new class of WNT inhibitors, which
utilize a distinct mechanism to negatively regulate the activation
of WNT signaling (10). The human
transcript has two TIKI orthologs, TIKI-1 and TIKI-2. TIKIs have
three separate functional domains: i) Signal peptide sequence; ii)
TIKI domain; and iii) transmembrane domain. The TIKI domain is
highly conserved between different organisms and exerts the
predominant protease activity for WNTs. This can result in the
removal of several N-terminal amino acids from the mature WNT
proteins, or its immature form in the endoplasmic reticulum (ER),
thereby inducing oligomerization of WNT proteins and minimizing
their activity. siRNA-mediated TIKI2 downregulation potentiates the
WNT-3a-induced WNT pathway in mammalian cells. Overexpression of
TIKI proteins has been shown to be able to suppress the
WNT/β-catenin pathway (10). This
indicated that adenoviral vector-based TIKI expression may be an
effective method to treat glioma. However, to date, the association
of TIKIs with glioma and the effect of TIKI gene therapy for glioma
have remained to be investigated.
In the present study, miRNA response elements (MREs)
of tumor suppressor miR-124, which have previously caused
reduced expression levels in a number of glioma cells (11), were constructed in an adenoviral
vector with glioma specificity. Subsequently, this vector was used
to express TIKI2 to suppress the aberrant WNT activation in glioma
cells, aiming to impair the proliferation, invasion and
tumorigenesis of glioma cells both in vitro and in
vivo.
Materials and methods
Cell lines
Human glioma, U-87 MG, U-251 MG, U-373 MG and M059J,
neuronal, HCN-2, fibroblast, MRC-5 and BJ, and endothelial,
HUV-EC-C, cell lines were purchased from the American Type Culture
Collection (ATCC, Manassas, VA, USA). Human L-02 normal liver cells
were obtained from the Shanghai Cell Collection (Shanghai, China).
Cells were cultured using recommended media supplemented with 10%
of fetal bovine serum (FBS; Invitrogen Life Technologies, Carlsbad,
CA, USA), 4 mm glutamine, 100 units/ml penicillin, and 100 μg/ml
streptomycin in a 5% CO2 and humidified atmosphere at
37°C. Media included the following: ATCC-formulated Eagle’s Minimum
Essential Medium for U-87 MG, U-251 MG, U-373 MG, MRC-5 and BJ;
Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F12 medium (1:1)
for M059J; DMEM for HCN-2; ATCC-formulated of F-12K Medium for
HUV-EC-C; and RPMI-1640 for L-02 (All media were purchased from
Invitrogen Life Technologies)
Primary culture
For primary astrocyte culture, cell samples were
obtained from aborted fetuses with informed consent from the
pregnant females, according to the previously published procedures
approved by the Ethical Review Board of Xinxiang Center Hospital
(Xinxiang, China). Briefly, brain tissue from the anterior
fontanelle was dissected into pieces with the removal of meninges,
followed by trypsin digestion. The digested cells were filtered
using a steel net and cultured in DMEM supplemented with 15% FBS.
Glial fibrillary acidic protein (GFAP) expression was confirmed by
immunofluorescent staining.
Quantitative polymerase chain reaction
(qPCR)
Fresh cancerous and noncancerous brain tissues (n=9)
were obtained with written informed consent from all patients
according to protocols approved by Ethical Review Board in Xinxiang
Center Hospital (Xinxiang, China), in order to analyze miR-124
expression. All patients underwent surgical resection of primary
glioma at Xinxiang Center Hospital (Xinxiang, China).
Total RNA was extracted from glioma and noncancerous
brain tissue as well as from the indicated cell lines using TRIzol™
solution (Sigma-Aldrich, St Louis, MO, USA). The reverse
transcription reaction was performed using the TaqMan®
MicroRNA Reverse Transcription kit (Applied Biosystems, Carlsbad,
CA, USA) following the manufacturer’s instructions. qPCR was
performed using TaqMan® 2× Universal PCR Master Mix
(Applied Biosystems) on a CFX96™ Real-Time PCR Detection System
(Bio-Rad Laboratories, Hercules, CA, USA) supplied with LightCycler
Data Analysis software, version 3.5 (Roche Diagnostics, Basel,
Switzerland).
To determine the abundance of TIKI2 mRNA in
adenovirus (Ad)-infected cells, a multiplicity of infection (MOI)
of 10 of the indicated adenoviruses were added to the cell
cultures. Following 48 h, cells were harvested for RNA extraction,
followed by reverse transcription into cDNAs using Rever Tra Ace
qPCR RT Kit (Toyobo, Osaka, Japan) according to the manufacturer’s
instructions. The 7.5-μl reactions were incubated for 30 min at
16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. qPCR was
performed using TaqMan® 2× Universal PCR Master Mix
(Applied Biosystems) using a CFX96™ Real-Time PCR Detection System
(Bio-Rad Laboratories) supplied with analytical software. The
following primers were used: TIKI2, forward 5′-GACCTGCGTGCTGATC-3′
and reverse 5′-TAAAAGAAGATGACAG-3′; Axin2, forward
5′-CCGGTGGACCAAGTCCTTAC-3′ and reverse 5′-TCCATTGCAGGCAAACCAGA-3′;
Lgr5, forward 5′-TGAACACCTGCTTGATGGCT-3′ and reverse
5′-TGCTGCGATGACCCCAATTA-3′. The housekeeping gene was GAPDH,
forward 5′-TCAGTGGTGGACCTG ACCTG-3′; reverse 5′-TGCTGTAGCCAAATT
CGTTG-3′.
Luciferase reporter assay
The sense and antisense sequences containing four
copies of the miR-124 MREs were synthesized, annealed and
inserted into the XhoI and NotI sites of psiCheck2
vectors (Promega Corp., Madison, WI, USA) immediately following the
Renilla luciferase gene, in order to construct an MRE-regulated
luciferase reporter, namely psiCheck2-124. The inserted MRE
sequence for the miRNAs used in the present study was as follows:
5′-TCGAGGATATCACAAACACCGTGCCTTAAACA AACA
CCGTGCCTTAAACAAACACCGTGCCTTAAACAAACA
CCGTGCCTTAAACAAACACCGATATCGC-3′; 5′-GGCC
GCGATATCGGTGTTTGTTTAAGGCACGGTGTTTGT
TTAAGGCACGGTGTTTGTTTAAGGCACGGTGTTTGT
TTAAGGCACGGTGTTTGTGATATCC-3′. Italicization indicates
endonuclease site.
Following 48 h of transfection of U-87 MG, U-251 MG,
astrocyte, MRC-5 and L-02 cells, the cells were harvested and
treated with lysis buffer. Firefly and Renilla luciferase
activities were determined with using the Dual-Luciferase reporter
system (Promega Corp.) following the manufacturers
instructions.
Adenovirus construction
Adenovirus Ad-enhanced green fluorescent protein
(EGFP) was kindly provided by Dr. Zhao (General Hospital of Chengdu
Military Area Command of Chinese PLA, Chengdu, China). Ad-TIKI2 was
constructed using the procedures described below. The sequence of
the human TIKI2 gene used in this study was available from GenBank
(accession no., JQ653416). The cDNA templates were obtained from
HeLa cells. A set of primers was used for PCR-based TIKI2 gene
amplification: Forward 5′-GCCGTCGACACCATGCACGCCGCCCTG-3′ and
reverse 5′-GCCGATATCTCAGGAGGGCCCAA-3′. The PCR product was digested
with SalI and EcoRV restriction endonucleases and
then inserted into the pShuttle-cytomegalovirus (CMV) vector at the
same site, to become pShuttle-CMV-TIKI2. For the construction of
the pShuttle-CMV-TIKI2-124, the DNA fragment containing four copies
of the miR-124 MREs was released by digestion with
EcoRV from the psiCheck2-124 vector, followed by insertion
into the same site of the pShuttle-CMV-TIKI2 to generate the
pShuttle-CMV-TIKI2-124 construct.
The pShuttle-CMV-TIKI2 and pShuttle-CMV-TIKI2-124
constructs were co-transfected using pAdEasy into human HEK-293
embryonic kidney cells with Lipofectamine 2000 transfection reagent
(Invitrogen Life Technologies). Following three rounds of plaque
purification and PCR-based identification, the adenoviruses
(Ad-TIKI2 and Ad-TIKI2-124) were harvested and purified by CsCl
gradient centrifugation. The titers of the adenoviruses were
determined using the TCID50 method (12) in HEK-293 cells and shown as
plaque-forming units per milliliter (pfu/ml).
Immunoblotting assay
To determine the expression levels of phosphorylated
and total β-catenin in adenovirus-infected cells, 10 MOIs of the
indicated adenoviruses were added to the cell cultures. After 48 h,
the proteins were harvested using an M-PER® Mammalian
Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA,
USA), separated by polyacrylamide gel electrophoresis and
transferred onto 0.45 μm nitrocellulose membranes. The membranes
were then blocked with 5% fat-free dry milk for 2 h. The membrane
was incubated with monoclonal rabbit IgG against p-β-catenin
(1:1,000), β-catenin (1:1,000) and GAPDH (1:2,000) primary
antibodies (Cell Signaling Technology, Inc., Danvers, MA, USA) for
2 h, and then incubated overnight with the corresponding secondary
antibody (polyclonal goat IgG against rabbit IgG, 1:10,000;
ZSGB-BIO, Beijing, China). The membranes were visualized using
SuperSignal West Dura Extended Duration Substrate (Thermo Fisher
Scientific) and the ChemiDoc XRS+ chemiluminescence
imaging system (Bio-Rad Laboratories).
T Cell Factor (TCF) reporter assay
The activation of endogenous WNT signaling was
further examined by a TCF reporter dual luciferase assay. Glioma
cells were seeded in 24-well tissue culture plates and transfected
with the TCF reporter vector (TOPflash) (Millipore, Billercia, MA,
USA) and control Renilla luciferase reporter vector (pRL-TK) (40:1)
(Promega Corportation), using Lipofectamine 2000 (Invitrogen Life
Technologies). Overnight, cells were infected with the indicated
adenoviruses of 10 MOI. Forty-eight hours following adenovirus
administration, the cells were collected and resuspended in lysis
buffer (Promega Corp.) and the levels of luciferase activity were
monitored for the adenovirus and Renilla expression levels using
the Dual-Luciferase Reporter Assay kit and a luminometer (Promega
Corp.) according to the manufacturer’s instructions. All
experiments were performed in triplicate.
MTT assay
Adenoviral vectors of indicated MOIs were added to
the cell cultures. Following transduction for the indicated times,
50 ml MTT (1 mg/ml) was added. The MTT reagent was removed after 4
h and 150 ml dimethyl sulfoxide was added. The spectrophotometric
absorbance was measured on a model 550 microplate reader (Bio-Rad
Laboratories) at 570 nm, with a reference wavelength of 655 nm.
Cell viability was calculated according to the following formula:
Cell viability = absorbance value of infected cells/absorbance
value of uninfected control cells.
Apoptosis analysis by cytometry
The cells were transduced with Ad-TIKI2,
Ad-TIKI2-124 or Ad-EGFP of 10 MOI for 48 h. The cells were then
harvested and fixed in 70% ethanol and stained with propidium
iodide (PI; 200 mg/ml) for cytometrical analysis using an Aria II
sorter (BD Biosciences, San Diego, CA, USA). For each group, 10,000
cells were counted to determine the percentages of the
sub-G0/G1 population.
Contact-independent colony formation
assay
To determine the contact-independent colony
formation efficiency, cells were seeded into low-attachment 96-well
plates (Corning Life Sciences, Cambridge, MA, USA) at a density of
10 cells per well. The cells were infected with 10 MOI of Ad-EGFP,
Ad-TIKI2 or Ad-TIKI2-124 and maintained in stem cell medium. Equal
volumes of fresh media were replaced to replenish the growth
factors and nutrients every three days. Methyl cellulose (1%)
(Sigma-Aldrich) was added to prevent cell aggregation. In this way,
an individual sphere derived from a single cell was produced. Cells
were incubated for 14 days and then the number of wells which
contained spheres with a diameter >75 μm were counted. The
experiments were performed in triplicate.
Transwell® assay
The effect of Ad-TIKI2 and Ad-TIKI2-124 on the
invasion of glioma cells was evaluated using the
Transwell® assay in a 24-well plate (Corning Life
Sciences). The Transwell membrane with 8.0 μm pores was coated with
or without Matrigel® (1 μg/μl; BD Biosciences). Cells
were infected with 10 MOI of adenoviruses and resuspended in
DMEM/F12 basic medium. The cells were then seeded in the upper
chambers (5×104 cells each). The nutritional attractant
in the lower chambers was DMEM/F12 medium containing 10% FBS.
Subsequently, the cells were allowed to migrate for 24 h. The
migrant cells on the membranes of the lower chambers were stained
with crystal violet and then the cells in ≥10 randomly selected
microscopic fields at 100× magnification were counted and expressed
as the invasive cells per microscopic field. All the experiments
were performed in triplicate.
Animal experiments
The procedures for animal experiments were approved
by the Committee for the Use and Care on Animals in the Xinxiang
Center Hospital (Xinxiang, China).
A U-87 MG tumor xenograft model was established by
injecting 2×106 cells into the right flanks of
five-week-old male BALB/c nude mice (obtained from the Shanghai
Laboratory Animal Center, Shanghai, China) The mice were kept at an
18–22°C aired condition with 50–60% humidity. As soon as tumors
grew to 6–8 mm in diameter, 24 mice were randomly and equally
divided into four groups (n=6). The mice were intratumorally
injected with 100 μl phosphate-buffered saline (PBS), with or
without 2×108 pfu of Ad-EGFP, Ad-TIKI2 and Ad-TIKI2-124,
respectively. The injections were repeated every other day for five
times, with a total dosage of 1×109 pfu of
adenoviruses.
The tumor diameter was measured by periodic
measurements using calipers and the volume was calculated using the
following formula: Tumor volume (mm3) = maximal length
(mm) × [perpendicular width (mm)]2/2. The tumor weights
were measured when the mice were sacrificed. Immunohistological
staining was used to determine the localization of β-catenin in the
sections of established glioma.
Statistical analysis
Each experiment was performed at least three times.
All values are expressed as the mean ± standard deviation, and
compared with the unpaired two-tailed t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
MREs of miR-124 mediate
glioma-specific TIKI2 expression
The first aim of the present study was to construct
a glioma-specific expression vector for exogenous genes, including
TIKI2. miR-124 has been shown to be underexpressed in glioma
cells as compared with noncancerous brain tissue; therefore, the
MREs of miR-124 were used to restrict exogenous gene
expression within glioma cells for subsequent experiments. The
levels of miR-124 were confirmed to be downregulated in
glioma cell lines (Fig. 1A) and
primary glioma from patients (Fig.
1B) by qPCR assays. Subsequently, a luciferase reporter assay
was used to verify the effectiveness of the miR-124 MREs in
the regulation of inserted genes (Fig.
1C). The data indicated that the MREs-regulated psiCheck2
vector expressed luciferase in a glioma-selective fashion (Fig. 1D).
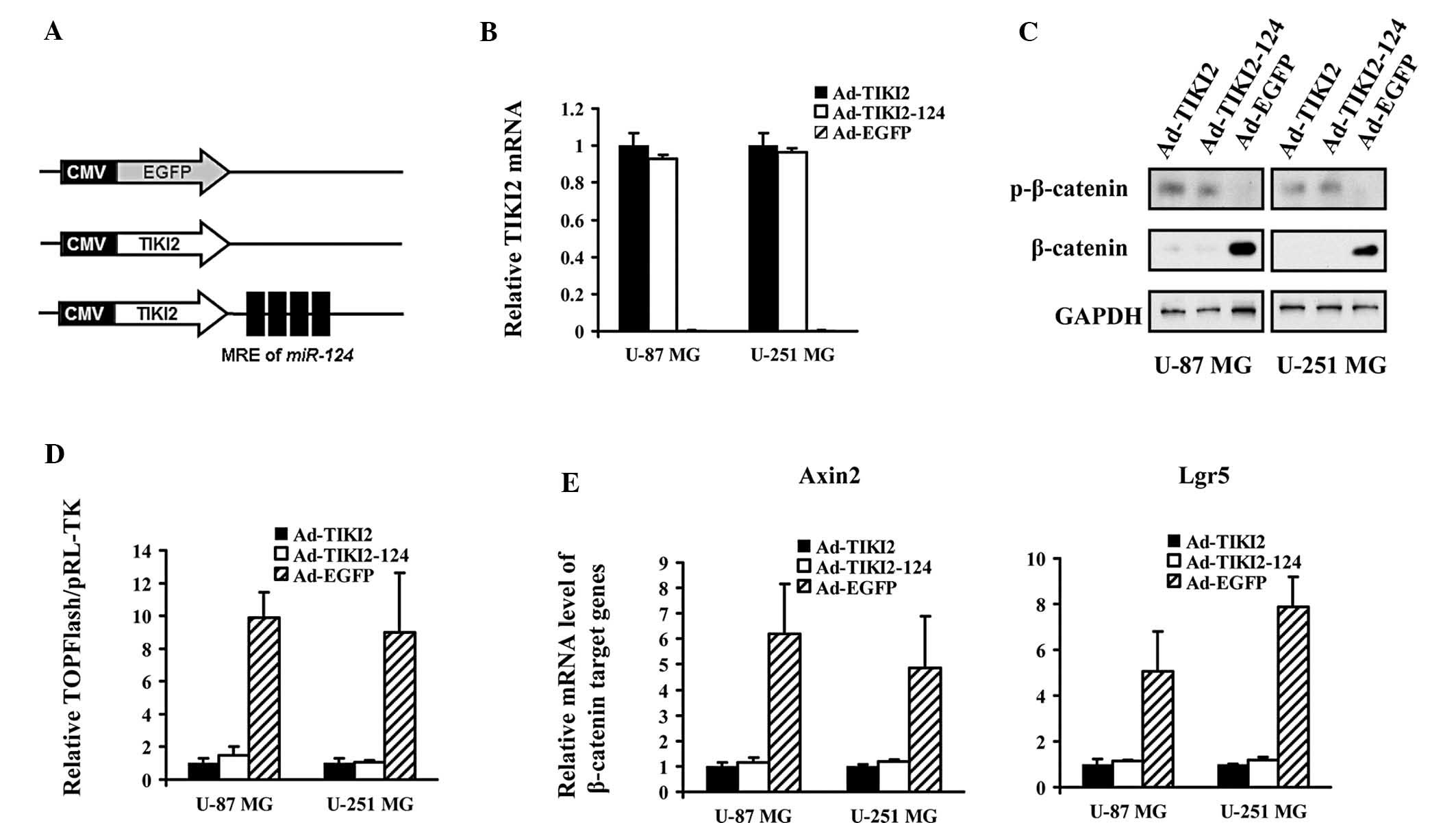
Adenovirus-mediated TIKI2 expression
suppresses the activation of the WNT pathway in glioma cells
An adenoviral vector expressing TIKI2 was
constructed, which harbored four copies of MREs of miR-124
following the coding region of TIKI2 (Ad-TIKI2-124) (Fig. 2A). The Ad-TIKI2-124, and Ad-TIKI2
and Ad-EGFP control vectors were transduced into U-87 MG and U-251
MG cells. qPCR assays confirmed that TIKI2 was highly expressed in
the tested glioma cells infected with both Ad-TIKI2-124 and
Ad-TIKI2 (Fig. 2B). Immunoblot
analysis of β-catenin expression revealed that the phosphorylation
of β-catenin was enhanced in Ad-TIKI2-124- and Ad-TIKI2-infected
glioma cells, with a reduced stabilization of β-catenin (Fig. 2C). TOPFlash reporter experiments
indicated that the transcriptional activity of transcription
factor/lymphoid enhancer-binding factor (TCF/LEF), the major
downstream effector in the WNT/β-catenin pathway, was potently
inhibited in the glioma cells infected with the two distinct
TIKI2-expressing adenoviruses (Fig.
2D). The mRNA abundance of the WNT signaling target genes was
markedly decreased in U-87 MG and U-251 MG cells upon infection
with Ad-TIKI2-124 and Ad-TIKI2 (Fig.
2E).
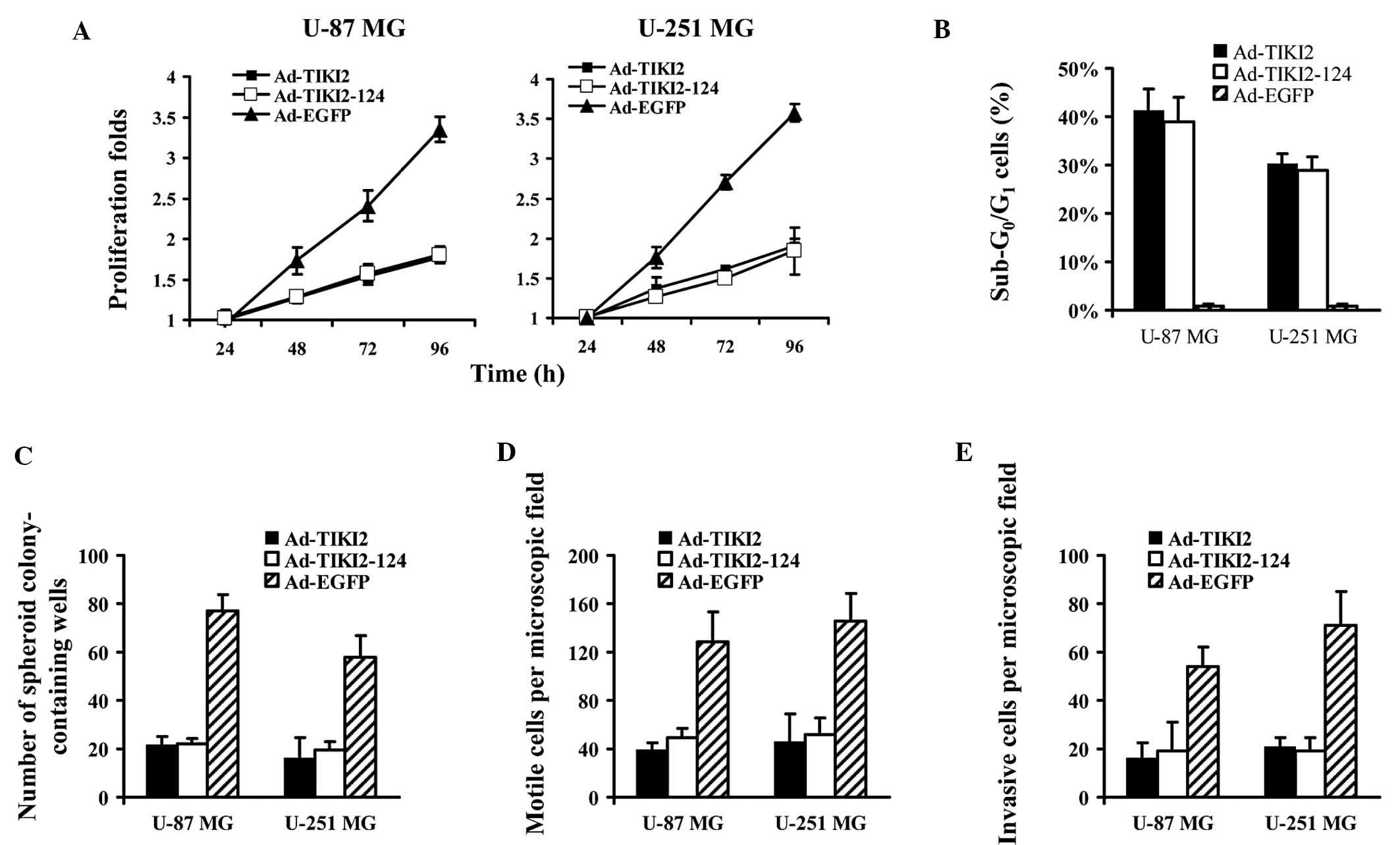
Ad-TIKI2-124 and Ad-TIKI2 exert high
anti-tumor activity on glioma cells
The effects of the TIKI2-expressing adenoviral
vector on the biological traits of glioma cells was analyzed. MTT
assays indicated that Ad-TIKI2-124 and Ad-TIKI2 was able to
decelerate the proliferation of glioma cells (Fig. 3A). The determination of the
sub-G0/G1 subpopulation revealed that TIKI2
expression lead to an increase in the percentage of apoptotic U-87
MG and U-251 MG cells (Fig. 3B).
Furthermore, soft agar assays showed that Ad-TIKI2-124 and Ad-TIKI2
infection of glioma cells greatly compromised the colony formation
ability, evidenced by a markedly lower number of colonies detected
in the group transduced with the two vectors as compared with the
group infected with Ad-EGFP (Fig.
3C). Finally, the motility and invasion capacity of glioma
cells were found to be highly impaired when TIKI2 expression
mediated by the infected adenoviruses occurred (Fig. 3D and E).
Growth of U-87 MG xenograft is suppressed
by TIKI2 expression in vivo
The antitumor activity of TIKI2 was further studied
in a mouse model. U-87 MG cells were subcutaneously injected into
the flanks of mice. When the tumor grew to 8–10 mm, Ad-TIKI2,
Ad-TIKI2-124, Ad-EGFP and PBS were intratumorally administrated.
The diameter of these tumors was periodically measured using a
caliper and their sizes were calculated (Fig. 4A). The weights of U-87 MG cells are
presented in Fig. 4B.
Immunohistological staining revealed that extranuclear
translocation of β-catenin frequently occurred in the tumors
treated with Ad-TIKI2 and Ad-TIKI2-124, while β-catenin accumulated
in the nuclei of Ad-EGFP and PBS-injected tumors (Fig. 4C).
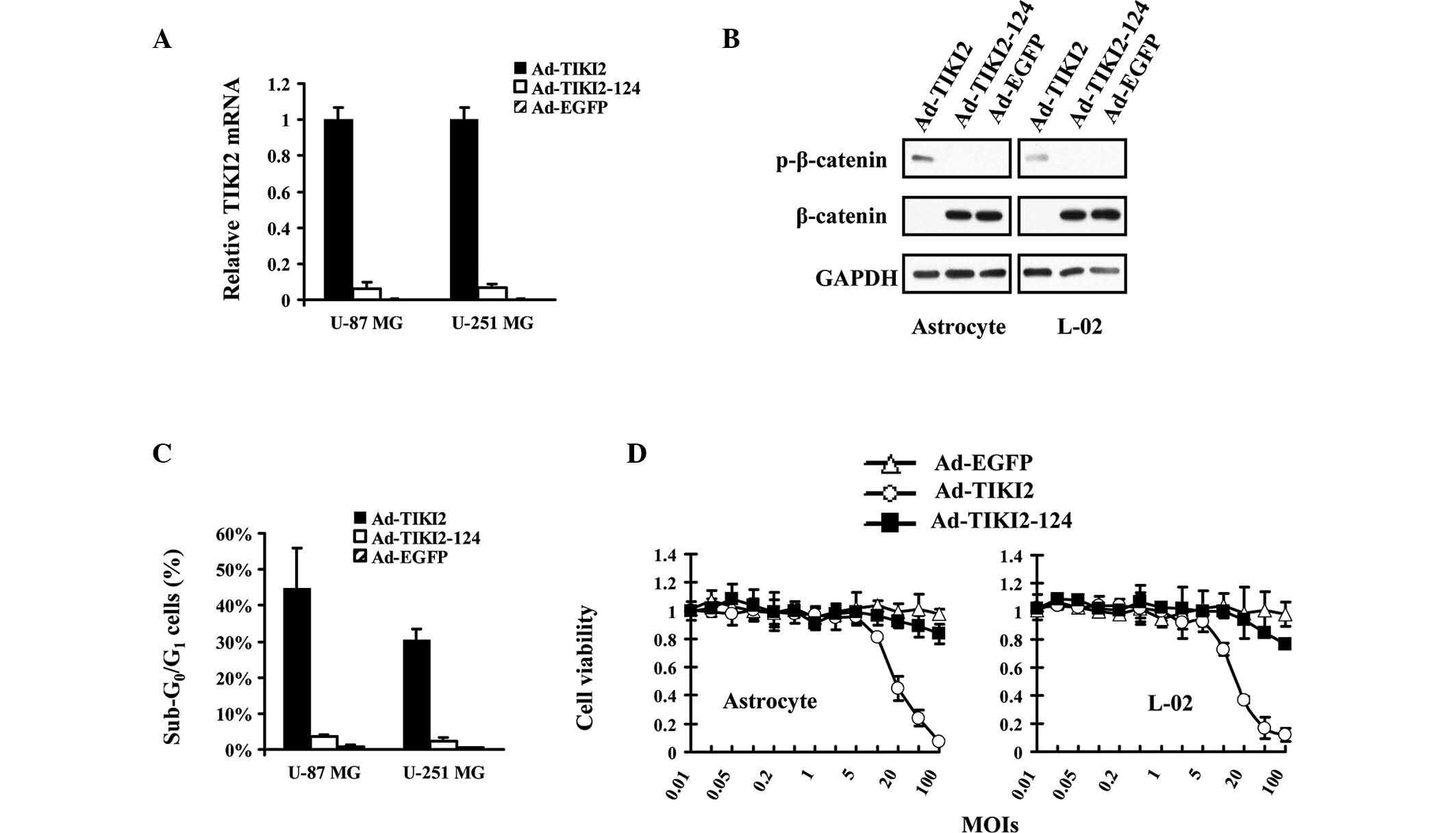
Ad-TIKI2-124 protects normal cells from
TIKI2-mediated suppression of WNT pathway
The effect of TIKI2 expression was a major concern
in the present study. It was therefore investigated whether TIKI2
is able to affect the activation of WNT signaling in normal cells
and induce cytotoxicity. qPCR assays revealed that TIKI2 expression
was suppressed in normal cells infected with Ad-TIKI2-124, which
may be due to the existence of miR-124 in these cell lines
(Fig. 5A). Immunoblot assays
indicated that β-catenin was stabilized in astrocyte and L-02 cells
infected with Ad-TIKI2-124 (Fig.
5B), suggesting that WNT signaling was not affected by the
treatment of Ad-TIKI2-124. In addition, the percentage of
sub-G0/G1 cells was not altered in
Ad-TIKI2-124-transduced normal cells, as compared with
Ad-EGFP-infected cells (Fig. 5C).
Finally, MTT assays revealed that there was no significant
cytotoxicity to normal cells when Ad-TIKI2-124 was added to the
cultures (Fig. 5D).
Discussion
There has been extensive research into glioma gene
therapy for many years. A recent phase III clinical trial revealed
that adenovirus-mediated herpes simplex virus thymidine kinase
(HSV-tk) expression significantly prolonged the survival of glioma
patients (13). This encouraging
result is expected to attract increasing attention from additional
researchers.
The suppressor selected for glioma gene therapy is
critical to ensure effective treatment. To date, numerous genes
have been trialled for glioma therapy (14). In addition to HSV-tk, key genes in
the glioma-specific molecular pathways have additionally been
studied for gene therapy. Adenovirus-mediated delivery of
calreticulin and melanoma-associated antigen 3 has been shown to
inhibit invasion and angiogenesis of U-87 MG glioblastoma cells
through suppression of the metastasis-associated pathway (15). TNF-related apoptosis-inducing
ligand and arresten expression mediated by adenoviral vectors have
also been demonstrated to potently suppress glioma cell growth by
targeting its survival pathway (16). Adenovirus-mediated delivery of
basic fibroblast growth factor small interfering RNA was found to
suppress the activation of signal transducer and activator of
transcription 3 signaling, and thus, induce the apoptosis in U-251
MG glioma cells (17).
Aberrant WNT/β-catenin signaling has been shown to
have a central function in the development of glioblastoma
(5). Proliferation, apoptosis and
invasion have been identified to be affected by the activation of
WNT/β-catenin signaling. Various miRNAs and small molecular
compounds have been shown to exert anti-glioma activity by
suppressing the activation of the WNT pathway (5), suggesting that targeting this pathway
may be an effective strategy for glioma gene therapy. The current
strategy used to suppress the activation of WNT signaling is by
transfection of glioma cells with β-catenin siRNA (9); however, the therapeutic effect is
limited by the low efficiency of transfection.
In the present study, a distinct method to suppress
the activation of the WNT pathway was employed. TIKI2, a protease
specific for WNT protein inactivation, was inserted into adenoviral
vectors for intracellular expression in glioma cells. Subsequent
experiments revealed that TIKI2 potently inhibited WNT/β-catenin
signaling in glioma cell lines.
To improve the biosafety of TIKI2 expression-based
gene therapy, MREs of miR-124 were utilized to confer its
expression with high selectivity for glioma cells. Previous studies
have demonstrated that the application miRNA MREs is able to
regulate exogenous gene expression in a glioma-specific fashion
(18–21). To further improve the selectivity
of gene expression mediated by adenoviral vectors, additional miRNA
MREs may be selected for glioma therapy. miR-122, a
liver-enriched miRNA, can be used to further reduce the
cytotoxicity induced by TIKI2 expression in hepatic cells (21).
In addition to the TIKI gene family, other WNT
inhibitors could also be used for glioma gene therapy. Dickkopf WNT
signaling pathway inhibitor 1 (DKK1) has been shown to suppress the
proliferation of glioma cells, secreted from mesenchymal stem cells
of the umbilical cord (22), and a
therapeutic effect of DKK1 expression has additionally been
verified in gastric cancers (23).
Therefore, DKK1 may also be selected as an exogenous gene for
MRE-regulated adenoviral vector overexpression in future
studies.
Finally, the fiber region of adenoviral vectors can
be reconstructed to enhance infectiveness for glioma cells.
Currently, a 5/35 chimeric adenovirus that harbors a fused fiber
protein from the two different serotypes has been tested for glioma
gene therapy (24), and this
recombinant adenovirus has been used in gene therapy treatment of a
wide range of cancers (23,12).
In future experiments, the 5/35 adenoviral vector may be used to
express TIKI2 in order to potently suppress WNT signaling.
In conclusion, a TIKI2 expressing adenovirus
regulated by MREs of miR-124 was investigated for its use in
glioma therapy. Experimental data confirmed that this strategy is
highly effective for glioma treatment and may be promising for
clinical application.
References
|
1
|
Reardon DA, Galanis E, DeGroot JF, et al:
Clinical trial end points for high-grade glioma: the evolving
landscape. Neuro Oncol. 13:353–361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Recławowicz D, Stempniewicz M, Biernat W
and Słoniewski P: Modern approach to WHO grade II glioma
classification and treatment – review of the literature. Neurol
Neurochir Pol. 42:536–545. 2008.PubMed/NCBI
|
|
3
|
Mitlianga PG, Sioka C, Vartholomatos G, et
al: p53 enhances the Delta-24 conditionally replicative adenovirus
anti-glioma effect. Oncol Rep. 15:149–153. 2006.PubMed/NCBI
|
|
4
|
Wang X, Han L, Zhang A, et al:
Adenovirus-mediated shRNAs for co-repression of miR-221 and miR-222
expression and function in glioblastoma cells. Oncol Rep.
25:97–105. 2011.PubMed/NCBI
|
|
5
|
Zhang K, Zhang J, Han L, Pu P and Kang C:
Wnt/beta-catenin signaling in glioma. J Neuroimmune Pharmacol.
7:740–749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012.
|
|
7
|
Kamino M, Kishida M, Kibe T, et al: Wnt-5a
signaling is correlated with infiltrative activity in human glioma
by inducing cellular migration and MMP-2. Cancer Sci. 102:540–548.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gotze S, Wolter M, Reifenberger G, Muller
O and Sievers S: Frequent promoter hypermethylation of Wnt pathway
inhibitor genes in malignant astrocytic gliomas. Int J Cancer.
126:2584–2593. 2010.PubMed/NCBI
|
|
9
|
Pu P, Zhang Z, Kang C, et al:
Downregulation of Wnt2 and beta-catenin by siRNA suppresses
malignant glioma cell growth. Cancer Gene Ther. 16:351–361. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Abreu JG, Yokota C, et al: Tiki1
is required for head formation via Wnt cleavage-oxidation and
inactivation. Cell. 149:1565–1577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia H, Cheung WK, Ng SS, et al: Loss of
brain-enriched miR-124 microRNA enhances stem-like traits and
invasiveness of glioma cells. J Biol Chem. 287:9962–9971. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He X, Liu J, Yang C, et al: 5/35
fiber-modified conditionally replicative adenovirus armed with p53
shows increased tumor-suppressing capacity to breast cancer cells.
Hum Gene Ther. 22:283–292. 2011. View Article : Google Scholar
|
|
13
|
Westphal M, Ylä-Herttuala S, Martin J, et
al: Adenovirus-mediated gene therapy with sitimagene ceradenovec
followed by intravenous ganciclovir for patients with operable
high-grade glioma (ASPECT): a randomised, open-label, phase 3
trial. Lancet Oncol. 14:823–833. 2013. View Article : Google Scholar
|
|
14
|
Tobias A, Ahmed A, Moon KS and Lesniak MS:
The art of gene therapy for glioma: a review of the challenging
road to the bedside. J Neurol Neurosurg Psychiatry. 84:213–222.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu XL, Zhao D, Sun DP, et al:
Adenovirus-mediated delivery of CALR and MAGE-A3 inhibits invasion
and angiogenesis of glioblastoma cell line U87. J Exp Clin Cancer
Res. 31:82012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X, Mao Q, Wang D, Zhang W and Xia H: A
fiber chimeric CRAd vector Ad5/11-D24 double-armed with TRAIL and
arresten for enhanced glioblastoma therapy. Hum Gene Ther.
23:589–596. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Xu X, Feng X, Zhang B and Wang J:
Adenovirus-mediated delivery of bFGF small interfering RNA reduces
STAT3 phosphorylation and induces the depolarization of
mitochondria and apoptosis in glioma cells U251. J Exp Clin Cancer
Res. 30:802011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bo Y, Guo G and Yao W: MiRNA-mediated
tumor specific delivery of TRAIL reduced glioma growth. J
Neurooncol. 112:27–37. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao Y, Li Y, Wang L, et al: microRNA
response elements-regulated TRAIL expression shows specific
survival-suppressing activity on bladder cancer. J Exp Clin Cancer
Res. 32:102013. View Article : Google Scholar
|
|
20
|
Liu J, Ma L, Li C, et al: Tumor-targeting
TRAIL expression mediated by miRNA response elements suppressed
growth of uveal melanoma cells. Mol Oncol. 2013.PubMed/NCBI
|
|
21
|
Ma L, Liu J, Shen J, et al: Expression of
miR-122 mediated by adenoviral vector induces apoptosis and cell
cycle arrest of cancer cells. Cancer Biol Ther. 9:554–561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma S, Liang S, Jiao H, et al: Human
umbilical cord mesenchymal stem cells inhibit C6 glioma growth via
secretion of dickkopf-1 (DKK1). Mol Cell Biochem. 2013.PubMed/NCBI
|
|
23
|
Wang B, Liu J, Ma LN, et al: Chimeric 5/35
adenovirus-mediated Dickkopf-1 overexpression suppressed
tumorigenicity of CD44+ gastric cancer cells via
attenuating Wnt signaling. J Gastroenterol. 48:798–808. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hoffmann D, Meyer B and Wildner O:
Improved glioblastoma treatment with Ad5/35 fiber chimeric
conditionally replicating adenoviruses. J Gene Med. 9:764–778.
2007. View
Article : Google Scholar
|