Introduction
Renal cell carcinoma (RCC) is a highly vascularized
tumor which accounts for ~3% of all malignancies in adults, and has
the highest mortality rate of all urological malignancies (1,2).
Recently estimated cancer statistics showed that both the
incidence, and the mortality rate of RCC were among the 10 leading
cancers in the USA, with >65150 new cases and >8,780
mortalities recorded in 2013 (3).
RCC is a relatively asymptomatic disease and~30% of all patients
exhibit metastatic disease at the time of initial presentation
(4). When left untreated, the
five-year survival rate for metastatic RCC (mRCC) is ~2%, and the
median survival time is ~8 months (5). It is therefore crucial to identify
novel molecular biomarkers which may be used for early detection
and targeted therapy, based on the developing knowledge of the
tumorigenesis mechanisms of RCC.
MicroRNAs (miRNAs) are a class of non-coding RNAs,
~22 nucleotides (nt) in length, which are involved in the control
of gene expression by targeting mRNAs for either translational
repression or degradation (6,7). A
previous study has shown that miRNAs have important roles in
various cellular processes, including proliferation,
differentiation, metabolism and survival (8). Deregulation of miRNAs can alter
normal cell growth and development, leading to numerous disorders,
including cancers (8). To date,
aberrant expression or mutation of miRNAs has been observed in
numerous types of human cancer. It is estimated that >60% of
human protein-coding genes are under selective pressure to maintain
pairing to miRNAs (9), confirming
the significance of miRNAs in the gene regulatory network.
Recently, miR-125a-5p has been reported to function
as a tumor suppressor in malignancies of the breast (10–12),
lung (12), liver (13) and the central nervous system
(14). However, the function and
clinical significance of miR-125a-5p in RCC has yet to be explored
(15). To determine whether
miR-125a-5p is important for the tumorigenesis of RCC, human renal
cancer and paired normal tissues were collected and compared for
miR-125a-5p expression levels, using quantitative polymerase chain
reaction (qPCR). Furthermore, the impact of miR-125a-5p on cell
proliferation, migration and apoptosis was analyzed by transfection
of synthesized miR-125a-5p mimics into RCC cell lines.
Materials and methods
Sample collection and RNA isolation
The RCC tissues and paired adjacent normal tissues
used throughout the present study were collected from hospitals of
the Guangdong, Hunan and Anhui provinces. Written informed consent
was obtained from all of the patients. Collection and use of the
samples was reviewed and approved by the Ethics Committees of the
local hospital and Peking University Shenzhen Hospital. Fresh RCC
and adjacent normal tissues, located 2.0 cm outside of visible RCC
lesions, were immersed in RNAlater® (Qiagen, Hilden,
Germany) for 30 min following dissection, and then stored at −80°C
until further use. All tissue samples were reviewed and classified
by hematoxylin & eosin (H&E) staining and the 2009 American
Joint Committee on Cancer (AJCC) staging system. Total RNA of each
sample was isolated using TRIzol® reagent (Invitrogen
Life Technologies, Carlsbad, CA, USA), and purified using an
RNeasy® Maxi kit (Qiagen), to the manufacturer’s
instructions. The clinical and pathological characteristics of the
48 RCC patients are shown in Table
1. The ages of the patients ranged from 29–76 years old, with a
median age of 52.
 | Table IClinical and pathological features of
patients with renal cell carcinoma. |
Table I
Clinical and pathological features of
patients with renal cell carcinoma.
| Variables | No. of cases |
|---|
| Total | 48 |
| Age (years) |
| >52 | 29 |
| <52 | 19 |
| Gender |
| Male | 30 |
| Female | 18 |
| Histological
type |
| Clear cell | 39 |
| Papillary | 9 |
| pT-stage |
| T1 | 27 |
| T2 | 19 |
| T3 and T4 | 2 |
| Fuhrman grade |
| I | 15 |
| II | 22 |
| III | 8 |
| IV | 3 |
| AJCC clinical
stages |
| I | 27 |
| II | 18 |
| III+IV | 3 |
Analysis of miR-125a-5p expression in RCC
tissues and paired normal tissues by qPCR
qPCR was used to detect the expression levels of
miR-125a-5p in the 48 RCC tissues and paired adjacent normal
tissues. A total RNA volume of 1 μg from each sample was isolated
for reverse transcription using an miScript Reverse Transcription
kit (Qiagen), according to the manufacturer’s instructions, in
order to obtain the cDNA template. The qPCR was performed using a
Light Cycle 480 Fluorescent Quantitative PCR System (Takara Bio,
Inc., Otsu, Japan) with the miScript SYBR Green PCR kit (Qiagen),
U6 was used as an internal control. The reaction mixture contained:
2 μl cDNA template, 1 μl specific microRNA primer, 2 μl 10×
miScript Universal Primer, 10 μl 2× QuantiTect SYBR Green PCR
Master Mix, and RNase-free water (Takara Bio, Inc.). The following
primers were used in the qPCR reaction: miR-125a-5p, forward
5′-TCCCTGAGACCCTTTAAACTGTGA-3′, and the reverse primer used was
provided by the miScript SYBR Green PCR kit; U6, forward
5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-ACGCTTCACGAATTTGCGT-3′. The
amplification conditions were: 95°C 2 min, followed by 95°C 15 sec,
58°C 30 sec and 72°C 30 sec, for 40 cycles.
Cell culture and transfection
ACHN and 786-O renal carcinoma cell lines obtained
from the laboratory of the Institute of Urology of Shenzhen
PKU-HKUST Medical Center (Shenzhen, China) were cultured in
Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific,
Waltham, MA, USA), supplemented with 10% fetal bovine serum
(Invitrogen Life Technologies), in a humidified incubator
containing 5% CO2. For restoration of miR-125a-5p in RCC
tissues with endogenously downregulated miR-125a-5p, synthesized
miR-125a-5p mimics (GenePharma, Shanghai, China) were transfected
into the cells using Lipofectamine® 2000 (Invitrogen
Life Technologies), according to the manufacturer’s instructions.
The cells were harvested and total RNA was isolated for qPCR
analysis 24 h after transfection.
Assessment of cellular proliferation by
MTT assay
Cellular proliferation was measured using an MTT
assay kit (Sigma, St Louis, MO, USA). The renal cancer cells
(~5×103 cells) were seeded into 96-well culture plates
and transfected with either miR-125a-5p mimics or a negative
control. The negative control was set up with medium only. At 0,
24, 48 or 72 h after transfection, the cells were incubated with 20
μl MTT reagent (5 mg/ml) for 4 h at 37°C. Following the removal of
MTT, the cells were incubated in dimethyl sulfoxide (150 μl; Sigma)
with shaking for 10 min at room temperature. The optical density
(OD) was measured using a microplate reader (Model 680; Bio-Rad,
Hercules, CA, USA) at a dual wavelength of 490/630 nm.
Wound scratch assay
The migratory ability of renal cancer cells was
assessed using a wound scratch assay. In this assay ~250,000 cells
were seeded in a 12-well dish followed by transfection after 24 h
with miR-125a-5p mimics or the negative control (both 80 pmol)
using Lipofectamine 2000. After 5 h of transfection, a vertical
wound was made using a sterile 10 μl pipette tip and markers were
assigned to allow for observation of the cells in the correct
location. Subsequently, the cells were rinsed three times with
phosphate-buffered saline and incubated at 37°C. Images of the
wound scratches were taken immediately as well as following a 24 h
incubation. The migration distance (μm) was assessed using a
standard caliper. The experiments were performed in triplicate, and
analyzed double-blind by at least two observers.
Flow cytometry
For the apoptosis assay, ~300,000 renal cancer cells
were cultured in six-well plates at 37°C and transfected with
either miR-125a-5p mimics or the negative control within 24 h.
After 48 h transfection, the cells, including floating cells, were
harvested, washed twice with cold PBS and resuspended in 100 μl 1x
binding buffer (Invitrogen Life Technologies). Subsequently, 3 μl
of propidium iodide (PI) and 5 μl of Annexin V-fluorescein
isothiocyanate (Invitrogen Life Technologies) were added to each
cell suspension. The fluorescence of the stained cells was analyzed
by flow cytometry (Beckman Coulter, Miami, FL, USA) within 30 min
following staining, according to the manufacturer’s
instructions.
Statistical analyses
SPSS version 17.0 statistical software (SPSS, Inc.,
Chicago, IL, USA) was used for statistical analyses. Statistical
significance was determined with the Student’s t-test and the
paired t-test. A P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-125a-5p expression is downregulated
in 48 paired RCC tissues, as compared with adjacent tissues
A previous study showed miR-125a-5p to be
downregulated in RCC tissues, as determined by miRNA expression
profiling (16). In order to
confirm the results of the former study, the expression levels of
miR-125a-5p, in 48 RCC tissues and paired normal tissues, were
analyzed using qPCR. The relative expression of miR-125a-5p [Log2
Ratio] is shown in Fig. 1A. The
present results confirmed that miR-125a-5p expression was
significantly downregulated in renal cancer tissues, as compared
with the expression levels in the paired adjacent normal tissues
(P<0.01, Fig. 1B).
Transfection efficiency
To analyze the function of miR-125a-5p in RCC,
miR-125a-5p mimics and a negative control were transfected into
ACHN and 786-O renal cancer cell lines. The relative expression of
miR-25a-5p, following transfection, was determined by qPCR. The
expression levels in the ACHN and 786-O cells transfected with the
mimics were 163.2 and 134.6, respectively (Fig. 1B).
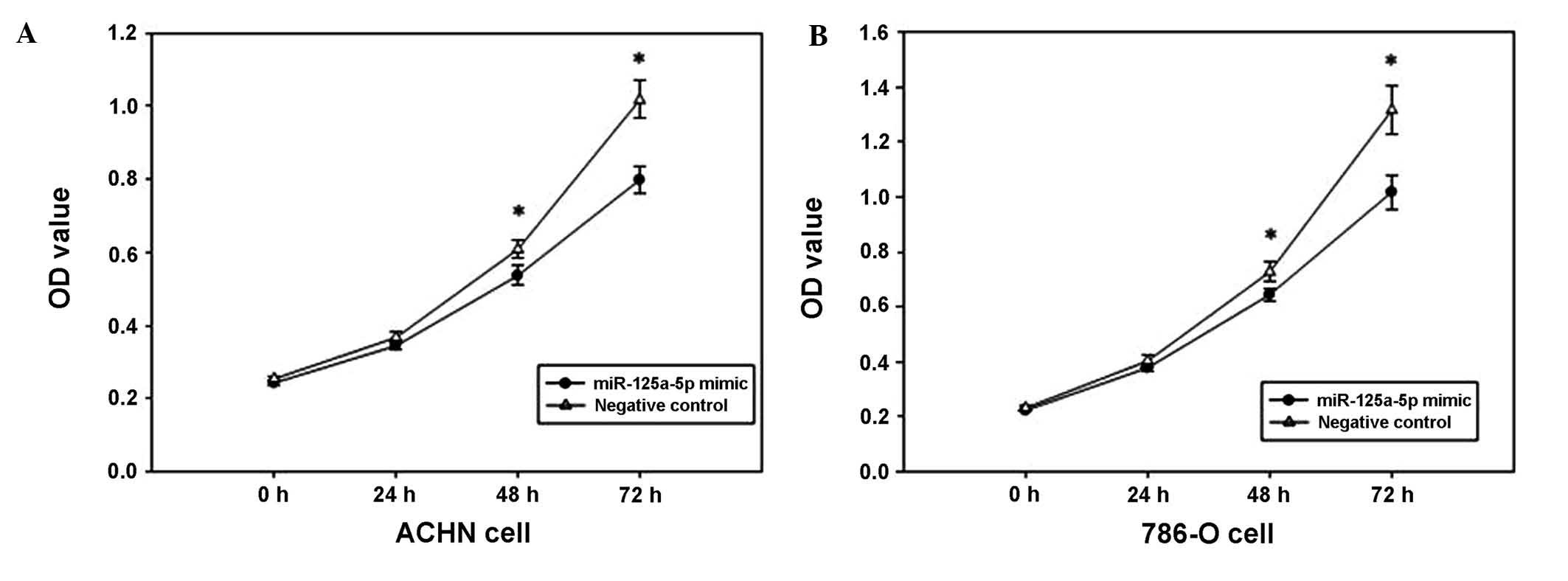
Overexpression of miR-125a-5p suppresses
cell proliferation
The MTT assay was used to determine the effect of
miR-125a-5p mimics on the proliferation of renal cancer cells. The
OD values of the miR-125a-5p mimics and the negative control
groups, were measured at 0, 24, 48 and 72 h after transfection. The
outcomes revealed that ACHN cell proliferation was decreased by
6.075, 13.907 and 22.461% (all P<0.05), and the proliferation of
786-O cells was decreased by 6.168, 11.438, and 20.642% (all
P<0.05) at 24, 48 and 72 h after transfection, respectively, as
compared with 0 h. The results indicated that the upregulation of
miR-125a-5p expression may suppress the proliferation of renal
cancer cells (Fig. 2).
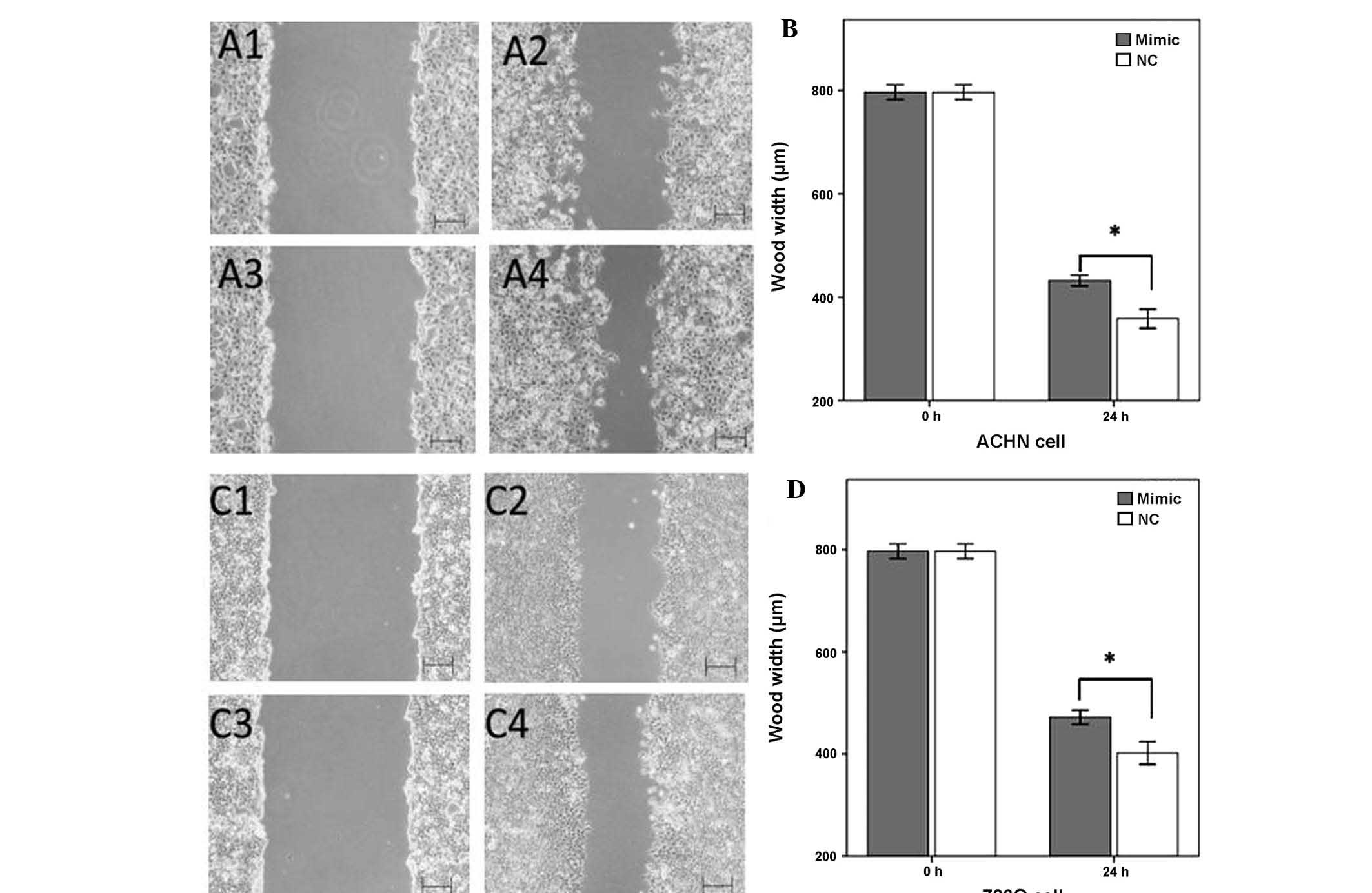
MiR-125a-5p mimics inhibit cell
migration
The effects of upregulation of miR-125a-5p
expression on cellular migration in renal cancer cells was observed
by a wound scratch assay. As shown in Fig. 3, the wound widths of the cells
transfected with a negative control were significantly narrower, as
compared with the wound widths in the miR-125a-5p mimics group
(P<0.05). These results suggested that the upregulation of
miR-125a-5p inhibited the migratory ability of the renal cancer
cells (Fig. 3).
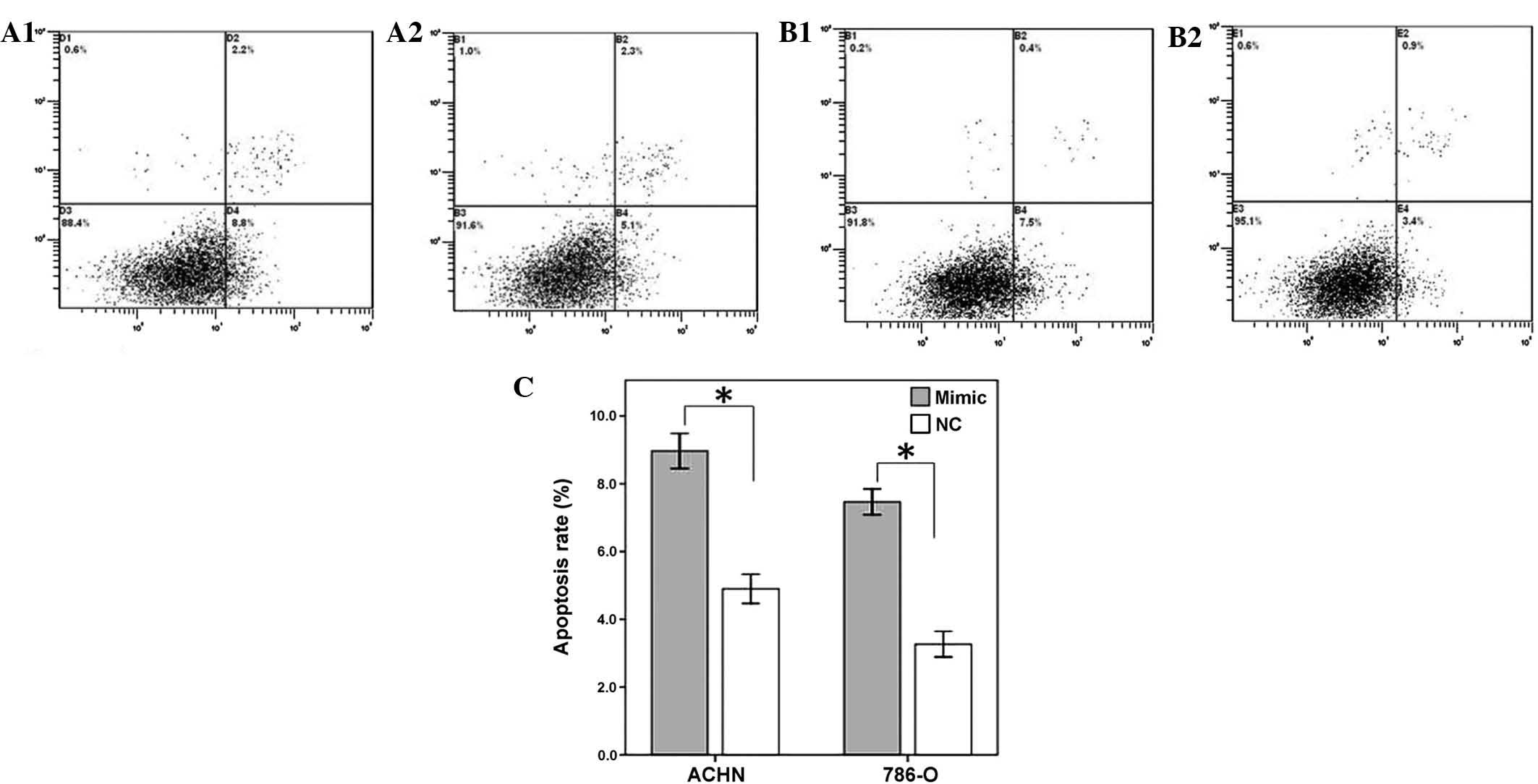
Restoration of miR-125a-5p induces
cellular apoptosis
To determine the impact of miR-125a-5p on renal
cancer cell apoptosis, a flow cytometry assay was performed to
detect the rate of apoptosis in the renal cancer cells 48 h after
transfection with miR-125-5p mimics or a negative control. The
results revealed that the apoptotic rates of ACHN transfected with
miR-125a-5p mimics and negative control group were 8.8 and 5.1%
respectively, whereas the apoptotic rates of 786-O cells were 7.5
and 3.4% (P<0.01), therefore suggesting that the restoration of
miR-125a-5p expression may accelerate the rate of apoptosis of
renal cancer cells (Fig. 4).
Discussion
miRNAs are short, single-stranded RNAs which are
associated with gene regulation, at both the transcriptional and
translational level, by base-pairing to complementary mRNA
sequences in their target genes (17). MiRNAs have diverse functions and
are involved in a variety of biochemical processes, including
cellular differentiation, proliferation, apoptosis, metabolism and
disease, including cancer (18).
Deregulation of miRNAs is emerging as a major aspect of cancer
etiology, and has previously been confirmed to be capable of
promoting cancer initiation and progression in vivo
(19), including in RCC. Numerous
miRNAs have been confirmed to be upregulated in RCC, including
miR-21, miR-210 and miR-155 (18,19),
while miR-127-3p, miR-145 and miR-126 have been shown to be
significantly downregulated and correlated with relapse-free
survival in patients with nonmetastatic RCC (20). Furthermore, serum miRNA levels have
been associated with RCC and may be used as potential biomarkers;
including miR-210 for early detection (19), miR-378 and miR-451 for diagnosis
(21), and miR-122, miR-514,
miR-192 and miR-215 for prognosis prediction and as therapeutic
targets (18,22).
MiR-125a-5p has been described as a tumor suppressor
in numerous human cancers (10–13).
Previous research has demonstrated that miR-125a expression is
downregulated in certain cancers and it may reduce the expression
levels of several oncogenes, for example, human epidermal growth
factor receptor-2 (HER-2), podoplanin membrane sialo-glycoprotein,
matrix metalloproteinase 11 and vascular endothelial growth factor
A (13). However, the role of
miR-125a in RCC remains unclear.
To determine whether miR-125a-5p is involved in RCC
tumorigenesis, qPCR was used to quantify the miR-125a-5p expression
levels in 48 RCC tissues and paired normal tissues. The effects of
miR-125a-5p on cell migration, proliferation and apoptosis were
also analyzed by transfection of RCC cell lines with synthetic
miR-125a-5p mimics.
The present results showed that miR-125a-5p
expression was significantly downregulated in RCC tissues, as
compared with the expression levels in paired normal renal tissues.
Transfection of miR-125a-5p mimics into 786-O and ACHN renal cancer
cell lines inhibited cellular proliferation, migration and induced
apoptosis, as compared with the negative control group, indicating
that miR-125a-5p may be characterized as a tumor suppressor in RCC
by inhibiting cellular proliferation and migration, and promoting
cellular apoptosis. The further study of miR-125a-5p target genes
may provide a deeper understanding of the molecular mechanisms of
tumorigenesis in RCC.
Previous studies have demonstrated that miRNAs can
be detected in the circulation and may serve as potential
biomarkers for numerous diseases, including cancer. It was
previously reported that miR-125a-5p could serve as a biomarker for
rheumatoid arthritis (23), and it
has been associated with a worsened disease progression of
Hepatitis B virus chronic infection (24). Alongside miR-125a-3p, miR-125a-5p
has shown to be a useful prognostic marker in gastric cancer
(25,26). Whether miR-125a-5p can be used as a
potential biomarker for the early detection and prognosis
prediction of RCC requires further investigation.
Due to the involvement of miRNAs in cancer
development, the manipulation of cellular miRNA levels has emerged
as a potential strategy for therapeutic intervention (10). Overexpressed HER-2 is observed in
breast and gastric cancers (27).
Previous studies have demonstrated the improvement of survival
rates in patients treated with HER-2-targeted therapy (27,28)
and the anti-HER-2 therapy trastuzumab has been used as a first
line treatment for HER-2-positive cancers in the clinical practice
(29). Previous research has
revealed HER-2 to be a direct target of miR-125a-5p. Nishida et
al (25) revealed that
transfection with miR-125a mimics suppressed the proliferation of
gastric cancer cells. The effects of the mimics were enhanced when
used in combination with trastuzumab. These results indicate that
miR-125a-5p mimics alone, or in combination with trastuzumab, may
be a novel therapeutic approach against gastric cancer. Further
research on the role of miR-125a-5p in the tumorigenesis of RCC may
explore its potential for targeted therapy.
In conclusion, the present study was, to the best of
our knowledge, the first to reveal the downregulation of
miR-125a-5p expression in RCC and to demonstrate its significant
role in affecting cellular proliferation, migration and apoptosis.
Further research is needed to define the target genes of
miR-125a-5p in RCC and to explore the potential use of miR-125a-5p
in early detection, prognosis prediction and as a targeted therapy
for RCC.
Acknowledgements
This work was supported by the National Natural
Science Foundation of China (no.81101922), Medical Scientific
Research Foundation of Guangdong Province of China (no. A2012584
and A2013606), Science and Technology Development Fund Project of
Shenzhen (no. JCYJ20130402114702124) and Shenzhen Science and
Technology Project (no. 201302050).
References
|
1
|
Cairns P: Renal cell carcinoma. Cancer
Biomark. 9:461–473. 2010.
|
|
2
|
McLaughlin JK, Lipworth L and Tarone RE:
Epidemiologic aspects of renal cell carcinoma. Semin Oncol.
33:527–533. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chow WH and Devesa SS: Contemporary
epidemiology of renal cell cancer. Cancer J. 14:288–301. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rasmussen F: Metastatic renal cell cancer.
Cancer Imaging. 13:374–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu L and Belasco JG: Micro-RNA regulation
of the mammalian lin-28 gene during neuronal differentiation of
embryonal carcinoma cells. Mol Cell Biol. 25:9198–9208. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vaz C, Ahmad HM, Sharma P, et al: Analysis
of microRNA transcriptome by deep sequencing of small RNA libraries
of peripheral blood. BMC Genomics. 11:2882010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin Y, Zeng Y, Zhang F, et al:
Characterization of microRNA expression profiles and the discovery
of novel microRNAs involved in cancer during human embryonic
development. PLoS One. 8:e692302013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
10
|
Wang S, Huang J, Lyu H, et al: Functional
cooperation of miR-125a, miR-125b, and miR-205 in
entinostat-induced dowregulation of erbB2/erbB3 and apoptosis in
breast cancer cells. Cell Death Dis. 4:e5562013. View Article : Google Scholar
|
|
11
|
Li W, Duan R, Kooy F, et al: Germline
mutation of microRNA-125a is associated with breast cancer. J Med
Genet. 46:358–360. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee CH, Kuo WH, Lin CC, et al:
MicroRNA-regulated protein-protein interaction networks and their
functions in breast cancer. Int J Mol Sci. 14:11560–11606. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bi Q, Tang S, Xia L, et al: Ectopic
expression of MiR-125a inhibits the proliferation and metastasis of
hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One.
7:e401692012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Karsy M, Arslan E and Moy F: Current
progress on understanding microRNAs in glioblastoma multiforme.
Genes Cancer. 3:3–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Osanto S, Qin Y, Buermans HP, et al:
Genome-wide microRNA expression analysis of clear cell renal cell
carcinoma by next generation deep sequencing. PLoS One.
7:e382982012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jing Q, Huang S, Guth S, et al:
Involvement of microRNA in AU-rich element-mediated mRNA
instability. Cell. 120:623–634. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo S, Bai H, Megyola CM, et al: Complex
oncogene dependence in microRNA-125a-induced myeloproliferative
neoplasms. Proc Natl Acad Sci USA. 109:16636–16641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zaman MS, Shahryari V, Deng G, et al:
Up-regulation of microRNA-21 correlates with lower kidney cancer
survival. PLoS One. 7:e310602012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao A, Li G, Péoc’h M, Genin C and
Gigante M: Serum miR-210 as a novel biomarker for molecular
diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol.
94:115–120. 2013. View Article : Google Scholar
|
|
20
|
Slaby O, Redova M, Poprach A, et al:
Identification of MicroRNAs associated with early relapse after
nephrectomy in renal cell carcinoma patients. Genes Chromosomes
Cancer. 51:707–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Redova M, Poprach A, Nekvindova J, et al:
Circulating miR-378 and miR-451 in serum are potential biomarkers
for renal cell carcinoma. J Transl Med. 10:552012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khella HW, Bakhet M, Allo G, et al:
miR-192, miR-194 and miR-215: a convergent microRNA network
suppressing tumor progression in renal cell carcinoma.
Carcinogenesis. 34:2231–2239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murata K, Furu M, Yoshitomi H, et al:
Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p
as plasma biomarkers for rheumatoid arthritis. PLoS One.
8:e691182013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Coppola N, Potenza N, Pisaturo M, et al:
Liver microRNA hsa-miR-125a-5p in HBV chronic infection:
correlation with HBV replication and disease progression. PLoS One.
8:e653362013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nishida N, Mimori K, Fabbri M, et al:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hashiguchi Y, Nishida N, Mimori K, et al:
Down-regulation of miR-125a-3p in human gastric cancer and its
clinicopathological significance. Int J Oncol. 40:1477–1482.
2012.PubMed/NCBI
|
|
27
|
Pazo Cid RA and Antón A: Advanced
HER2-positive gastric cancer: current and future targeted
therapies. Crit Rev Oncol Hematol. 85:350–362. 2013. View Article : Google Scholar
|
|
28
|
Smith MB, Reardon J and Olson EM:
Pertuzumab for the treatment of patients with previously untreated
HER2-positive metastatic breast cancer. Drugs Today (Barc).
48:713–722. 2012.
|
|
29
|
Sendur MA, Aksoy S and Altundag K:
Pertuzumab in HER2-positive breast cancer. Curr Med Res Opin.
28:1709–1716. 2012. View Article : Google Scholar : PubMed/NCBI
|