Introduction
Resveratrol is a naturally occurring phytoalexin,
abundant in red grapes, which possesses antioxidant and
anti-inflammatory properties. Previous studies have demonstrated
that resveratrol in association with the consumption of red wine
exhibits cardioprotective effects (1). In addition, several studies have
demonstrated that resveratrol is important in the prevention of
skin cancer (2), human breast
cancer (3), oral squamous cell
carcinoma (4) and the inhibition
of angiogenesis (5), thereby
suggesting that resveratrol has anticancer properties. However, the
molecular mechanisms underlying the biological effects conferred by
resveratrol have not been fully defined.
The signal transducer and activator of transcription
(STAT) family is important in cells and is able to promote cell
proliferation and other biological processes, which can be
triggered by cytokines or growth factors (6,7).
STAT is activated by phosphorylation of a critical tyrosine
residue, which then forms dimers between two phosphorylated STAT
monomers. Following this, the dimers are translocated into the
nucleus where STAT regulates the expression of its target genes.
Within the STAT family, Stat3 is constitutively activated in
diverse types of human tumor. Constitutively active Stat3 is able
to induce oncogenic processes, growth, survival and angiogenesis
(8–10), while the suppression of
phosphorylated Stat3 (p-Stat3) induces the suppression of tumor
growth and apoptosis (11,12). Previous studies have demonstrated
that p-Stat3 is able to be downregulated by gene associated with
retinoid-IFN-induced mortality 19 (GRIM-19) (13,14).
Several studies have reported that the GRIM-19
protein can interact with the Stat3 signaling pathway. GRIM-19 has
become a novel anticancer target in cancer cells that have
constitutively active Stat3 (15,16).
Given that GRIM-19 and Stat3 are present and active in many types
of human tumor (17–19), there is considerable potential for
resveratrol to modulate signal transduction pathways involved in
tumor progression. The present study investigated t(he effects of
resveratrol on GRIM-19-Stat3 signaling in HeLa cells, which were
derived from a cervical tumor.
Materials and methods
Cell lines, reagents and treatment
conditions
The HeLa cell line was obtained from the American
Type Culture Collection (Manassas, VA, USA). Resveratrol and
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
were purchased from Sigma (St. Louis, MO, USA). Antibodies against
Stat3 and p-Stat3 (phosphorylated at tyrosine 705) were purchased
from Cell Signaling Technology (Beverly, MA, USA). Antibodies
against β-actin were purchased from Santa Cruz Biotechnology, Inc.
(Santa Cruz, CA, USA). Antibodies against GRIM-19 were purchased
from eBioscience (San Diego, CA, USA). Secondary antibodies were
purchased from Beijing Biosynthesis Biotechnology Co., Ltd.
(Beijing, China). Penicillin, streptomycin, Dulbecco’s modified
Eagle’s medium (DMEM) and fetal bovine serum (FBS) were obtained
from Gibco-BRL (Grand Island, NY, USA). HeLa cells were grown in
DMEM supplemented with 10% heat-inactivated FBS, 100 U/ml
penicillin and 100 mg/ml streptomycin for 48 h at 37°C/5%
CO2. The HeLa cells were divided into experimental
groups and each treatment condition was a single dose of
resveratrol at the indicated concentration. The vehicle control was
0.1% dimethyl sulfoxide (DMSO).
Western blot analysis
Western blotting was used to analyze the expression
levels of p-Stat3, Stat3, GRIM-19 and β-actin. The cells were lysed
with RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% sodium
deoxycholate, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride and 1 mM
EDTA) for 45 min at 4°C. Approximately 30 μg of total protein was
loaded into each lane of 10 and 15% polyacrylamide gels and
subjected to sodium dodecyl sulfate polyacrylamide gel
electrophoresis. The proteins were then transferred onto
nitrocellulose membranes (Novex; Invitrogen Life Technologies,
Carlsbad, CA, USA) and the blots were blocked in 5% (w/v) non-fat
milk for 1 h at room temperature. The blots were incubated with the
appropriate primary antibodies overnight at 4°C (GRIM-19 polyclonal
mouse, 1:1,000 dilution; p-Stat3 monoclonal rabbit, 1:2,000; Stat3
monoclonal rabbit, 1:2,000; and β-actin polyclonal mouse, 1:3,000).
The blots were washed three times for 5 min per wash and exposed to
horseradish peroxidase-conjugated secondary antibodies
(Biosynthesis Biotechnology Co., Ltd.) for 2 h. The blots were then
examined using enhanced chemiluminescence reagent (Thermo Fisher
Scientific, Waltham, MA, USA) and the band intensities were
measured and quantified using Quantity One software (Bio-Rad,
Hercules, CA, USA).
Reverse-transcription polymerase chain
reaction (RT-PCR) assays
Total RNA was isolated from HeLa cells using
TRIzol® reagent (Life Technologies, Rockville, MD, USA)
following 24 h treatment with resveratrol or 48 h after
transfection with GRIM-19 or a GRIM-19 short interfering RNA
(siRNA). cDNA was generated from 1 μg of total RNA using a cDNA
synthesis kit (Takara Biotechnology Co., Ltd., Dalian, Lianning,
China) according to the manufacturer’s instructions. Primer
sequences (GeneCore, Shanghai, China) specific for cyclin B1,
cyclin D1, B-cell lymphoma 2 (Bcl-2), vascular endothelial growth
factor (VEGF), Stat3 and GRIM-19 were used (Table I). β-actin was used for
normalization of the cDNA input levels. Following cDNA synthesis,
the PCR thermal cycling profile comprised 32 cycles of denaturation
(95°C for 30 sec), annealing (56°C for 30 sec) and extension (72°C
for 30 sec). The reaction was terminated with a final extension
step (72°C for 5 min) following 32 cycles. The amplicons were
separated by electrophoresis on 2% (w/v) agarose gel and visualized
by staining with ethidium bromide. Three biological replicates were
analyzed for each sample point and at least three reactions were
used to calculate the expression levels. Relative expression was
quantified using densitometry and the Gel Image Version 3.74 System
(Tianon, Shanghai, China).
 | Table IOligonucleotide primer sequences used
in the present study. |
Table I
Oligonucleotide primer sequences used
in the present study.
| Name | Sequence (5′→3′) | Amplicon (bp) |
|---|
| Cyclin B1 | F:
GCAGCACCTGGCTAAGAATGT
R: GCCTTGGCTAAATCTTGAACT | 147 |
| Cyclin D1 | F:
GCGAGGAACAGAAGTGCG
R: AGGCGGTAGTAGGACAGGAA | 484 |
| Bcl-2 | F:
AGGATTGTGGCCTTCTTTGA
R: CCTACCCAGCCTCCGTTAT | 155 |
| VEGF | F:
ACGGACAGACAGACAGACACC
R: CCCAGAAGTTGGACGAAAAGT | 176 |
| β-actin | F:
AGCCTCGCCTTTGCCGATCC
R: ACATGCCGGAGCCGTTGTCG | 100 |
| Stat3 | F:
AGTCAGTGACCAGGCAGAAGA
R: ATTTGTTGACGGGTCTGAAGT | 265 |
| GRIM-19 | F:
CGGGACCGGAAGTGTGGGATAC
R: GCAGAGCATTTATTCCGTCCCAG | 435 |
Transient RNA interference and
transfections
GRIM-19 was knocked down using small siRNAs, with a
non-targeting siRNA used in parallel as a negative control
(GenePharma Co., Shanghai, China). Primary cultures were
transfected with a GRIM-19 siRNA or an irrelevant siRNA (as a
control) using the X-tremeGENE HP DNA transfection reagent (Roche
Diagnostics GmbH, Mannheim, Germany). After 2 days, the protein
expression levels of GRIM-19, p-Stat3, Stat3 and β-actin in HeLa
cells were analyzed.
Plasmid construction and DNA
transfection
The human GRIM-19 sequence was amplified from HeLa
cells using RT-PCR and cloned between the NotI and
EcoRV sites of the Pflag-CMV™-4 mammalian expression vector.
The pFLAG tag was added to the N-terminus of the GRIM-19 sequences
in all the constructs. The transfection of plasmids into the cells
was conducted using X-tremeGENE HP DNA transfection reagent
according to the manufacturer’s instructions. Following 2 days, the
protein expression of GRIM-19, p-Stat3, Stat3 and β-actin in HeLa
cells was analyzed.
Cell viability assay
Cell viability was determined using MTT assays
according to the manufacturer’s instructions. HeLa cells were
seeded in 96-well culture plates at an optimal density of
1×104 cells/well. Briefly, phosphate-buffered saline
containing MTT at a final concentration of 0.5 mg/ml was added to
each well following treatment with resveratrol for 24 h, and then
incubated at 37°C for 4 h. The medium was gently aspirated and 150
μl DMSO was added to each well. The plates were agitated for 10 min
on a shaker to dissolve the formazan product. A well containing
DMSO without cells was used as a blank control and the optical
density at 490 nm in each well was determined using a
spectrophotometer (BioTek Instruments Inc., Winooski, VT, USA).
Statistical analysis
The data were expressed as the mean ± standard
deviation for three or more independent experiments. Statistical
significance was estimated using one-way analysis of variance
followed by the Student-Newman-Keuls test for comparison of several
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
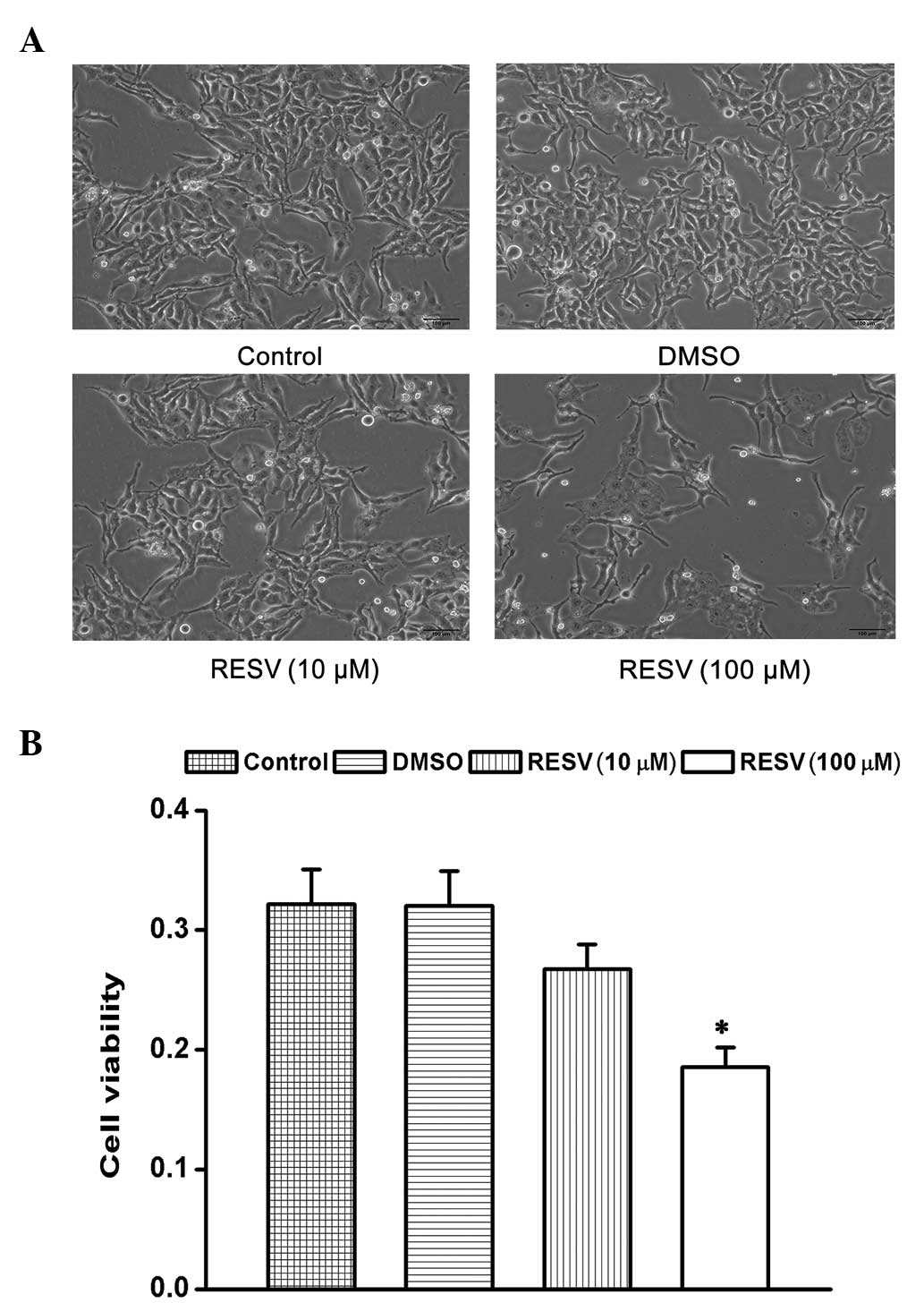
Effect of resveratrol on the
proliferation and viability of HeLa cells
The present study examined the effects of
resveratrol on the proliferation and cell viability of HeLa cells.
Analyses by MTT assays demonstrated that the treatment of HeLa
cells with resveratrol (10 and 100 μM) induced cell shrinkage
(Fig. 1A) and decreased cell
viability in a dose-dependent manner compared with the control and
DMSO groups (Fig. 1B).
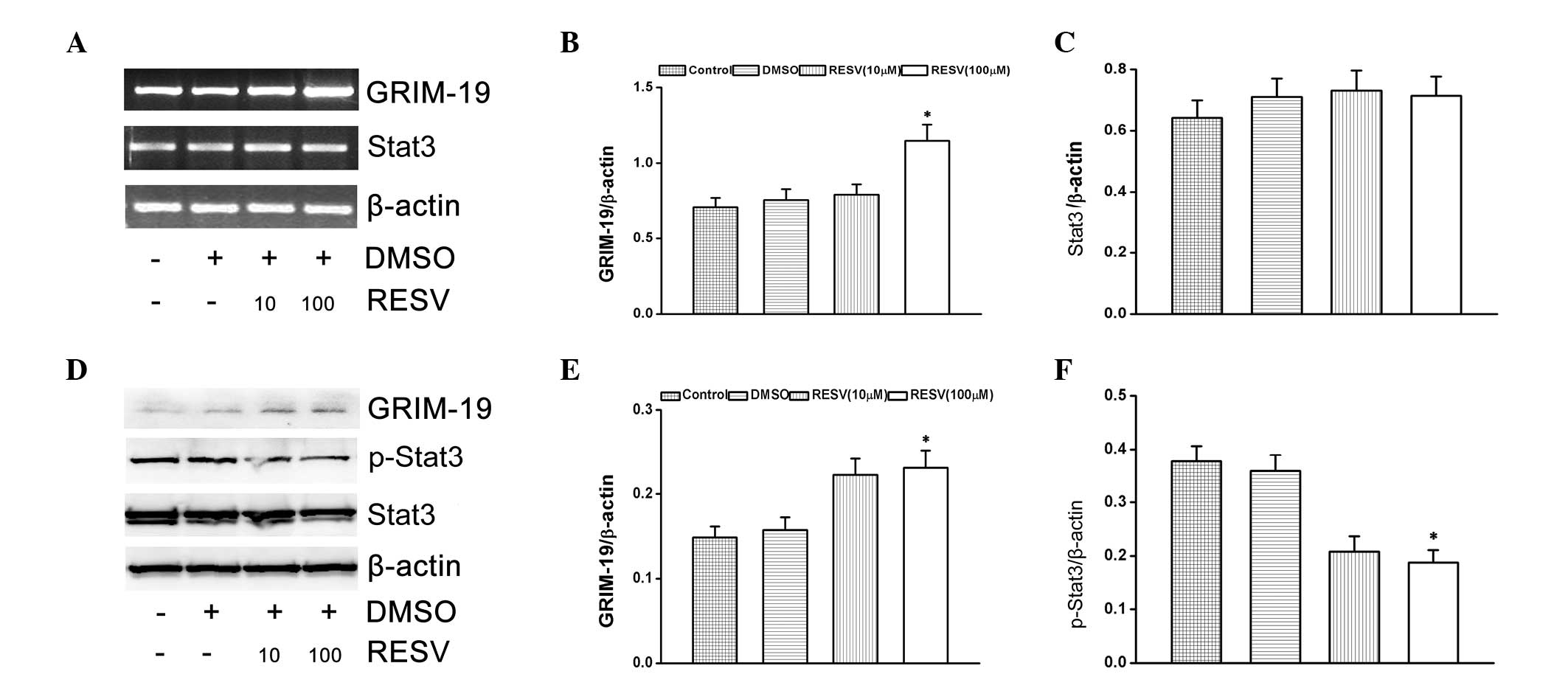
Effect of resveratrol on p-Stat3 and
GRIM-19
The present study investigated the effects of
resveratrol on Stat3 activation and GRIM-19 expression in HeLa
cells. The cells were treated with resveratrol (10 and 100 μM) for
24 h. HeLa cells treated with resveratrol significantly induced the
mRNA and protein expression of GRIM-19 (Fig. 2A–E). At the same time, p-Stat3
protein expression levels were downregulated (Fig. 2F), however, Stat3 mRNA expression
levels were unaltered (Fig.
2C).
Effect of GRIM-19 on p-Stat3 and
Stat3-associated genes
To understand the function of GRIM-19 on the Stat3
signaling pathway, GRIM-19 with a FLAG tag was overexpressed. The
levels of p-Stat3 were decreased by the overexpression of GRIM-19
(Fig. 3A). The relative expression
level of p-Stat3 is shown as ratio to β-actin (Fig. 3B). The expression of GRIM-19 is
shown in Fig. 3C. Stat3-associated
genes (cyclin B1, VEGF and Bcl-2) were downregulated by the
overexpression of GRIM-19. However, GRIM-19 had no effect on the
transcription levels of cyclin D1 (Fig. 3D).
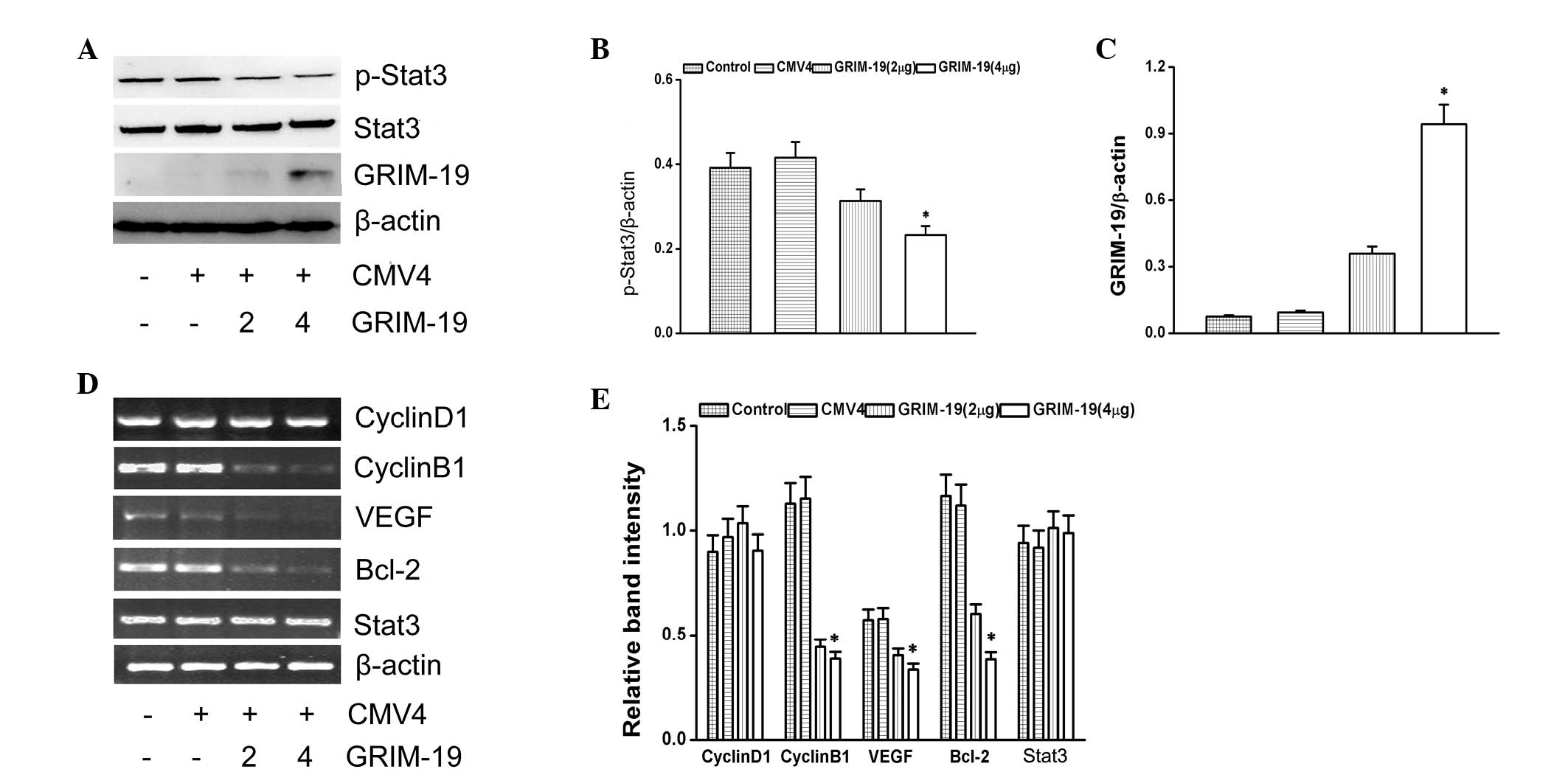 | Figure 3Effects of GRIM-19 on p-Stat3
expression. (A) Expression of p-Stat3 and GRIM-19 was detected by
western blotting. The abundance of (B) p-Stat3 and (C) GRIM-19
proteins as a ratio to β-actin. (D) Representative ethidium
bromide-stained gels showing Stat3, cyclin D1, cyclin B1, VEGF,
Bcl-2 and β-actin amplicons. (E) Abundance of cyclin D1, cyclin B1,
VEGF, Bcl-2 and Stat3 mRNAs is shown as a ratio to β-actin. Data
are expressed as the mean ± standard deviation from three
independent experiments (*P<0.05, compared with the
control group). GRIM-19, retinoid-IFN-induced mortality 19; Stat3,
signal transducer and activator of transcription 3; p-Stat3,
phosphorylated Stat3; VEGF, vascular endothelial growth factor;
Bcl-2, B-cell lymphoma 2; CMV4, cytomegalovirus resistance 4. |
GRIM-19 affects the function of
resveratrol on the Stat3 signaling pathway
The cells transfected with GRIM-19 siRNA clearly
downregulated the cell cytotoxicity induced by resveratrol
(Fig. 4F). Furthermore, the
expression of GRIM-19, p-Stat3 and Stat3 was detected (Fig. 4A–C). Resveratrol downregulated
p-Stat3 expression, while transfection with GRIM-19 siRNA resulted
in the suppression of p-Stat3 downregulation induced by resveratrol
(Fig. 4C and D). The results
confirmed that GRIM-19 expression, induced by resveratrol, is
involved in p-Stat3 suppression induced by resveratrol. GRIM-19 was
also involved in the proliferation and cytotoxicity induced by
resveratrol in HeLa cells.
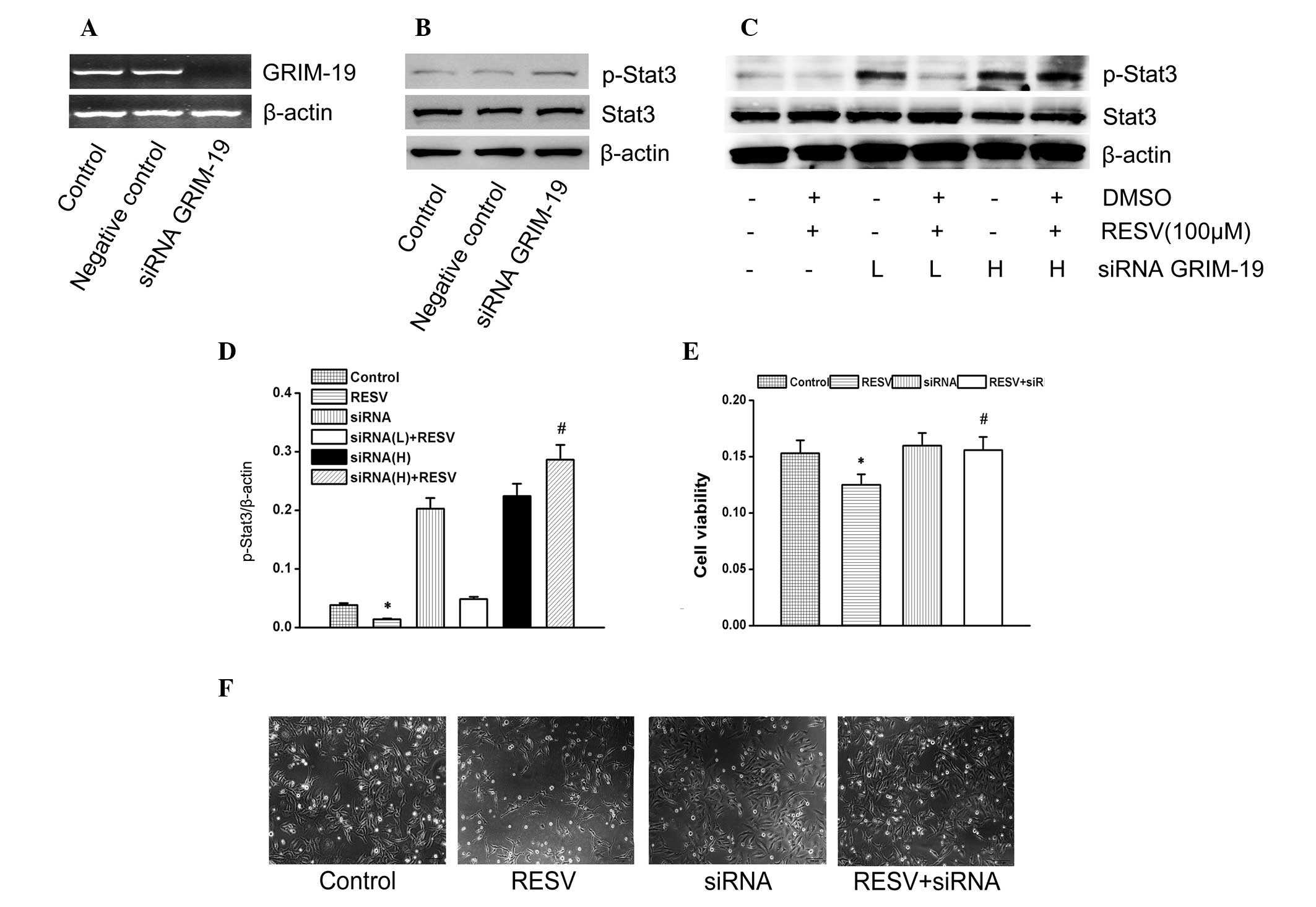 | Figure 4Effects of GRIM-19 siRNA on the
function of resveratrol. (A) Expression of GRIM-19 and Stat3 as
shown by reverse-transcription polymerase chain reaction methods
and the quantity of GRIM-19 siRNAs. (B) Upregulation of the
expression of the p-Stat3 protein is shown as a ratio to β-actin.
(C) Representative western blots showing p-Stat3, Stat3 and β-actin
proteins in the same samples. (D) Expression of the p-Stat3 protein
is shown as a ratio to β-actin. (E) HeLa cells transfected with
GRIM-19 siRNA were incubated with the indicated concentrations of
resveratrol for 24 h at 37°C. Morphological alterations in HeLa
cells following treatment with resveratrol were observed following
24 h. (F) Effects of the GRIM-19 siRNA on the function of
resveratrol in HeLa cells. Cell viability was determined by the MTT
bromide reduction assay. Data are presented as the mean ± standard
deviation of three independent experiments (*P<0.05,
compared with the control group; #P<0.05, compared
with the resveratrol groups). GRIM-19, retinoid-IFN-induced
mortality 19; Stat3, signal transducer and activator of
transcription 3; DMSO, dimethyl sulfoxide; RESV, resveratrol;
siRNA, short interfering RNA; L, low (20 μm); H, high (40 μm). |
Discussion
Findings of previous studies have provided new
insights into the biological mechanisms of resveratrol and its
associated stilbene compounds have also been investigated (20). To the best of our knowledge, the
present study has demonstrated for the first time that resveratrol
is able to induce the expression of GRIM-19. GRIM-19 is important
in the function of resveratrol on the Stat3 signaling pathway. As
previously indicated, resveratrol induced a downregulation in cell
viability and induced cell-cycle arrest (21,22).
Furthermore, resveratrol was able to induce aberrant downstream
Stat3 signaling (23,24). The results of the present study
have shown that resveratrol downregulated cell viability and
inhibited p-Stat3 in HeLa cells.
Stat3 is important in cancer development. STAT
family members are phosphorylated by receptor-associated kinases in
response to cytokines or growth factors. The phosphorylated STATs
then translocate to the cell nucleus where they act as
transcription activators and regulate the expression of target
genes. Previous studies have demonstrated that Stat3 has an
oncogenic function and that chronic Stat3 activation is important
in gastric cancer (25). Increased
Stat3 activity is able to upregulate the survival signal in cancer
cells (26) and specific
inhibition of Stat3 is a potentially useful therapy against various
types of cancer (27). The present
study found that resveratrol suppressed the expression of p-Stat3
and inhibited the proliferation of cancer cells. These results
suggest that resveratrol suppresses HeLa cell proliferation and
survival, and the anticancer function of resveratrol is partially
dependent on the inhibition of Stat3 activation.
The overexpression of GRIM-19 downregulates p-Stat3
levels. At the same time GRIM-19 suppresses the transcription
levels of cyclin B1, VEGF and Bcl-2. These are all downstream genes
associated with cell proliferation and survival (13,14).
Although the function of GRIM-19 in several types of cancer and the
Stat3 signaling pathway have been previously reported (15,19),
the effect of GRIM-19 on the function of resveratrol and its
association with the Stat3 signaling pathway remains to be
elucidated. The results from the present study clearly demonstrate
that resveratrol induced the expression of GRIM-19 and suppressed
the expression of p-Stat3. The GRIM-19 siRNA inhibited the
suppressive effects of resveratrol on the Stat3 signaling pathway,
while upregulating cell survival compared with the resveratrol
group. The association between resveratrol and the Stat3 signaling
pathway remains to be elucidated. However, there is a clear
correlation between the role of Stat3 in cancer development
(8,9) and the function of resveratrol
(28,29). To the best of our knowledge, the
present study has reported for the first time that resveratrol
induced the expression of GRIM-19, and that GRIM-19 is important in
the effects of resveratrol on HeLa cells via the abrogation of
Stat3 signaling and further investigation is required in other
types of cancer. In the present study, resveratrol at a high
concentration (100 μM) suppressed the proliferation of HeLa cells
and the Stat3 signaling pathway. Previous studies have also
demonstrated that resveratrol has an effect on the src-Stat3
signaling pathway (22) and on
tumor development (4). These
results, in association with the results from the present study,
elucidate the importance of resveratrol and its antitumor cell
activities, which are partially dependent on the concentration of
resveratrol (30–33).
In conclusion, the present study has reported that
GRIM-19 expression, induced by resveratrol, affects the Stat3
signaling pathway. Modulation of this signal transduction pathway
contributes to the resveratrol-induced biological effects on
various types of cancer (34). The
present study highlights a new mechanism through which resveratrol
inhibits the Stat3 signaling pathway. However, further
investigation is required in order to fully elucidate the
anti-tumorigenic effects of resveratrol.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (nos. 81070110 to M. Wei and
81100099 to P. Xin) and the Shanghai Science and Technology
Innovation Research Program (no. 11410701900 to M. Wei).
Abbreviations:
|
DMSO
|
dimethyl sulfoxide
|
|
DMEM
|
Dulbecco’s modified Eagle’s medium
|
|
GRIM-19
|
gene associated with retinoid-
IFN-induced mortality 19
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-
2,5-diphenyltetrazolium bromide
|
|
Stat3
|
signal transducer and activator of
transcription 3
|
|
FBS
|
fetal bovine serum
|
References
|
1
|
Kopp P: Resveratrol, a phytoestrogen found
in red wine. A possible explanation for the conundrum of the
‘French paradox’? Eur J Endocrinol. 138:619–620. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jang M, Cai L, Udeani GO, et al: Cancer
chemopreventive activity of resveratrol, a natural product derived
from grapes. Science. 275:218–220. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mgbonyebi OP, Russo J, Russo IH, et al:
Antiproliferative effect of synthetic resveratrol on human breast
epithelial cells. Int J Oncol. 12:865–869. 1998.PubMed/NCBI
|
|
4
|
Elattar TM and Virji AS: The effect of red
wine and its components on growth and proliferation of human oral
squamous carcinoma cells. Anticancer Res. 19:5407–5414. 1999.
|
|
5
|
Srivastava RK, Unterman TG and Shankar S:
FOXO transcription factors and VEGF neutralizing antibody enhance
antiangiogenic effects of resveratrol. Mol Cell Biochem.
337:201–212. 2010. View Article : Google Scholar
|
|
6
|
Bromberg J and Darnell JE Jr: The role of
STATs in transcriptional control and their impact on cellular
function. Oncogene. 19:2468–2473. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu H and Jove R: The STATs of cancer - new
molecular targets come of age. Nat Rev Cancer. 4:97–105. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Macias E, Rao D and Digiovanni J: Role of
stat3 in skin carcinogenesis: insights gained from relevant mouse
models. J Skin Cancer. 2013:6840502013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cho KH, Jeong KJ, Shin SC, et al: STAT3
mediates TGF-beta1-induced TWIST1 expression and prostate cancer
invasion. Cancer Lett. 336:167–173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
You W, Tang Q, Zhang C, et al: IL-26
promotes the proliferation and survival of human gastric cancer
cells by regulating the balance of STAT1 and STAT3 activation. PLoS
One. 8:e635882013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han Z, Feng J, Hong Z, et al: Silencing of
the STAT3 signaling pathway reverses the inherent and induced
chemoresistance of human ovarian cancer cells. Biochem Biophys Res
Commun. 435:188–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu W, Xiao H, Lin J and Li C: Discovery of
novel STAT3 small molecule inhibitors via in silico site-directed
fragment-based drug design. J Med Chem. 56:4402–4412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okamoto T, Inozume T, Mitsui H, et al:
Overexpression of GRIM-19 in cancer cells suppresses STAT3-mediated
signal transduction and cancer growth. Mol Cancer Ther.
9:2333–2343. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nallar SC, Kalakonda S, Lindner DJ, et al:
Tumor-derived mutations in the gene associated with retinoid
interferon-induced mortality (GRIM-19) disrupt its anti-signal
transducer and activator of transcription 3 (STAT3) activity and
promote oncogenesis. J Biol Chem. 288:7930–7941. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bu X, Zhao C, Wang W and Zhang N: GRIM-19
inhibits the STAT3 signaling pathway and sensitizes gastric cancer
cells to radiation. Gene. 512:198–205. 2013. View Article : Google Scholar
|
|
16
|
Lufei C, Ma J, Huang G, et al: GRIM-19, a
death-regulatory gene product, suppresses Stat3 activity via
functional interaction. Embo J. 22:1325–1335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou T, Chao L, Rong G, et al:
Down-regulation of GRIM-19 is associated with STAT3 overexpression
in breast carcinomas. Hum Pathol. 44:1773–1779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nallar SC, Kalakonda S, Sun P, et al:
Identification of a structural motif in the tumor-suppressive
protein GRIM-19 required for its antitumor activity. Am J Pathol.
177:896–907. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Hao H, Zhao S, et al:
Downregulation of GRIM-19 promotes growth and migration of human
glioma cells. Cancer Sci. 102:1991–1999. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robertson CN, Roberson KM, Padilla GM, et
al: Induction of apoptosis by diethylstilbestrol in
hormone-insensitive prostate cancer cells. J Natl Cancer Inst.
88:908–917. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wen S, Li H, Wu ML, et al: Inhibition of
NF-kappaB signaling commits resveratrol-treated medulloblastoma
cells to apoptosis without neuronal differentiation. J Neurooncol.
104:169–177. 2011. View Article : Google Scholar
|
|
22
|
Kotha A, Sekharam M, Cilenti L, et al:
Resveratrol inhibits Src and Stat3 signaling and induces the
apoptosis of malignant cells containing activated Stat3 protein.
Mol Cancer Ther. 5:621–629. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Capiralla H, Vingtdeux V, Zhao H, et al:
Resveratrol mitigates lipopolysaccharide- and Abeta-mediated
microglial inflammation by inhibiting the TLR4/NF-kappaB/STAT
signaling cascade. J Neurochem. 120:461–472. 2012. View Article : Google Scholar :
|
|
24
|
Scuto A, Kirschbaum M, Buettner R, et al:
SIRT1 activation enhances HDAC inhibition-mediated upregulation of
GADD45G by repressing the binding of NF-kappaB/STAT3 complex to its
promoter in malignant lymphoid cells. Cell Death Dis. 4:e6352013.
View Article : Google Scholar
|
|
25
|
Giraud AS, Menheniott TR and Judd LM:
Targeting STAT3 in gastric cancer. Expert Opin Ther Targets.
16:889–901. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deng J, Liu Y, Lee H, et al: S1PR1-STAT3
signaling is crucial for myeloid cell colonization at future
metastatic sites. Cancer Cell. 21:642–654. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tkach M, Coria L, Rosemblit C, et al:
Targeting Stat3 induces senescence in tumor cells and elicits
prophylactic and therapeutic immune responses against breast cancer
growth mediated by NK cells and CD4+ T cells. J Immunol.
189:1162–1172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sheth S, Jajoo S, Kaur T, et al:
Resveratrol reduces prostate cancer growth and metastasis by
inhibiting the Akt/MicroRNA-21 pathway. PLoS One. 7:e516552012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang H, Zhang H, Tang L, et al:
Resveratrol inhibits TGF-beta1-induced epithelial-to-mesenchymal
transition and suppresses lung cancer invasion and metastasis.
Toxicology. 303:139–146. 2013. View Article : Google Scholar
|
|
30
|
Aziz MH, Kumar R and Ahmad N: Cancer
chemoprevention by resveratrol: In vitro and in vivo studies and
the underlying mechanisms (Review). Int J Oncol. 23:17–28.
2003.PubMed/NCBI
|
|
31
|
Jeong WS, Kim IW, Hu R and Kong AN:
Modulation of AP-1 by natural chemopreventive compounds in human
colon HT-29 cancer cell line. Pharm Res. 21:649–660. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu Y and Liu F: Targeting mTOR: Evaluating
the therapeutic potential of resveratrol for cancer treatment.
Anticancer Agents Med Chem. 13:1032–1038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chung MY, Lim TG and Lee KW: Molecular
mechanisms of chemopreventive phytochemicals against
gastroenterological cancer development. World J Gastroenterol.
19:984–993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kong AN, Yu R, Hebbar V, et al: Signal
transduction events elicited by cancer prevention compounds. Mutat
Res. 480–481:231–241. 2001. View Article : Google Scholar
|