Introduction
Hepatocellular carcinoma (HCC) is a highly prevalent
malignancy that is the third leading cause of cancer-related
mortality worldwide (1). Although
significant progress has been made in understanding the cellular
and molecular biology of HCC, the prognosis for HCC patients
remains poor. In China, the majority of HCC cases are secondary to
hepatitis B and C, and are often accompanied by hepatic
fibrosis/cirrhosis. Fibrosis occurs in response to hepatocyte
injury and the subsequent activation of hepatic stellate cells
(HSCs) (2). The combined effects
of hepatocyte injury and HSC activation are required for the
development of hepatic fibrosis/cirrhosis. Hepatic cirrhosis
enhances hepatocyte sensitivity to carcinogenic factors in the
environment and to liver cancer associated pro-inflammatory
cytokines. Hepatitis, hepatic fibrosis/cirrhosis and liver cancer
are thus regarded as the ‘inflammation-fibrosis-cancer axis’ in the
pathogenesis of liver carcinoma (3,4).
Activated HSCs are a major component of the liver
interstitium in HCC. Chau et al (5) investigated HCC liver tissue using
immunohistochemistry and electron microscopy, and identified
numerous myofibroblasts (activated HSCs) among the cancer cells.
Activated HSCs also secrete large quantities of collagen, providing
a collagen source for HCC capsule formation and the accumulation of
excess collagen in HCCs without a capsule (6,7).
Several studies have confirmed that activated HSCs
promote the proliferation and development of HCC cells in
vitro and in vivo (8,9).
However, the mechanism by which activated HSCs affect HCC cells
remains elusive. In the present study, we examined whether
activated HSCs induce specific changes in HCC gene expression, and
began preliminary investigations into the resulting differences in
protein expression. In addition, the effects of activated HSCs on
the proliferation, migration, and invasiveness of HCCs was
investigated in vitro and on tumor growth in
vivo.
Materials and methods
Animals
Specific pathogen-free Buffalo rats (male and
female) were purchased from the Charles River Laboratory
(Wilmington, MA, USA) and maintained at the Shanghai Laboratory
Animals Co., Ltd. (SLAC; Shanghai, China). Male Buffalo rats
weighing 200–250 g were provided by SLAC and housed in the
Experimental Animal Center of Affiliated Zhongshan Hospital of
Fudan University (Shanghai, China). Animals were provided access to
water and food ad libitum. The present study was approved by
the Ethics Committee of the Affiliated Zhongshan Hospital of Fudan
University.
Cell lines
McA-RH7777 liver cancer cells from Buffalo rats were
purchased from American Type Culture Collection (Mannassas, VA,
USA).
McA-RH7777/hepatic stellate cell
co-culture and examination
Preparation of liver cancer
cell-conditioned medium (CM)
McA-RH7777 liver cancer cells were maintained in
DMEM containing 10% fetal bovine serum (FBS) at 37°C in an
environment with 5% CO2. As described in previous
studies (10), when the cells were
~100% confluent, they were washed and maintained in serum-free DMEM
for 2 h. This medium was then discarded and the cells were
incubated in serum-free DMEM for 24 h. The medium from the second
incubation was obtained and centrifuged, and the supernatant was
collected, filtered through a 0.22 μm filter and stored at −20°C
until use.
Preparation of induction-activated
HSCs (iHSCs)
HSCs were separated from the rat liver using a
modified Friedman method (11).
Newly separated HSCs were maintained in DMEM containing 10% FBS for
two days and then in McA-RH7777-CM (50% CM, 10% FBS and 40% DMEM)
for 5–7 days to produce iHSCs (10). Several methods were employed to
determine the purity of HSCs. The freshly separated cells were
observed and identified. Under a light microscope, lipid droplets
have high refractivity and spontaneous fluorescence. Following two
days in culture, the purity of the HSCs was evaluated based on
autofluorescence intensity and the fluorescence intensity of
desmin-positive cells. HSC purity was demonstrated to be >95%.
Immunofluorescence staining of alpha-smooth muscle actin (α-SMA)
was used to detect activated HSCs.
HCC cell culture
McA-RH7777 cells in the exponential growth phase
were digested with trypsin solution containing EDTA and then
suspended in DMEM containing 10% FBS at a cell density of
2×105/ml. In the control group, McA-RH7777 cells (2
ml/well) were cultured alone, while in the experimental group, the
cells were co-cultured with iHSCs in a transwell chamber (diameter,
24 mm; pore size, 0.4 μm; Corning, Shanghai, China). McA-RH7777
cells were seeded into the lower chamber (cell density similar to
that of the controls) and the iHSC suspension (1 ml/well) was added
to the upper chamber (2×105/ml). McA-RH7777 cells or
iHSCs were maintained in DMEM containing 10% FBS for 24 h and then
in DMEM containing 1% FBS for three days.
cDNA microarray analysis
Total RNA extracted from the control McA-RH7777
cells cultured alone or co-cultured with iHSCs was analyzed using a
cDNA microarray. Two independent isolations and microarray analyses
(experiments 1 and 2) were performed using 4×44K Rat Genome Array
chips (Agilent Technologies, Santa Clara, CA, USA) according to the
manufacturer’s instructions. Data analysis was performed using
Feature Extraction and GeneSpring 10.0 software (Agilent
Technologies).
Quantitative (q)PCR analysis of gene
expression
To quantify selected gene expression, McA-RH7777
cells or McA-RH7777 cells co-cultured with iHSCs were lysed with
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) at
a concentration of 100 μl/106 cells and the total RNA
was extracted. The mRNA was reverse-transcribed using an oligo (dT)
18–25 primer and Omniscript reverse transcriptase (Toyobo, Co.,
Ltd., Osaka, Japan). The selected gene transcripts were quantified
by qPCR using gene-specific primers (Table I) and SYBR Green Realtime PCR
Master Mix (Toyobo, Co., Ltd.). The expression of glyceraldehyde
3-phosphate dehydrogenase (GADPH) was used as an internal control.
PCR amplification conditions were as follows: 95°C for 10 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 20 sec.
Samples were run in triplicate using a real-time PCR thermocycler
(ABI PRISM 7000 Sequence Detection System; Applied Biosystems,
Foster City, CA, USA) and the results were analyzed using matched
software. Relative gene expression was determined by normalizing to
GAPDH expression in each set of the samples according to the
manufacturer’s instructions. According to the sequences of the
target gene and GAPDH in GenBank, the corresponding primers were
designed using Premier 5.0 and BLAST was used to exclude the
homologous sequences (Table
II).
 | Table IPCR primer sequences. |
Table I
PCR primer sequences.
| Gene | Primer pairs | Accession no. |
|---|
| Ccl2 | sense:
AGTCGGCTGGAGAACTACAAG
anti-sense: TGAAGTCCTTAGGGTTGATGC | NM_031530 |
| Cxcl1 | sense:
ACCCAAACCGAAGTCATAGC
anti-sense: GGGACACCCTTTAGCATCTT | NM_030845 |
| Cxcl10 | sense:
CATGAACAGACGCTGAGACC
anti-sense: TGCGGACAGGATAGACTTGC | NM_139089 |
| Irf1 | sense:
TGAAGGACCAGAGCAGGAA
anti-sense: GTCAGATAAGGTGTCAGGGCTA | NM_012591 |
| Irf7 | sense:
TCTGGAGAACAGGGAAGAAGT
anti-sense: GTGGCTGTATTGCAGAACCT | NM_001033691 |
| Irf9 | sense:
GCCATTCAAGCGAAGTATCAG
anti-sense: CCGCCATAGATGAAGGTGAG | NM_001012041 |
| Icam1 | sense:
CAAACGGGAGATGAATGGT
anti-sense: CTCTGGCGGTAATAGGTGTAA | NM_012967 |
| Itgae | sense:
CTGCCTTATGAAGTGGAGCG
anti-sense: TGGAGATGAGCCCGAAGTGT | AF020046 |
| Junb | sense:
TAAAGAGGAACCGCAGACC
anti-sense: GCTTTCGCTCCACTTTGAT | NM_021836 |
| Pdgfra | sense:
GCCAGGAGACGAGGTATCAA
anti-sense: TCCCAGAGCAGAACGCCATA | M63837 |
| Sept4 | sense:
CAAGTTGAGGACAATGCTGGTG
anti-sense: GCGATTCCGTTCCTTCACTA | NM_001011893 |
| Tapbp | sense:
CTTGGGATGACGACAATGAT
anti-sense: AATGGTGACGGACAGTGGAGAC | NM_033098 |
| Gapdh | sense:
TACAAGGAGTAAGAAACCGTGGAC
anti-sense: GTTATTATGGGGTCTGGGATGG | NM_017008 |
 | Table IIqPCR was performed to confirm the
results from the DNA microarray assay. |
Table II
qPCR was performed to confirm the
results from the DNA microarray assay.
| | McA-RH7777 | McA-RH7777 +
iHSC | |
|---|
| |
|
| |
|---|
| Gene | Abbreviation | Exp. 1 | Exp. 2 | Exp. 1 | Exp. 2 | log2 iHSCc |
|---|
| TAP binding
protein | Tapbp | 9.609 | 9.868 | 11.842 | 11.797 | 2.081 |
| Intercellular
adhesion molecule 1 | Icam1 | 12.916 | 12.918 | 14.14 | 14.053 | 1.1795 |
| Chemokine (C-X-C
motif) ligand 1a | Cxcl1 | 13.161 | 13.185 | 14.45 | 14.499 | 1.3015 |
| Chemokine (C-C
motif) ligand 2 | Ccl2 | 4.617 | 4.38 | 6.42 | 6.411 | 1.917 |
| Chemokine (C-X-C
motif) ligand 10 | Cxcl10 | 12.13 | 12.008 | 15.67 | 15.741 | 3.6365 |
| Jun B
proto-oncogene | Junb | 9.242 | 9.274 | 10.43 | 10.415 | 1.1645 |
| Interferon
regulatory factor 7 | Irf7 | 7.439 | 8.38 | 15.506 | 15.499 | 7.593 |
| Interferon
regulatory factor 9 | Irf9 | 8.061 | 8.32 | 10.499 | 10.497 | 2.3075 |
| Interferon
regulatory factor 1 | Irf1 | 6.754 | 7.285 | 9.867 | 9.843 | 2.8355 |
| Septin 4 | 4-Sep | 8.874 | 8.854 | 7.771 | 7.7 | −1.1285 |
| Platelet-derived
growth factor receptorb | Pdgfra | 6.51 | 7.079 | 5.499 | 5.458 | −1.316 |
| Integrin, alpha
E | Itgae | 11.244 | 11.021 | 10.081 | 10.021 | −1.0815 |
Western blot analysis of interferon
regulatory proteins and integrin αE
McA-RH7777 cells (1×106) cultured alone
or co-cultured with iHSCs were lysed with 1 ml RIPA buffer
containing 10 μl PMSF. Total protein was prepared by standard
procedures. The total protein concentration was estimated by the
Bradford assay, with BSA as the standard. The proteins were
separated by SDS-PAGE and transferred to polyvinylidene difluoride
membranes. Membranes were blocked for 1 h at room temperature in 5%
non-fat milk in 0.1% Tween-20, then washed and incubated overnight
at 4°C with an anti-rat antibody against one of the following
proteins: interferon regulatory factor 1 (Irf1; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), interferon regulatory
factor 9 (Irf9; Santa Cruz Biotechnology, Inc.), integrin alpha E
(1:1500; ITGAE; Biolegend, San Diego, CA, USA) or β-actin (1:1000;
BD Pharmingen, San Jose, CA, USA). Immunodetection was performed
using the ECL blotting detection system (Bio-Rad, Hercules, CA,
USA).
McA-RH777 cells incubated with
iHSC-CM
Preparation of iHSC-CM (iHSC-CM)
Newly separated HSCs were maintained in DMEM
containing 10% FBS for two days and then in CM-containing medium
(50% CM, 10% FBS and 40% DMEM) for 5–7 days to induce activation of
the cells. The medium was then refreshed and the cells maintained
in serum-free DMEM for 24 h, following which the supernatant was
collected, centrifuged and used as iHSC-CM.
HCC cell proliferation assay
McA-RH7777 cells in the exponential growth phase
were digested with trypsin and re-suspended in DMEM containing 20%
FBS (1×105/ml). These cells were seeded into 96-well
plates (50 μl/well) and divided into two groups (6 wells/group). In
the treatment group, 50 μl of iHSC-CM was added to each well, while
in the control group, 50 μl of DMEM was added to each well. Cell
proliferation was detected with CCK-8 (Cell counting kit-8; Nanjing
KeyGen Biotech Co., Ltd., Nanjing, China) according to the
manufacturer’s instructions.
HCC migration assay (scratch
test)
McA-RH7777 cells were digested with trypsin and
suspended in DMEM containing 10% FBS (2×105/ml). These
cells were then seeded onto 6-well plates (3 ml/well). When cells
reached 70% confluence, they were maintained in serum-free medium
for 12 h and then divided into two groups (3 wells/group). The
supernatant was removed and the cell layer was scratched with a
10-μl pipette tip. Cells were then washed with PBS to remove
shedding cells. In the experimental group, cells were maintained in
DMEM (1.5 ml) containing 2% FBS and 1.5 ml of iHSC-CM. In the
control group, cells were grown in DMEM (3 ml) containing 1% FBS.
Cell migration was observed under a light microscope (DFC500;
Leica, Wetzlar, Germany).
Detection of HCC invasion
A transwell chamber was prepared by adding Matrigel
to the chamber, followed by incubation at 4°C overnight. Serum-free
DMEM was diluted at a ratio of 1:7 and then added to the upper
chamber (80 μl). The transwell chamber was placed on a plate and
incubated at 37°C for 12 h. A suspension of McA-RH7777 cells with a
density of 1×105 cells/ml was prepared. In the control
group, cells were suspended in serum-free DMEM, while in the
experimental group, cells were suspended in a solution with a 1:1
ratio of serum-free DMEM and iHSC-CM. The cell density in the two
groups was identical. These cells (200 μl) were added to the upper
chamber of the transwell chamber. In the control group, DMEM
containing 10% FBS (600 μl) was added to the lower chamber. In the
experimental group, DMEM containing 20% FBS (300 μl) and iHSC-CM
(300 μl) was added to the lower chamber. Cells were then incubated
for 36 h and fixed in neutral formalin. The Matrigel and cells in
upper chamber were removed, followed by Giemsa staining. Several
fields were randomly selected under a light microscope followed by
cell counting to determine the extent of invasion.
Changes in protein expression in
co-cultured cells
McA-RH7777 cells in the exponential growth phase
were digested with trypsin and suspended in DMEM containing 10% FBS
at a density of 4×105/ml. These cells were seeded into
6-well plates (2 ml/well) and divided into two groups (3
wells/group). In the control group, McA-RH7777 cells were cultured
alone, while in the experimental group, cells were co-cultured with
iHSCs in a transwell chamber (diameter, 24 mm; pore size, 0.4 μm).
McA-RH7777 cells were seeded into the lower chamber (cell density
was similar to that of controls) and iHSC suspension (1 ml) was
added to the upper chamber (2×105/ml). One day later,
the medium was refreshed with serum-free DMEM and incubated for two
days. The supernatant was then collected from the two groups and
ELISA (R&D Labs, Minneapolis, MN, USA) was performed to detect
the hepatic growth factor (HGF), interleukin 6 (IL-6), matrix
metalloprotein-2 (MMP-2), matrix metalloprotein-9 (MMP-9), tumor
necrosis factor-α (TNF-α) and transforming growth factor-β1
(TGF-β1).
In vivo study of tumorigenicity
Rats were divided into two groups (n=6/group). In
the control group, 200 μl of McA-RH7777 cells (2×106
cells) were subcutaneously inoculated into the limbs of Buffalo
rats. In the experimental group, 200 μl of McA-RH7777 cells
(2×106 cells) and 200 μl of iHSC (1×106
cells) were subcutaneously inoculated into the limbs of these rats.
Three weeks following inoculation, rats were sacrificed with an
anesthesia overdose and the subcutaneous tumor was collected,
weighed, embedded in paraffin and sectioned, followed by H&E
staining. The dimensions of the tumor were then recorded.
Statistical analysis
Data were presented as the mean ± standard
deviation. Two-sided values of P<0.05 were considered to
indicate a statistically significant result. Differences in the
quantitative data between McA-RH7777+iHSC and the control groups
were examined using the independent two samples t-test. Statistical
analyses were performed using SPSS 15.0 statistics software (SPSS,
Inc, Chicago, IL, USA).
Results
Identification of HSCs and iHSCs
Fig. 1 demonstrates
the histological characteristics of resting and activated HSCs. In
resting HSCs, refraction of intracellular lipid droplets (Fig. 1A) and blue-green fluorescence
following excitation at 328 nm (Fig.
1B) were observed. Desmin staining demonstrated the purity of
HSCs to be >95%. HSCs activated by incubation with HCC-CM
revealed positive staining for α-smooth muscle actin (α-SMA), a
marker for activated cells (Fig.
1F).
Co-culture with iHSCs alters gene
expression in McA-RH7777 cells via a paracrine mechanism
Microarray results
cDNA microarray examination detected 28,728 genes,
573 of which were up- or downregulated >2-fold over the control
levels (HCC cells cultured alone). Of the genes with altered
expression levels, 432 were upregulated and 141 were downregulated
(Fig. 2A). Upregulated genes
include Tapbp, Ccl2, Cxcl1, Cxcl10, Junb, Igf1, Stat1, Irf7, Irf9
and Csf1. 4-Sep was among the downregulated genes. Based on GO
enrichment analysis, upregulated genes were classified into ten
groups (Fig. 2B) and downregulated
genes were classified into five groups (Fig. 2C). The genes with altered
expression encoded cell surface receptors, proteins involved in
cell metabolism, cell adhesion molecules, cell signaling pathway
molecules, chemokines and immune-associated factors.
Confirmation of cDNA microarray assay
results by qPCR
qPCR was used to detect the expression of selected
genes, to validate the findings from the cDNA microarray assay. The
results demonstrated a significant increase in the expression of
Tabpb, Icam1, Cxcl1, Ccl2, CXCL10, Junb, Irf7, Irf9 and Irf1
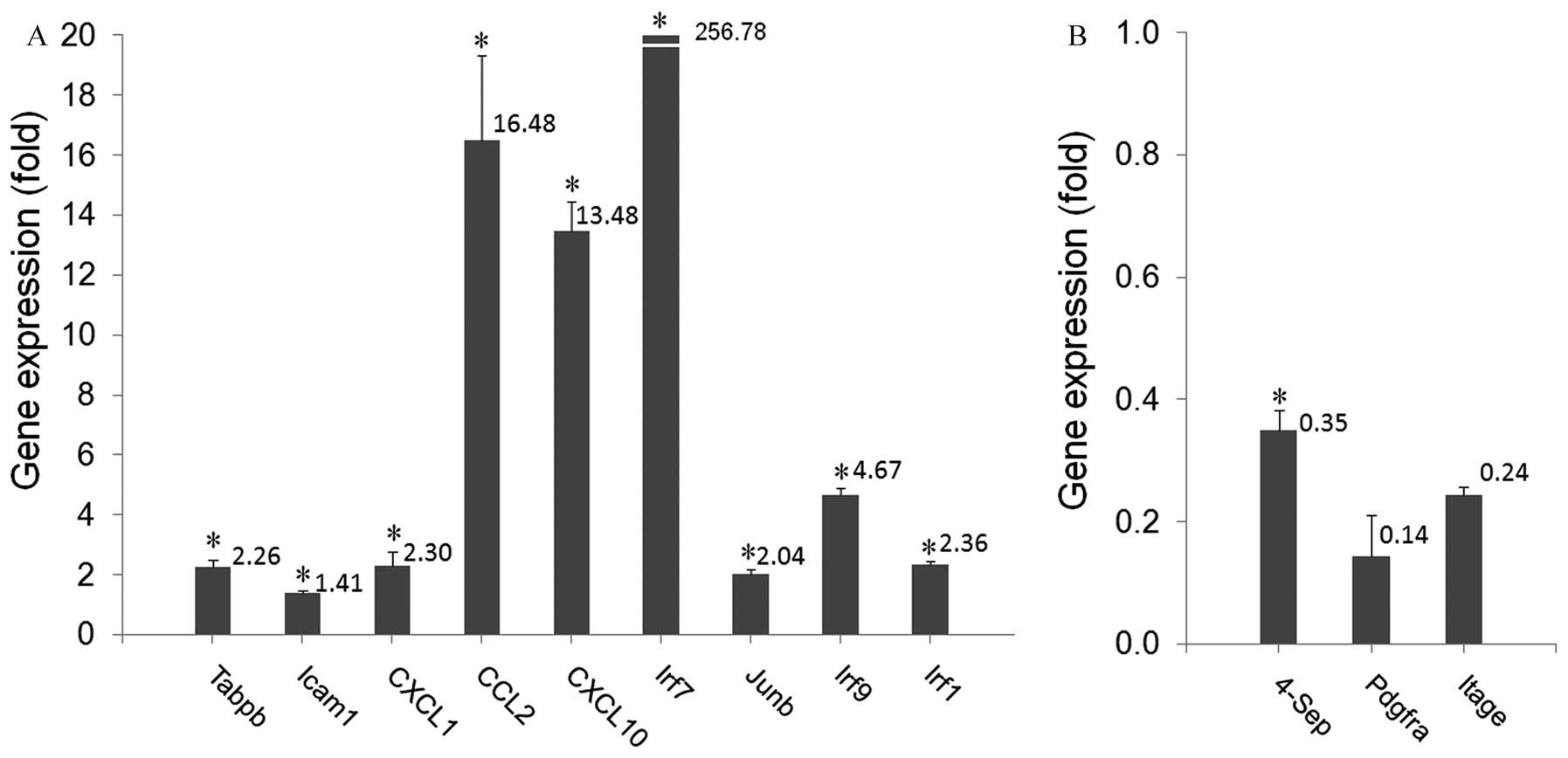
(Fig. 3A) in the McA-RH7777-iHSC
cells compared with the controls (range, 1.4–256. 8-fold; P≤0.016).
The expression level for 4-Sep (Fig.
3B) decreased significantly compared with the controls (0.35
fold; P<0.001). The expression levels of PDGFRA and ITGAE also
decreased, however these differences did not reach statistical
significance (P>0.05). These qPCR findings confirmed the results
of the cDNA microarray assay.
Confirmation by western blot
analysis
Western blot analysis was used to detect the protein
expression of three genes. Co-culture of McA-RH7777 with iHSC
downregulated the expression of integrin αE protein and upregulated
the protein expression of Irf1 and Irf9 as compared with the
protein expression levels in McA-RH7777 control cells. These
results were consistent with those of the qPCR and cDNA microarray
assays (Fig. 4).
iHSCs promote proliferation, migration
and invasion of HCC cells via paracrine mechanisms
CM from iHSCs promotes HCC cell proliferation. In
the presence of iHSC-CM, HCC cells proliferated more rapidly than
the control cells. Increased cell counts were observed in the
McA-RH7777 and the McA-RH7777 + iHSC groups. However the cell count
[Optical density (OD) value)] increased more rapidly in the
McA-RH7777 + iHSC co-culture than in McA-RH7777 alone. This cell
count difference reached significance at 72 h (OD value, 0.62 vs.
0.50; P<0.001) and remained at 96 h (OD value, 0.82 vs. 0.72;
P=0.001; Fig. 5A).
CM from iHSCs promotes migration of
HCC cells
To investigate the effect of iHSC paracrine
mechanisms on the migration of HCC cells, the scratch test was
employed. In the presence of iHSC-CM, the migration rate of HCC
cells increased markedly (Fig.
5B).
CM of iHSCs increased the invasiveness
of HCC cells
The transwell assay demonstrated that the number of
invasive cells in the McA-RH7777 + iHSC-CM preparation was
significantly higher than in the controls (42.6 vs. 18.8, P=0.001;
Fig. 5C).
Co-culture induces changes in protein
secretion from HCC cells
ELISA of cell culture medium revealed that the
concentrations of HGF (150.4 vs. 122.8 ng/ml), IL-6 (56.3 vs. 32.8
ng/ml), MMP-2 (48.3 vs. 37.3 ng/ml) and MMP-9 (20.6 vs. 11.7 ng/ml)
in the McA-RH7777 + iHSC group were significantly higher than in
the control group (all P≤0.007; Fig.
6). No significant difference was observed in TNF-α and TGF-β
concentrations between McA-RH7777 + iHSC and the control
groups.
Tumorigenicity test in Buffalo rats
By three weeks following inoculation, the cancer
cells had invaded the skin of rats co-injected with McA-RH777 cells
and iHSCs. Three of the six rats developed skin ulcers and skin
invasion was pathologically confirmed in all three rats. However,
in the rats injected with HCC cells alone, the skin was not
involved and the cancers had complete capsules. The tumors weighed
significantly more in the McA-RH7777 + iHSC group than in controls
(2.18 vs. 1.04 g; P<0.01; Fig.
7).
Discussion
In the present study, we observed that the
incubation of HCC cells in iHSC-CM increased the rate of HCC cell
proliferation and migration, and the number of invasive HCC cells.
Co-culture of iHSCs and HCC cells induced extensive changes in the
gene expression profile and increased the expression of HGF, IL-6,
MMP-2 and MMP-9 in HCC cells. In addition, co-injection of HCC and
activated HSCs into rats significantly increased the weight of the
resulting HCC tumors.
HSCs are located in Disse’s space and are rich in
cytoplasmic lipid droplets containing vitamin A. HSCs account for
~3% of non-parenchymal cells (12). In the presence of liver injury,
HSCs become activated and transform into myofibroblasts. These
activated HSCs are characterized by changes in morphology, a
reduction in lipid droplets, the presence of α-SMA and active
proliferation (13,14). As observed in the current study,
these activated HSCs have been demonstrated to increase the growth
and proliferation of HCCs in vitro and in vivo
(9,15).
Previously, we demonstrated that treatment of
stellate cells with HCC-CM resulted in a gene expression profile
that differed from that observed in stellate cells activated by
culture medium alone (10). In the
present study, the converse experiment was conducted. The
co-culture of activated stellate cells and HCC cells under
non-contact conditions was observed to alter the gene expression
profile of the HCC cells. Thus, the stellate cells secreted
substances that triggered gene expression alterations in the HCC
cells.
Results of the microarray assay demonstrated changes
in the expression of 573 HCC cell genes following co-culture with
stellate cells. There are numerous known functions of these genes,
including as cell surface receptors, proteins which involved in
cell metabolism, adhesion molecules, signaling pathway molecules,
chemokines and immune associated factors. These genes have
functions similar to those with altered expression in HSCs
activated by HCC-CM. We therefore hypothesized that iHSCs may have
an extensive effects on McA-RH7777 HCC cells by impacting
numerousgenes involved in cell survival, proliferation, metabolism
and immunity. The extent to which these changes in gene expression
actually alter protein expression are not addressed in the present
study and require further investigation.
The present study also investigated the effects of
iHSC-CM on the proliferation, migration and invasiveness of HCC
cells. It was observed that iHSC-CM promoted the proliferation,
migration and invasion of HCC cells in vitro, a result
consistent with previous studies (9,15,16).
Our in vivo results were similar. The tumorigenicity test
demonstrated that cancer occurred earlier in HCC cells
co-inoculated with iHSCs. Four days following co-inoculation, a
rice-sized mass was identified under the skin in the co-injected
rats. However, in the group inoculated with HCC cells alone, such a
mass was not identified until one week after inoculation. This
in vivo evidence further confirmed that iHSCs promoted the
growth and invasion of HCC cells, similar to results reported by
Amann et al (15).
The possible mechanism underlying the paracrine
effects of iHSCs was investigated by examining the expression of
key regulatory proteins in HCC cells in response to iHSC paracrine
factors. TGF-β has been demonstrated to be increased in liver
injury and to be important in activating HSCs and transforming them
into myofibroblasts (17).
However, HCC cells co-cultured with iHSCs demonstrated no change in
TGF-β1 or TNF-α production. These results were unexpected given the
known activities of TGF-β. We suggested that while TGF-β may
promote HSC activation, the HSC-induced changes observed here were
independent of TGF-β activity.
Our preliminary protein studies investigated the
expression levels of proteins encoded by six of the genes
overexpressed in HCC cells following co-culture with iHSCs. Marked
increases in the expression of HGF, IL-6, MMP-2 and MMP-9 were
observed (P<0.05). HGF promotes the endothelial mesenchymal
transition of HCC cells and cancer formation, and has been reported
to act through the Akt pathway (18). However, HCC migration in the
presence of HSCs has been reported by Santomoto et al to
depend on the MEK/ERK pathway and not on the P13K/Akt pathway
(19). IL-6 may facilitate
angiogenesis, vascularization and tumorigenicity (20), and may have an important role in
tumor progression (21). MMPs
degrade the extracellular matrix and have been demonstrated to
promote the invasion and metastasis of cancer cells (22). Therefore, the increase in MMP-2 and
MMP-9 observed in our study may be associated with the increase
invasiveness of HCC cells following exposure to iHSCs. Evidence
from other studies on the mechanisms underlying how iHSCs increase
HCC growth and invasiveness vary. The NFκB and ERK pathways have
been reported by a number of studies to be involved (15,19).
In addition, HSCs have been reported to have a systemic
immunosuppressive effect, and to promote angiogenesis and
proliferation (9).
In summary, the in vitro alterations in the
gene expression profile of HCC cells in response to co-cultivation
with iHSCs were extensive. While the significance of changes in the
expression of specific genes has not been addressed here, we
hypothesize that the interaction between HSCs and HCC cells in
vivo may be multifaceted, and that activated HSCs may have an
important role in the progression of HCC. Alterations in the
expression of a number of the genes observed in our microarray
results may be involved in the regulation of invasion/metastasis
and may contribute to the accelerated tumor growth noted in the
in vivo experiments. Investigation of these changes and of
strategies to inhibit them may have considerable clinical
applications in the future.
Acknowledgements
This study was supported by the National Key Basic
Research Project, (no. 2004CB518708A) China. Support for
third-party writing assistance for this manuscript was provided by
Shanghai Roche Pharmaceuticals Ltd. (Shanghai, China). All
decisions regarding the final content were conducted by the
authors. As such, the authors take full responsibility for the
content and expression of the submitted manuscript.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
2011.61(2): 69–90
|
|
2
|
Friedman SL: Molecular mechanisms of
hepatic fibrosis and principles of therapy. J Gastroenterol.
32:424–430. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elsharkawy AM and Mann DA: Nuclear
factor-kappa B and the hepatic inflammation-fibrosis-cancer axis.
Hepatology. 46:590–597. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong CM and Ng IO: Molecular pathogenesis
of hepatocellular carcinoma. Liver Int. 28:160–174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chau KY, Lily MA, Wu PC and Yau WL:
Myofibroblasts in hepatitis B related cirrhosis and hepatoellular
carcinoma. J Clin Pathol. 45:446–448. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bridle KR, Crawford DH, Powell LW and Ramm
GA: Role of myofibroblasts in tumor encapsulation of hepatocelluar
carcinoma in haemochromatosis. Liver. 21:96–104. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Terada T, Makimoto K, Terayama N, Suzuki Y
and Nakanuma Y: Alpha-smooth muscle actin-positive stroma cells in
cholangiocarcinomas, hepatocellular carcinomas and metastatic liver
careinomas. J Hepatol. 24:706–712. 1996. View Article : Google Scholar
|
|
8
|
Olaso E, Salado C, Egilegor E, et al:
Proangiogenic role of tumor-activated HSCs in experimental melanoma
metastasis. Hepatology. 37:674–685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao W, Zhang L, Yin Z, et al: Activated
HSCs promote hepatocellular carcinoma development in
immunocompetent mice. Int J Cancer. 129:2651–2661. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia Y, Chen R, Song Z, Ye S, Sun R, Xue Q
and Zhang Z: Gene expression profiles during activation of cultured
rat HSCs by tumoral hepatocytes and fetal bovine serum. J Cancer
Res Clin Oncol. 136:309–321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weiskirchen R and Gressner AM: Isolation
and culture of HSCs. Methods Mol Med. 117:99–113. 2005.PubMed/NCBI
|
|
12
|
Geerts A: On the origin of stellate cells:
mesodermal, endodermal or neuro-ectodermal? J Hepatol. 40:331–334.
2004. View Article : Google Scholar
|
|
13
|
Bachem MG, Meyer D, Melchior R, Sell KM
and Gressner AM: Activation of rat liver perisinusoidal lipocytes
by transforming growth factors derived from myofibroblastlike
cells. A potential mechanism of self perpetuation in liver
fibrogenesis. J Clin Invest. 89:19–27. 1992. View Article : Google Scholar
|
|
14
|
Rockey DC, Boyles JK, Gabbiani G and
Friedman SL: Rat hepatic lipocytes express smooth muscle actin upon
activation in vivo and in culture. J Submicrosc Cytol Pathol.
4:193–203. 1992.PubMed/NCBI
|
|
15
|
Amann T, Bataille F, Spruss T, et al:
Activated HSCs promote tumorigenicity of hepatocellular carcinoma.
Cancer Sci. 100:646–653. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Neaud V, Faouzi S, Guirouilh J, Le Bail B,
Balabaud C, Bioulac-Sage P and Rosenbaum J: Human hepatic
myofibroblasts increase invasiveness of hepatocellular carcinoma
cells: evidence for a role of hepatocyte growth factor. Hepatology.
26:1458–1466. 1997. View Article : Google Scholar
|
|
17
|
Meindl-Beinker NM, Katsuzaki K and Dooley
S: TGF-β signaling in the onset and progression of hepatocellular
cancer. Dig Dis. 30:514–523. 2012.
|
|
18
|
Ogunwobi OO and Liu C: Hepatocyte growth
factor upregulation promotes carcinogenesis and
epithelial-mesenchymal transition in hepatocellular carcinoma via
Akt and COX-2 pathways. Clin Exp Metast. 28:721–731. 2011.
View Article : Google Scholar
|
|
19
|
Santomato A, Franseva E, Dituri F, et al:
HSCs stimulate HCC cell migration via laminin-5 production. Clin
Sci (Lond). 121:159–168. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim SW, Kim JS, Papadopoulos J, et al:
Consistent interactions between tumor cell IL-6 and macrophage
TNF-alpha enhance the growth of human prostate cancer cells in the
bone of nude mouse. Int Immunopharmacol. 11:862–872. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kao JT, Lai HC, Tsai SM, et al: Rather
than interleukin-27, interleukin-6 expresses positive correlation
with liver severity in naïve hepatitis B infection patients. Liver
Int. 32:928–936. 2012.PubMed/NCBI
|
|
22
|
Okazaki I and Inagaki Y: Novel strategies
for hepatocellular carcinoma based on MMPs science. Anticancer
Agents Med Chem. 12:753–763. 2012. View Article : Google Scholar : PubMed/NCBI
|