Introduction
Activated leukocyte cell adhesion molecule (ALCAM),
also known as cluster of differentiation 166 (CD166), is a highly
conserved 110 kDa multidomain transmembrane type 1 glycoprotein of
the immunoglobulin superfamily (1). ALCAM consists of a NH2-terminal
hydrophobic signal peptide, followed by extracellular,
transmembrane and cytoplasmic domains (2). ALCAM was identified as a CD6 ligand
and is expressed on activated leukocytes, fibroblasts, epithelia
and neurons (3). ALCAM is involved
in osteogenesis (4), neurite
extension (5), hematopoiesis
(6) and embryonal implantation in
the uterus (7), and mediates
homotypic/heterotypic interactions between tumor cells and between
endothelial and tumor cells (8).
Previously, alterations in the expression of ALCAM mRNA and protein
have been reported in several human tumors, including prostate
cancer (9), colon cancer (10), breast cancer (11), glioblastoma (12) and non-small-cell lung cancer
(13). In metastatic melanoma,
ALCAM controls the transition from local cell proliferation to
tissue invasion and functions as a cell surface sensor for cell
density in metastatic melanoma (14).
The soluble isoform of ALCAM (sALCAM) is an
alternative short transcript that comprises only the first 3′-exon.
The transcript contains a single Ig domain and lacks a
transmembrane domain.: In addition to a regulatory effect on ALCAM
function, sALCAM has an ALCAM independent effect in endothelial
cell assays (15). sALCAM serum
levels have been shown to be elevated in patients with pancreatic
(16) and esophageal (17) cancer, and elevated sALCAM levels in
peripheral blood are independent prognostic markers of poor
survival for patients (17).
microRNAs (miRNAs) mediate translational repression
or direct mRNA cleavage by binding complementary sequences of the
mRNA 3′ untranslated region (3′ UTR) (18). Candidate miRNAs of target mRNAs
have been computationally predicted using several bioinformatic
tools (19,20). In human hepatoma cell lines, ALCAM
was verified as a target gene of miR-9 by luciferase reporter and
western blot assays (21). miR-9
is significantly reduced in highly invasive uveal melanoma cell
lines, and has been demonstrated to suppress migration and invasion
partly through downregulation of the NF-κB1 signaling pathway
(22). The anti-proliferative and
pro-apoptotic activity of miR-9 via the direct targeting of MTHFD2
may contribute to tumor suppressor-like activity (23).
Gastric cancer (GC) is one of the most common
malignancies and a major leading cause of cancer-related mortality
worldwide (24). Subsequent to
curative surgery and multimodal therapeutic approaches, certain
patients with GC undergo an unexpected postoperative course,
largely due to a lack of adequate predictive biomarkers and valid
therapeutic targets. Ishigami et al (25) suggested that membranous
CD166-positivity may be a promising prognostic marker in GC
(25). However, ALCAM and sALCAM
expression profiles have not previously been investigated in GC.
The current study aimed to investigate the expression patterns of
ALCAM mRNA and protein in GC tissues and the serum sALCAM levels,
and to correlate ALCAM expression with clinicopathological
parameters, including survival duration. It was hypothesized that
miRNAs are associated with ALCAM overexpression in GC, therefore
the functions of computationally predicted miR-9 in GC cells were
analyzed.
Materials and methods
Patients
All blood and tissue samples were obtained from
patients who had presented to the Guangzhou First People’s hospital
(Guangzhou, China) and the study protocol was approved by the
Ethics Committee. Written consent was obtained from all
participants prior to surgery or blood sample collection.
Primary GC (n=66) and adjacent non-tumor tissues
(>5 cm away from the margin of the tumor) were snap-frozen in
liquid nitrogen immediately subsequent to surgery, and then stored
at −80°C until use. The patients were composed of 41 (62.12%) males
and 25 (37.88%) females, with ages ranging from 31 to 78 years
(median 63 years). None of the patients had received chemotherapy
and/or radiotherapy prior to surgery. The tissue samples were
formalin (10%;Shjisbio, Shanghai, China)-fixed and paraffin
(Shjisbo)-embedded, then sliced into 4-μm sections for
immunohistochemistry. For serum sALCAM quantification, peripheral
blood samples were obtained from 72 patients with GC (26 females
and 46 males, median age 67 years), 82 patients with precancerous
lesion of gastric cancer (PLGC; 30 females and 52 males, median age
62 years) and 73 blood-bank donors (28 females and 45 males, median
age 64 years) as healthy controls. Exclusion criteria included
patients with other malignant tumors and severe diseases in other
organs. Lauren’s pathohistological classification (26) of GC was used, and PLGC was
diagnosed according to the World Health Organization classification
system (27). The histological
diagnosis of each case was confirmed by two experienced
pathologists independently.
For patients with GC, data were collected on age,
gender and tumor histology, and the tumor stage was determined
according to the 2010 tumor-node-metastasis (TNM) classification of
the Union for International Cancer Control and American Joint
Committee on Cancer (27). The
clinical follow-up data were obtained from outpatient medical
records and phone interviews with patients and their family
members. Overall survival (OS) was defined as the duration between
the date of diagnosis and the date of mortality or last follow-up
appointment. Patients with GC who died of unrelated diseases were
regarded as censored cases.
Cell culture
The human GC cell lines MGC-803, BGC-823, HGC-27 and
SGC-7901, and the normal human gastric epithelial cell line GES-1
were purchased from Landerbio (Guangzhou, China). HGC-27 cells were
cultured in DMEM/F12 complete medium (Hyclone Laboratories, Inc.,
Logan, UT, USA), while the other cell lines were cultured in RPMI
1640 (Gibco Life Technologies, Grand Island, NY, USA) medium
supplemented with 10% heat-inactivated fetal bovine serum plus 1%
penicillin and streptomycin. The cell lines were kept in a
humidified atmosphere with 5% CO2 at 37°C.
Transfection
Synthesized miR-9 mimic and negative control (NC)
were purchased from Guangzhou RiboBio Co., Ltd.(Guangzhou, China).
The cells were transfected with the mimic or the NC at 50 nM,
according to the manufacturer’s instructions. The cells were
transfected using Lipofectamine 2000 (Invitrogen Life Technologies,
USA) at 30–50% confluence, according to the manufacturer’s
instructions and were harvested following 12, 24 and 48 h of
transfection.
RNA extraction, reverse transcription
(RT) and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from the tissues and cell
lines using TRIzol® (Ambion Life Technologies, Carlsbad,
CA, USA) according to the manufacturer’s instructions. Total cDNA
was obtained from the RT reaction using PrimeScript RT Reagent Kit
(Takara Bio, Inc., Otsu, Japan). ALCAM expression was anlayzed by
qPCR using a Stratagene SYBR Green I assay (Agilent Technologies,
Inc., Santa Clara, CA, USA) and normalized to that of GAPDH. The
sequences of the primers (Invitrogen, Lige Technologies) were as
follows: ALCAM F 5′-TTTTACTTACCAGGACAGC-3′ and R
5′-GACATAGTTTCCAGCATC-3′; GAPDH F 5′-GCA CCGTCAAGGCTGAGAA C-3′ and
R 5′-TGGTGAAGAC GCCAGTGGA-3′. The expression levels of miR-9 in the
cell lines and tissues were determined using miRACLE cDNA Synthesis
Kit and miRACLE qPCR miRNA Master Mix (Genetimes Technology, Inc.,
Shanghai, China). The miR-9 levels were normalized to the U6 snRNA
levels.
Immunohistochemical staining
For 66 pairs of GC and adjacent non-tumor tissues,
antigen retrieval was achieved by pressure cooking in citric acid
antigen repairing buffer (pH 6.0; Shjisbio). The sections were
treated with 3% H2O2, followed by treatment
with bovine serum (Hyclone Laboratores, Inc.,). The primary
mouse-derived monoclonal anti-CD166 antibody (Abcam, Cambridge, MA,
USA) was diluted to 1:100 and incubated. The EnVision system (Dako,
Glostrup, Denmark) was used to visualize the immunostaining. The
tissue sections were independently evaluated by two researchers
blinded to the patient characteristics and outcomes.
Sandwich ELISA
For the detection of sALCAM, Costar flexible 96-well
microtiter plates (Corning Incorporated, New York, NY, USA) were
coated with a monoclonal human anti-mouse ALCAM antibody (#MAB6561;
R&D Systems, Inc., Minneapolis, MN, USA). Bound protein was
detected using a biotinylated polyclonal human anti-goat antibody
(#BAF656; R&D Systems, Inc.), followed by
streptavidin-horseradish peroxidase (#4800-30-06; R&D Systems,
Inc.) using tetramethylbenzidine as the substrate. The color
reaction was stopped, then analyzed at 450 nm using a Dynatech
MR5000 Plate Reader (Dynex Technologies, Vienna, VA, USA). Human
ALCAM-Fc protein (R&D systems, Inc.) was used as an internal
standard control. Reproducibility and linearity were examined to
ensure that the immunoassay was suitable for measuring clinical
serum samples.
Statistical analysis
Group differences were calculated with a t-test or
Mann-Whitney test. The Fisher’s exact test or χ2 test
was used for the comparison of frequencies. The correlation between
two factors was evaluated with the Spearman’s rank correlation
test. Univariate survival analysis was performed using the
Kaplan-Meier method and the differences were assessed with the log
rank statistic. Independent prognostic factors were estimated by
the Cox proportional hazards stepwise regression model. The cut-off
level for sALCAM quantification was determined using the Youden
index. P<0.05 was considered to indicate a statistically
significant difference. All data were analyzed using the SPSS
software, version 16.0 (SPSS, Inc., Chicago, IL, USA).
Results
ALCAM mRNA expression is upregulated in
GC, and associated with advanced TNM stage and lymphatic
invasion
ALCAM mRNA was upregulated in 72.73% (48/66) of GC
tissues, with an average of a five-fold increase compared with the
matched non-tumor (NT) tissues (P=0.013; Fig. 1A). Elevated ALCAM mRNA in GC
tissues was significantly associated with an advanced TNM stage
(P=0.027) and lymphatic invasion (P=0.010) (Table I). No association was observed with
other clinicopathological variables, including gender, age and
histology.
 | Figure 1Analysis of ALCAM mRNA and miR-9
expression levels in gastric tissues and cell lines. (A) Comparison
of ALCAM mRNA expression levels in 66 paired GC and NT tissues by
qPCR. GC tissues (median 5.49) presented significantly higher
expression levels than the adjacent NT tissues (median 1.30)
(P=0.013). (B) ALCAM mRNA and miR-9 expression levels in four human
GC cell lines (MGC-803, BGC-823, HGC-27, SGC-7901) and one normal
human gastric epithelium cell line (GES-1). Compared with GES-1,
ALCAM mRNA expression levels in the gastric cancer cell lines
BGC-823, HGC-27 and SGC-7901 were significantly elevated, whilst
the mRNA expression level in MGC-803 cells was significantly
reduced. The expression levels of mature miR-9 in BGC-823, HGC-27
and SGC-7901 cell lines were significantly reduced, while the
levels significantly increased in MGC-803 cells
(*P<0.05 vs. GES-1 cells). (C) Comparison of miR-9
expression levels, which were significantly reduced in GC tissues
(median 0.55) compared with NT tissues (median 2.87) (P=0.023). (D)
Linear regression analysis indicated a significant negative
correlation between miR-9 and ALCAM mRNA (r=−0.382, P=0.014). (E)
miR-9 and ALCAM mRNA expression level at 0, 12, 24 and 48 h
following transfection of miR-9 mimic. The expression of miR-9 was
significantly downregulated, while the level of ALCAM mRNA was
markedly upregulated, at 12, 24 and 48 h (*P<0.05 vs.
0 h). ALCAM, activated leukocyte cell adhesion molecule; GC,
gastric cancer; NT, non-tumor. |
 | Table ICorrelations between mRNA expression
of activated leukocyte cell adhesion molecule and
clinicopathological features of gastric cancer. |
Table I
Correlations between mRNA expression
of activated leukocyte cell adhesion molecule and
clinicopathological features of gastric cancer.
| Characteristics | No. of patients | No. of patients with
elevated expression in cancer (%) | No. of patients with
expression in cancer (%) | P-valuec |
|---|
| Gender | | | | 0.300 |
| Male | 41 | 28 (42.42) | 13 (19.70) | |
| Female | 25 | 20 (30.30) | 5 (7.58) | |
| Age (years) | | | | 0.920 |
| ≤60 | 30 | 22 (33.33) | 8 (12.12) | |
| >60 | 36 | 26 (39.40) | 10 (15.15) | |
| Tumor
histologya | | | | 0.156 |
| Intestinal | 54 | 37 (56.06) | 17 (25.76) | |
| Diffuse | 12 | 11 (16.67) | 1 (1.51) | |
| TNM stageb | | | | 0.027 |
| I/II | 18 | 9 (13.64) | 9 (13.64) | |
| III/IV | 48 | 39 (59.08) | 9 (13.64) | |
| Lymphatic
invasion | | | | 0.010 |
| Yes | 49 | 40 (60.60) | 9 (13.64) | |
| No | 17 | 8 (12.12) | 9 (13.64) | |
ALCAM expression is under the regulation
of miR-9
Based on the Sanger miRNA database (http://www.mirbase.org) and TargetScan (http://www.targetscan.org) software, miR-9 was
selected, due to a predicted high complementarity between the seed
sequence of miR-9 and the two potential binding sites in the 3′-UTR
of ALCAM.
Compared with GES-1 cells, ALCAM mRNA expression
levels in the gastric cancer cell lines BGC-823, HGC-27 and
SGC-7901 were elevated, however, miR-9 levels in these three cell
lines were reduced (Fig. 1B). The
expression levels of miR-9 were reduced in 62.12% (41/66) of GC
tissues compared with the level in NT tissues (P=0.023; Fig. 1C). The expression levels of miR-9
were inversely correlated with those of ALCAM mRNA in GC tissues
(r=−0.293, P=0.014; Fig. 1D).
miR-9 and ALCAM mRNA levels were quantified
following transfection of SGC-7901 cells with the miR-9 mimic and
NC. SGC-7901 cells were selected due to having the lowest
endogenous miR-9 expression level among the four GC cell lines. At
12, 24 and 48 h subsequent to transfection of the miR-9 mimic, the
expression of miR-9 was significantly decreased, while the level of
ALCAM mRNA was markedly increased (P<0.05, Fig. 1E). However, there were no
significant differences in the miR-9 or ALCAM mRNA levels in the
NC-transfected cells (data not shown).
Clinical significance of ALCAM protein
expression and subcellular localization
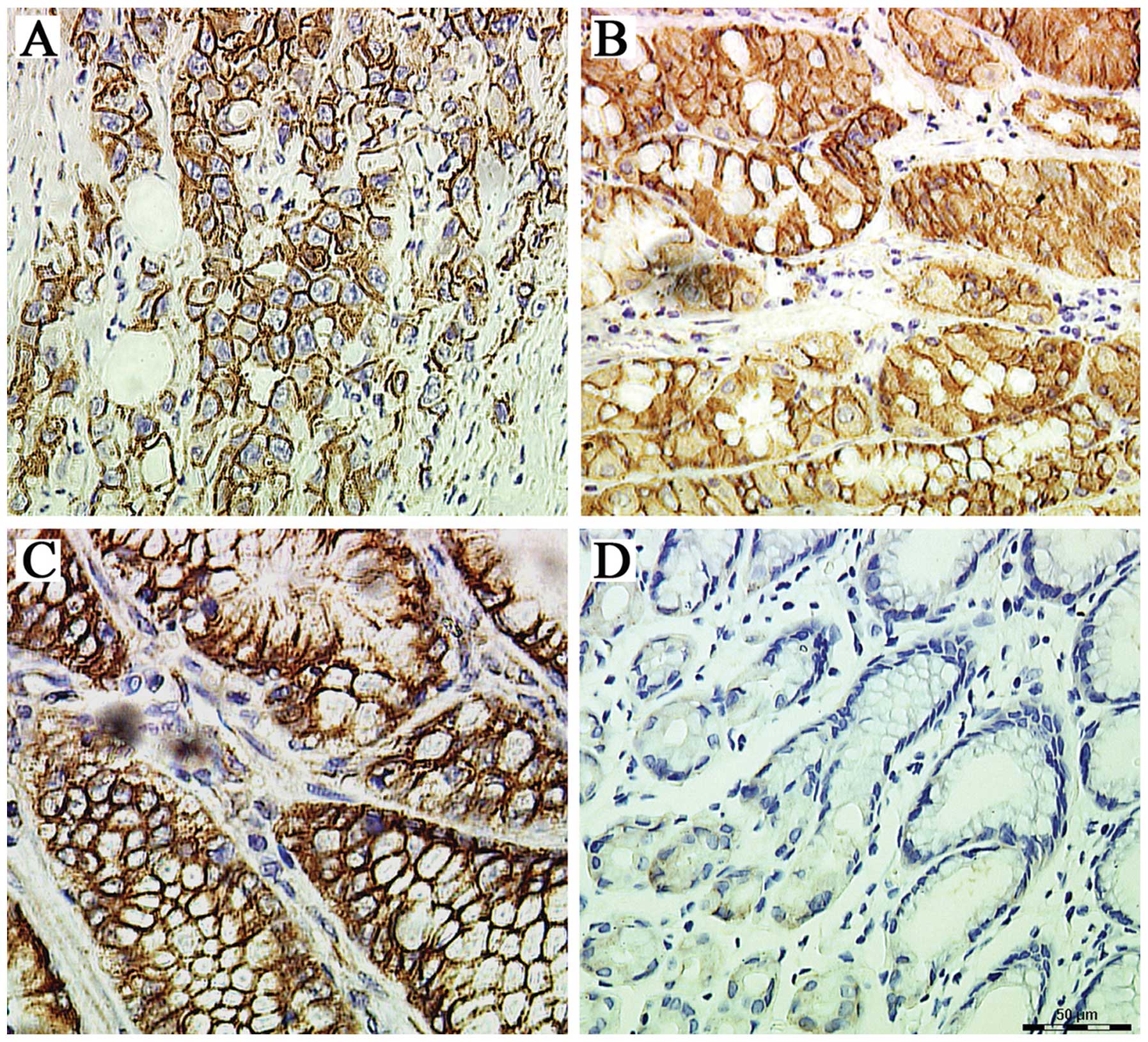
ALCAM protein expression was identified in the
cellular membrane and cytoplasm (Fig.
2). In GC tissues, the positive rates of membranous and
cytoplasmic ALCAM were 59.1% (39/66) and 48.48% (32/66),
respectively (Table II). In the
NT tissues the rates were 27.27% (18/66) and 22.72% (15/66),
respectively. A total of 23 GC tissues were positive for membranous
and cytoplasmic ALCAM expression simultaneously.
 | Table IIAssociation between activated
leukocyte cell adhesion molecule subcellular localization in cancer
tissues and clinicopathological factors. |
Table II
Association between activated
leukocyte cell adhesion molecule subcellular localization in cancer
tissues and clinicopathological factors.
| | Membranous ALCAM in
cancer tissues | Cytoplasmic ALCAM
in cancer tissues |
|---|
| |
|
|
|---|
|
Characteristics | No. of
patients | Positive (%) | Negative (%) | P-value | Positive (%) | Negative (%) | P-value |
|---|
| Gender | | | | 0.526 | | | 0.569 |
| Male | 41 | 23 (34.85) | 18 (27.27) | | 21 (31.82) | 20 (30.30) | |
| Female | 25 | 16 (24.24) | 9 (13.64) | | 11 (16.67) | 14 (21.21) | |
| Age (years) | | | | 0.715 | | | 0.208 |
| ≤60 | 30 | 17 (25.76) | 13 (19.70) | | 12 (18.18) | 18 (27.27) | |
| >60 | 36 | 22 (33.33) | 14 (21.21) | | 20 (30.30) | 16 (24.24) | |
| Tumor
histology | | | | 0.953 | | | 0.450 |
| Intestinal | 54 | 32 (48.48) | 22 (33.33) | | 25 (37.88) | 29 (43.94) | |
| Diffuse | 12 | 7 (10.61) | 5 (7.58) | | 7 (10.61) | 5 (7.58) | |
| TNM stage | | | | 0.041 | | | 0.880 |
| I/II | 18 | 7 (10.61) | 11 (16.67) | | 9 (13.64) | 9 (13.64) | |
| III/IV | 48 | 32 (48.48) | 16 (24.24) | | 23 (34.85) | 25 (37.88) | |
| Lymphatic
invasion | | | | 0.040 | | | 0.891 |
| Yes | 49 | 34 (51.52) | 15 (22.73) | | 24 (36.36) | 25 (37.88) | |
| No | 17 | 5 (7.58) | 12 (18.18) | | 8 (12.12) | 9 (13.64) | |
The upregulated membranous ALCAM expression in
cancer tissues was significantly associated with advanced tumor TNM
stage (P=0.041) and lymphatic invasion (P=0.040); however, the
cytoplasmic ALCAM was not associated with any of the
clinicopathological variables (P>0.05) (Table II). The log-rank test indicated
that the median OS was significantly reduced in tumors positive for
membranous ALCAM (20 months, n=39) compared with tumors negative
for membranous ALCAM (41 months, n=27) (P=0.012; Fig 3A). OS was not significantly
associated with cytoplasmic ALCAM in cancer tissues, and the OS
median durations in tumors positive for cytoplasmic ALCAM and
negative for cytoplasmic ALCAM were 21 (n=32) and 31 (n=34) months,
respectively (P=0.117; Fig.
3B).
Serum sALCAM level and its clinical
relevance
No significant difference was observed between the
genders or ages of the three groups (P>0.05; data not shown).
The serum level of sALCAM was significantly elevated in GC patients
(50.79±25.86 ng/ml), compared with PLGC patients (41.10±16.30
ng/ml) and healthy blood donors (37.54±24.42 ng/ml) (P<0.05;
Fig. 4A). The
sensitivity-specificity associations for sALCAM to discriminate
between GC patients and healthy donors was established using a
receiver operating characteristic curve. The optimal cut-off value
determined by the Youden index was 44.89 ng/ml, and the area under
curve was 0.618 (P=0.008; Fig.
4B). The specificity of sALCAM >44.89 ng/ml in diagnosing GC
was 69.33% with a sensitivity of 41.67% compared with healthy
donors (data not shown). According to the cut-off value, 37
patients with GC were defined as the high-level group, and the
other 35 were considered as the low-level group. The Kaplan-Meier
survival curve demonstrated a significant difference (P=0.031)
indicating that OS duration was significantly shorter in the
high-level group compared with the low-level group (Fig. 4C).
Regarding the OS, the Cox proportional hazards model
(Table III) identified TNM stage
(P=0.001), lymphatic invasion (P=0.000), membranous ALCAM
expression in the primary tumor (P=0.015) and high serum sALCAM
level (P=0.036) as independent prognostic indicators. Gender, age
and tumor histology did not present significance following
multivariate analyses (P>0.05).
 | Table IIIMultivariate analysis of factors
contributing to overall survival in patients with gastric
cancer. |
Table III
Multivariate analysis of factors
contributing to overall survival in patients with gastric
cancer.
|
Characteristics | RR | 95% CI | P-valuea |
|---|
| Gender | 1.280 | 0.702–2.332 | 0.420 |
| Age | 1.763 | 0.960–3.238 | 0.068 |
| Tumor
histology | 1.289 | 0.876–1.896 | 0.198 |
| TNM stage | 5.149 | 2.003–13.233 | 0.001 |
| Lymphatic
invasion | 5.562 | 2.164–14.300 | 0.000 |
| ALCAM membrane
expression | 2.251 | 1.170–4.330 | 0.015 |
| ALCAM cytoplasmic
expression | 1.612 | 0.877–2.964 | 0.124 |
| sALCAM high
level | 1.909 | 1.043–3.493 | 0.036 |
Discussion
In the current study, the results demonstrated that
the ALCAM mRNA expression level is elevated in GC tissues and cell
lines. In addition, an inverse correlation was observed between
miR-9 and ALCAM mRNA expression in the GC tissues and cell lines,
and enforced expression of miR-9 led to a reduction of ALCAM mRNA
level in the cellular model. Membranous ALCAM was associated with
an advanced TNM stage, lymphatic invasion and poor prognosis. The
serum sALCAM levels were significantly elevated in GC patients in
addition to the membranous ALCAM expression, and sALCAM was
identified as an independent prognostic indicator.
The association of ALCAM with disease progression in
human malignancies was first suggested in malignant melanoma
(28). To the best of our
knowledge, the current study is the first to present data with
respect to ALCAM expression in gastric cancer. The results
demostrated that ALCAM levels were significantly increased in
cancer tissues with advanced TNM stages and lymphatic invasion. The
presence of membranous ALCAM may serve as an indicator of poor
prognosis in GC. Consistent with these results, elevated expression
levels of ALCAM have previously been detected in all 20 malignant
mesothelioma (MM) cell lines, and overexpression of ALCAM was
demonstrated to contribute to tumor progression in MM (29). Ishigami et al (25) reported the rates of membranous and
cytoplasmic ALCAM expression as 25.4 and 34.4%, respectively, which
are lower than the present findings. This may be due to the
inclusion of a greater number of patients with an advanced TNM
stage (72.72 vs. 24.64%) in the present cohort, or the innate
heterogeneity of GC pathogenesis.
ALCAM is bimodular, and the two modules are required
for stable cell adhesion and aggregation (30). ALCAM clustering is essential to
obtain stable adhesion and, at the cell surface, actin
cytoskeleton-dependent clustering of ALCAM molecules regulate ALCAM
mediated cell adhesion (31). In
metastatic melanoma BLM cells, ALCAM acts as a cell density sensor
and initiates a signal to induce MMP-2 activation (14). According to the type of cadherin
adhesion complexes and the cadherin status of the tumor cells,
ALCAM may differentially enhance or reduce invasiveness (32). Membranous and cytoplasmic ALCAM
expression patterns were identified in GC tissues, as reported in
other types of malignancies; however, the biological relevance of
these patterns of ALCAM distribution is largely unclear. With
respect to subcellular localization, Tomita et al (33) hypothesized that membranous ALCAM
may be an activated form that can interact with extracellular
components, as it consists of five extracellular domains, which
fulfill its original function in homophilic interactions (33). By contrast, loss of ALCAM membrane
expression has been implicated as an independent factor of
unfavorable prognosis in epithelial ovarian cancer patients
(34). The above evidence
indicates that the evaluation of the prognostic impact of ALCAM
expression should be considered for each type of tumor, based on
the subcellular localization and expression levels of ALCAM. The
molecular mechanisms by which ALCAM participates in tumor
proliferation, motility and metastatic invasion require further
investigation.
The extracellular domain of ALCAM (sALCAM) is shed
by metalloproteases and functions as an active messenger
interacting with surrounding tissues (35). It has been described as a promoter
of endothelial cell migration and an inhibitor of endothelial tube
formation. sALCAM modulates endothelial cell function through
ALCAM-dependent and -independent pathways, and its expression is
differentially regulated upon inflammatory stimulation (15). Several studies have reported its
potential as a biomarker for different tumor entities. Concurrent
with the present study, patients with elevated sALCAM level were
identified in a previous study to have significantly worse OS
durations, thus it may serve as a potential diagnostic and
prognostic serum marker for esophageal cancer (17). The sensitivity and specificity of
sALCAM was clearly inferior to the tumor marker most frequently
used for pancreatic cancer, CA19-9 (16).
No significant association was identified between
the elevated tissue expression and serum level in the patients. The
tissue levels of ALCAM may not have correlated with the serum
values for several reasons. For example, TNF-α has been
demonstrated to induce moderate and persistent upregulation of
sALCAM expression in HMVECs, although it exhibits minimal early
effects (15). The expression of
ALCAM protein does not necessarily result in increased cleavage of
sALCAM by proteases such as ADAM17 (36). The flushing of sALCAM into the
vascular system may be a consequence of the disruption of
anatomical barriers between tumor cells and the bloodstream. In
view of these studies (32,37),
it is clear that the mechanisms regulating the shedding of ALCAM,
its dissemination into the surrounding tissue and its entry into
the blood system are not clearly understood.
The exact mechanisms involved in the regulation of
ALCAM have yet to be elucidated, particularly the mechanism
underlying the aberrant expression of ALCAM during malignant
transformation. In a study by Wang et al (21), ALCAM mRNA and protein expression
levels were upregulated following serum deprivation (SD) in HepG2
and GQY-7701 cells, partly due to the SD-mediated NF-κB P50/P65
increase, which enhances open chromatin accessibility around the κB
motif independent of SWI/SNF complexes. miRNAs bind the 3′-UTR of
target mRNAs and direct mRNA cleavage (38). miR-9 was selected as a candidate in
the current study, as two potential binding sites in the 3′-UTR of
ALCAM were identified by computational prediction, and there is
phylogenic conservation of the binding site sequence among mammals.
In human hepatoma cell lines, it was verified that miR-9 targets
ALCAM, as ALCAM protein levels were suppressed by miR-9. Following
miR-9 mimic transfection, the level of ALCAM mRNA was significantly
reduced, indicating that miR-9 is able to downregulate the ALCAM
mRNA expression level in GC. Further studies are required to
investigate whether miR-9 translationally represses ALCAM protein
expression in GC cell lines.
Acknowledgements
The present study was supported by the Science and
Technology Development Plan Project of Guangzhou (no.
2011J4100025), the Key Project of Guangzhou Health Bureau (no.
20121A021004) and the Natural Science Foundation of Guangdong (no.
101510060 010 00016), China.
References
|
1
|
Ofori-Acquah SF and King JA: Activated
leukocyte cell adhesion molecule: a new paradox in cancer. Transl
Res. 151:122–128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bowen MA, Patel DD, Li X, et al: Cloning,
mapping, and characterization of activated leukocyte-cell adhesion
molecule (ALCAM), a CD6 ligand. J Exp Med. 181:2213–2220. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patel DD, Wee SF, Whichard LP, et al:
Identification and characterization of a 100-kD ligand for CD6 on
human thymic epithelial cells. J Exp Med. 181:1563–1568. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruder SP, Ricalton NS, Boynton RE, et al:
Mesenchymal stem cell surface antigen SB-10 corresponds to
activated leukocyte cell adhesion molecule and is involved in
osteogenic differentiation. J Bone Miner Res. 13:655–663. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sekine-Aizawa Y, Omori A and Fujita SC:
MuSC, a novel member of the immunoglobulin superfamily, is
expressed in neurons of a subset of cranial sensory ganglia in the
mouse embryo. Eur J Neurosci. 10:2810–2824. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cortés F, Deschaseaux F, Uchida N, et al:
HCA, an immunoglobulin-like adhesion molecule present on the
earliest human hematopoietic precursor cells, is also expressed by
stromal cells in blood-forming tissues. Blood. 93:826–837.
1999.PubMed/NCBI
|
|
7
|
Fujiwara H, Tatsumi K, Kosaka K, et al:
Human blastocysts and endometrial epithelial cells express
activated leukocyte cell adhesion molecule (ALCAM/CD166). J Clin
Endocrinol Metab. 88:3437–3443. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weidle UH, Eggle D, Klostermann S and
Swart GW: ALCAM/CD166: cancer-related issues. Cancer Genomics
Proteomics. 7:231–243. 2010.PubMed/NCBI
|
|
9
|
Kristiansen G, Pilarsky C, Wissmann C, et
al: ALCAM/CD166 is up-regulated in low-grade prostate cancer and
progressively lost in high-grade lesions. Prostate. 54:34–43. 2003.
View Article : Google Scholar
|
|
10
|
Weichert W, Knösel T, Bellach J, Dietel M
and Kristiansen G: ALCAM/CD166 is overexpressed in colorectal
carcinoma and correlates with shortened patient survival. J Clin
Pathol. 57:1160–1164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hein S, Müller V, Köhler N, et al:
Biologic role of activated leukocyte cell adhesion molecule
overexpression in breast cancer cell lines and clinical tumor
tissue. Breast Cancer Res Treat. 129:347–360. 2011. View Article : Google Scholar
|
|
12
|
Kijima N, Hosen N, Kagawa N, et al:
CD166/activated leukocyte cell adhesion molecule is expressed on
glioblastoma progenitor cells and involved in the regulation of
tumor cell invasion. Neuro Oncol. 14:1254–1264. 2012. View Article : Google Scholar :
|
|
13
|
Ishiguro F, Murakami H, Mizuno T, et al:
Membranous expression of activated leukocyte cell adhesion molecule
contributes to poor prognosis and malignant phenotypes of
non-small-cell lung cancer. J Surg Res. 179:24–32. 2013. View Article : Google Scholar
|
|
14
|
Lunter PC, van Kilsdonk JW, van Beek H, et
al: Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD),
a novel actor in invasive growth, controls matrix metalloproteinase
activity. Cancer Res. 65:8801–8808. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ikeda K and Quertermous T: Molecular
isolation and characterization of a soluble isoform of activated
leukocyte cell adhesion molecule that modulates endothelial cell
function. J Biol Chem. 279:55315–55323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tachezy M, Zander H, Marx AH, et al: ALCAM
(CD166) expression and serum levels in pancreatic cancer. PLoS One.
7:e390182012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tachezy M, Effenberger K, Zander H, et al:
ALCAM (CD166) expression and serum levels are markers for poor
survival of esophageal cancer patients. Int J Cancer. 131:396–405.
2012. View Article : Google Scholar
|
|
18
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sethupathy P, Megraw M and Hatzigeorgiou
AG: A guide through present computational approaches for the
identification of mammalian microRNA targets. Nat Methods.
3:881–886. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mendes ND, Freitas AT and Sagot MF:
Current tools for the identification of miRNA genes and their
targets. Nucleic Acids Res. 37:2419–2433. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang J, Gu Z, Ni P, et al: NF-kappaB
P50/P65 hetero-dimer mediates differential regulation of
CD166/ALCAM expression via interaction with micoRNA-9 after serum
deprivation, providing evidence for a novel negative
auto-regulatory loop. Nucleic Acids Res. 39:6440–6455. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu N, Sun Q, Chen J, et al: MicroRNA-9
suppresses uveal melanoma cell migration and invasion through the
NF-κB1 pathway. Oncol Rep. 28:961–968. 2012.PubMed/NCBI
|
|
23
|
Selcuklu SD, Donoghue MT, Rehmet K, et al:
MicroRNA-9 inhibition of cell proliferation and identification of
novel miR-9 targets by transcriptome profiling in breast cancer
cells. J Biol Chem. 287:29516–29528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Martel C, Forman D and Plummer M:
Gastric cancer: epidemiology and risk factors. Gastroenterol Clin
North Am. 42:219–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ishigami S, Ueno S, Arigami T, et al:
Clinical implication of CD166 expression in gastric cancer. J Surg
Oncol. 103:57–61. 2011. View Article : Google Scholar
|
|
26
|
Lauren P: The two histological main types
of gastric carcinoma: Diffuse and so-called intestinal-type
carcinoma. An attempt at a histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965.PubMed/NCBI
|
|
27
|
Lauwers GY, Carneiro F and Graham DY:
Gastric carcinoma. WHO Classification of tumors of the digestive
system. Bosman FT, Carneiro F and Hruban RH: IARC Press; Lyon: pp.
48–58. 2010
|
|
28
|
van den Oord CJJ, van Muijen GN, Weidle
UH, Bloemers HP and Swart GW: Activated leukocyte cell adhesion
molecule/CD166, a marker of tumor progression in primary malignant
melanoma of the skin. Am J Pathol. 156:769–774. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishiguro F, Murakami H, Mizuno T, et al:
Activated leukocyte cell-adhesion molecule (ALCAM) promotes
malignant phenotypes of malignant mesothelioma. J Thorac Oncol.
7:890–899. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
van Kempen LC, Nelissen JM, Degen WG, et
al: Molecular basis for the homophilic activated leukocyte cell
adhesion molecule (ALCAM)-ALCAM interaction. J Biol Chem.
276:25783–25790. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nelissen JM, Peters IM, de Grooth BG, van
Kooyk Y and Figdor CG: Dynamic regulation of activated leukocyte
cell adhesion molecule-mediated homotypic cell adhesion through the
actin cytoskeleton. Mol Biol Cell. 11:2057–2068. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jannie KM, Stipp CS and Weiner JA: ALCAM
regulates motility, invasiveness, and adherens junction formation
in uveal melanoma cells. PLoS One. 7:e393302012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tomita K, van Bokhoven A, Jansen CF,
Bussemakers MJ and Schalken JA: Coordinate recruitment of
E-cadherin and ALCAM to cell-cell contacts by alpha-catenin.
Biochem Biophys Res Commun. 267:870–874. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mezzanzanica D, Fabbi M, Bagnoli M, et al:
Subcellular localization of activated leukocyte cell adhesion
molecule is a molecular predictor of survival in ovarian carcinoma
patients. Clin Cancer Res. 14:1726–1733. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rosso O, Piazza T, Bongarzone I, et al:
The ALCAM shedding by the metalloprotease ADAM17/TACE is involved
in motility of ovarian carcinoma cells. Mol Cancer Res.
5:1246–1253. 2007. View Article : Google Scholar
|
|
36
|
Weidle UH, Eggle D and Klostermann S:
L1-CAM as a target for treatment of cancer with monoclonal
antibodies. Anticancer Res. 29:4919–4931. 2009.
|
|
37
|
Kahlert C, Weber H, Mogler C, et al:
Increased expression of ALCAM/CD166 in pancreatic cancer is an
independent prognostic marker for poor survival and early tumour
relapse. Br J Cancer. 101:457–464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|