Introduction
Colon cancer is one of the most common types of
cancer in the world, yet its underlying development mechanism is
unknown. However, there are a number of indications regarding the
progression from colonic epithelial cells to carcinoma. Unhealthy
eating habits and environmental pollution aggravate the mutation of
genes controlling cell adhesion and proliferation and consequently
play significant roles in cancer development (1).
microRNAs (miRNAs) are a class of approximately
22-nt, non-coding RNAs, that generally suppress gene expression
through binding to the 3′ untranslated region (3′UTR) of target
genes (2–4). microRNA-155 (miR-155) participates in
various aspects of cell physiology: it plays a critical role in the
innate and acquired immune response and is associated with various
diseases including autoimmune disorders, leukemia, atherosclerosis
and tumors including breast and lung (5).
In previous studies, high expression of miR-155 in
colorectal cancer has been linked to lymph node metastases
(6,7). Furthermore, lymph node metastases
have long been associated with higher proliferation and invasion
abilities. At the same time, a type of RNA-binding protein named
quaking (QKI), which suppresses gastric cancer cell proliferation
by decreasing β-catenin expression (8), has been detected in colon cancer
(9). The phenomenon of high
miR-155 expression and low QKI expression in colon cancer prompted
us to investigate the correlation between them and further
ascertain whether miR-155 affects the differentiation of colon
cancer by targeting QKI directly.
Materials and methods
Tissues
The study was approved by the ethics committee of
Wenzhou Medical University (Wenzhou, China). After obtaining their
informed consent, colon tumors and adjacent normal tissues were
collected from surgical patients at the First Affiliated Hospital
of Wenzhou Medical University prior to the operation. When
cancerous tissues were removed, they were frozen immediately at
−80°C. Next, the samples were divided into two groups depending on
the presence of lymphatic metastasis. The non-invasion group refers
to primary colon cancer samples without lymphatic metastasis, while
the invasion group refers to cancer samples with lymphatic
metastasis.
Cell culture
Human colon cancer cell lines SW480 and COLO205, and
the non-malignant gastric intestinal epithelial cell line GES-1
were all cultured in high-glucose Dulbecco’s modified Eagle’s
medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum (Gibco). Cells were maintained at 37°C under a 5%
CO2 atmosphere. SW480 was selected as the cell type for
the remaining experiments.
Quantitative polymerase chain reaction
(qPCR)
qPCR for mRNA or miRNA analyses was performed using
ABI PRISM® 7900HT (Applied Biosystems, Carlsbad, CA,
USA), with the following reactions: 95° for 30 sec, 40 cycles of
95° for 5 sec and 65° for 34 sec. The mRNA levels of 18S and U6
were used as an internal control for mRNA or miRNA, respectively.
Relative expression of RNA was measured using the 2−ΔΔCt
method. The primers were as follows: QKI: F-AAGCCC ACCCCAGATTACCT,
R-ACTCTGCTAATTTCTTC GTCCAG; β-catenin: F-AAAGCGGCTGTTAGTCACTGG,
R-CGAGTCATTGC ATACTGTCCAT; e-cadherin: F-CGAGAGCTACACGTTCACGG,
R-GGGTGTCGAGGG AAAAATAGG; lactase: F-TCCACCGCTGGTCCTCTAA,
R-CACTGTTTTCTCGT CTGGATTCT.
We first analyzed the dissolution curve to determine
the specificity of the qPCR amplification. Quantification of the
relative expression levels of these target genes was obtained by
the formula 2−ΔΔCt, where ΔΔCt = (Ct of the target gene
− Ct of U6)treatment − (Ct of the target gene − Ct of
U6)control. Each treatment condition was evaluated in
five wells. Data were given in arbitrary units relative to the
control, which was assigned a value of 1.
Western blot analysis
Total protein was harvested by lysate cells with
RIPA buffer (Cell Signaling Technologies, Beverly, MA, USA). The
bicinchoninic acid assay was used to determine the protein
concentration. Proteins were separated by SDS-PAGE gel, and then
transferred to a PVDF membrane (Millipore, Billerica, MA, USA).
After blocking with 5% non-fat milk, the membranes were incubated
with corresponding primary antibodies in a TBS/Tween-20 buffer
overnight at 4°C. The primary antibodies were against QKI (Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA), β-catenin,
e-cadherin and lactase (all from Cell Signaling Technologies).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an
endogenous control. The next day, after washing three times with
TBS/Tween-20, the PVDF membranes were incubated with appropriate
secondary antibodies for 1 h at room temperature. Finally, they
were washed another three times, then the membranes were soaked in
an enhanced chemiluminescence (ECL) reagent for 5 min.
Immunoreactivity was detected with the ECL reaction (Pierce
Biotechnology, Rockford, IL, USA). All experiments were repeated
three times.
Luciferase reporter assay
The wild-type (WT) 3′UTR and mutant (Mut) 3′UTR of
the human QKI gene were cloned according to the instructions from
http://www.targetscan.org/. The primers
used to amplify the QKI gene were as follows: QKI-3′UTR
F-5′-CACGTCTAGAGCATGTGTTT GACCTG-3′ and QKI-3′UTR R-5′-GGACTCCGGAGC
GCATACCAGTAA GCACG-3′. Then the reporter vector was constructed to
the PGL3-basic vector (Promega Corporation, Madison, WI, USA), and
the cloning site was XbaI/XbaI. The SW480 cells were
cultured in 12-well plates. When the cells reached 80–90%
confluence, the cells were transfected with 0.75 μg
PGL3-WT-3′UTR-QKI and PGL3-Mut-3′UTR-QKI plasmids, and then 10 pmol
mimic-miR-155 or mimic-control (Ruibobio, Guangzhou, China) was
added using Lipofectamine 2000 (Invitrogen Life Technologies,
Carlsbad, CA, USA). After 24 h, the cells were harvested by
digestion. The luciferase activity was measured according to the
manufacturer’s instructions. The relative luciferase activity was
expressed as the ratio of firefly luciferase fluorescence to
renilla luciferase fluorescence.
Cell cycle assay
The SW480 cells were cultured in DMEM in 12-well
plates. When they reached 80% confluence, miR-155 and QKI vector
was added to the supernatant. Following stimulation for 24 h, the
SW480 cells were harvested through centrifugation, and then fixed
with a 70% ethanol ice bath for 24 h. The following day, the cells
were washed with cold phosphate-buffered saline twice, and then
stained with propidium iodide (PI) protected from the light at 37°C
for 30 min. Each treatment condition was investigated in three
wells. Then flow cytometry (BD Biosciences, San Jose, CA, USA) was
used to measure the cell number in the G1, S and G2 phases.
Cell scratch assay
For scratch assays, the SW480 cells (cell density
5×105/cm2) were spread in a capsule. Then the
cells were stimulated with various treatments. After overspreading
the entire field of the microscope without overlapping, certain
cells were wiped off to leave a cross-shaped area without cells.
After 24 h, the results were evaluated according to the coverage of
cells.
Statistical analysis
GraphPad 5.0 software (GraphPad Software Inc., San
Diego, CA, USA) was used to process the data, which are presented
as the means ± standard deviation. Comparisons between two groups
were performed with a two-tailed t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
High expression of miR-155 and low
expression of QKI in colon cancer tissues
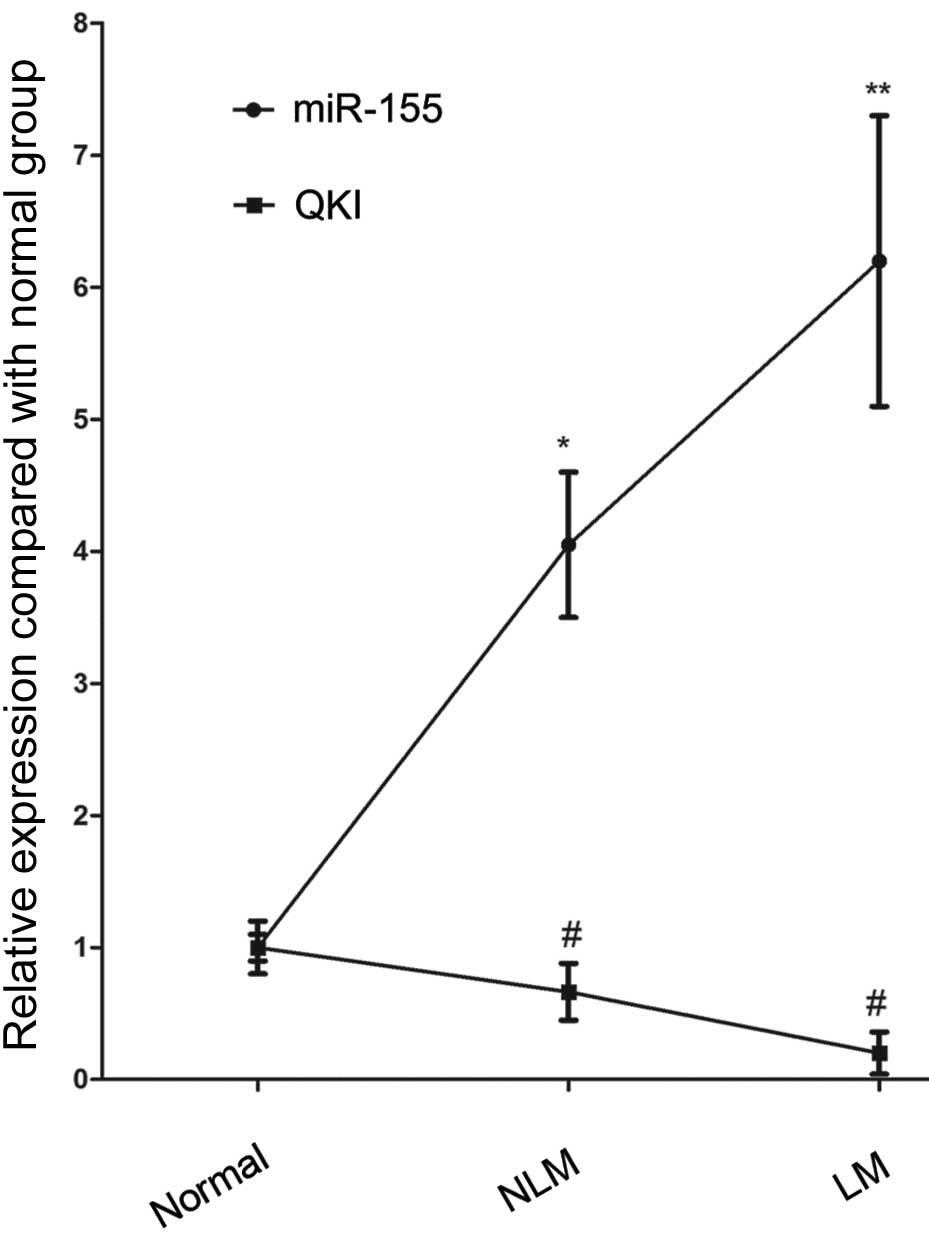
The qPCR findings revealed higher miR-155 expression
and lower QKI expression in colon cancer tissue than in normal
gastric intestinal epithelial tissue (Fig. 1). Moreover, colon cancer tissues
with lymph node metastasis had higher miR-155 expression than the
cancer tissues without lymph node metastasis (Fig. 1). Correspondingly, in comparison
with the primary colon cancer tissues, lower QKI expression was
observed in the colon cancer tissues with lymph node metastasis
(Fig. 1).
High expression of miR-155 and low
expression of QKI in colon cancer cell lines
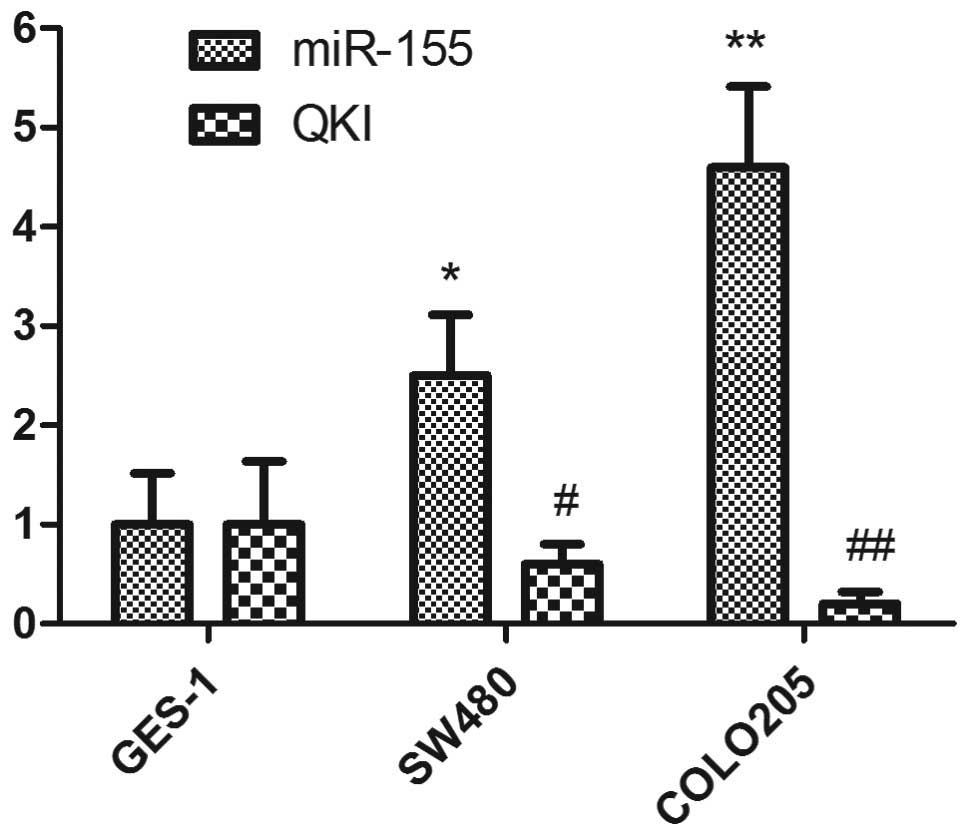
As expected, between the SW480 and COLO205 colon
cancer cell lines, COLO205 had the stronger invasion and metastasis
potential. GES-1 was considered to be the normal control of the two
cell lines. qPCR was also performed to investigate the expression
level of miR-155 and QKI in colon cancer cell lines. Consistent
with the results in cancer tissues, we demonstrated that miR-155
and QKI had a similar expression profile to the colon cancer
tissues (Fig. 2), with COLO205
cells having a higher expression level of miR-155 and lower
expression level of QKI (Fig.
2).
miR-155 decreases production of QKI by
acting on 3′UTR of QKI gene
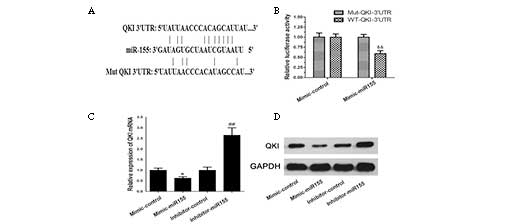
Due to the phenomenon of the simultaneous existence
of a high expression level of miR-155 and a low expression level of
QKI in colon cancer, we speculated that QKI was the target of
miR-155. By using computational deductions (10), we identified that one miR-155
binding site was included in the 3′UTR of QKI mRNA. We then
designed luciferase reporters with the WT 3′UTR and Mut 3′UTR of
QKI (Fig. 3A). The relative
luciferase activity of the 3′UTR was decreased by ~45% when SW480
cells were transfected with mimic-miR-155 (Fig. 3B). Furthermore, when the cells were
treated with mimic-miR-155, the QKI expression was significantly
decreased at both the mRNA and protein level (Fig. 3C and D). When they were treated
with miR-155 inhibitor, the expression of QKI increased
significantly (Fig. 3C and D).
miR-155 affects the cell cycle of colon
cancer cells in a QKI-dependent manner
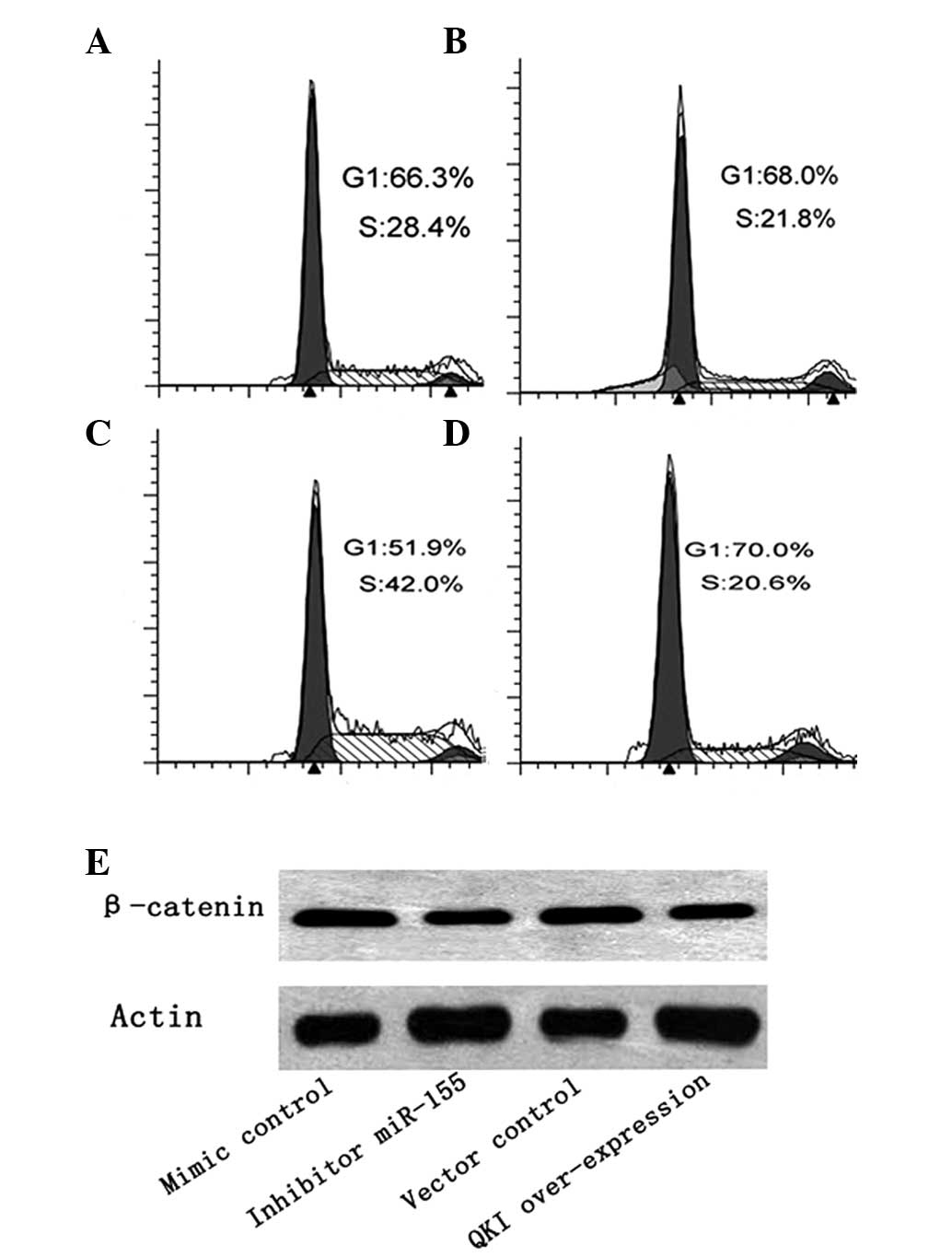
In order to verify whether miR-155 affects the colon
cancer cell cycle by targeting QKI we firstly performed PI staining
of SW480 cells, and the flow cytometry findings revealed that the
ratio of G1/S was increased in SW480 cells when treated with
miR-155 (Fig. 4B). When treated
with the QKI expression vector, the G1/S ratio also increased
significantly (Fig. 4D).
Then, we measured the expression of β-catenin, which
plays a critical role in the cell cycle of cancer cells by
promoting the expression of the majority of cell cycle-related
genes (11,12). We observed that miR-155 inhibitor
decreased the expression of β-catenin, which was consistent with
the results of overexpression of QKI stimulation (Fig. 4E).
miR-155 suppresses the invasion
capabilities of SW480 cells and inhibits the expression of
e-cadherin and lactase by targeting QKI
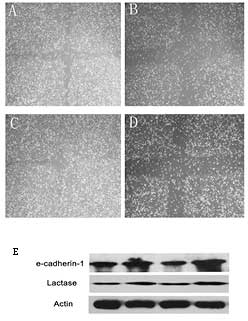
A cell scratch assay was carried out to confirm that
miR-155 indeed promotes the invasion capabilities of colon cancer
cells (Fig. 5B). E-cadherin, a
cell adhesion-related protein (13) whose expression is suppressed in
colon cancer cells, and lactase, a differentiation marker of colon
cells which is expressed at a low level in colon cancer cells, both
play significant roles in the differentiation of colon cancer cells
(9,14). Treatment with the miR-155 inhibitor
and QKI overexpression vector increased the expression of
e-cadherin and lactase, which promoted colon cancer cell
differentiation; the results obtained through transfection of
miR-155 inhibitor were similar (Fig.
5E).
Discussion
In a previous study, high expression of miR-155 was
detected in colon cancer and this was considered to be associated
with lymph node metastases (6). To
our knowledge, the cellular and molecular effects of miR-155 on
colon cancer cells have not previously been reported in detail. In
this study, we demonstrated that the RNA-binding protein QKI is a
target of miR-155 in colon cancer cells. miR-155 promotes the
proliferation and invasion abilities of colon cancer cells by
targeting QKI.
As is already known, miRNA is mostly relevant to
inflammation, and establishes an association between inflammation
and cancer (15). Recent studies
have shown that high expression of miR-155 was detected in various
cancers including breast (16,17),
lung (18), lymphoma and leukemia
(19). By inducing mutator
activity, miR-155 intensified the progression process from normal
epithelia cells to cancer cells (20). The present study demonstrated that
miR-155 expression is positively correlated with lymph node
metastasis in colon cancer, which is consistent with the results of
a previous study (6). Furthermore,
our study clarified the connection between miR-155 and QKI, which
exerts anti-proliferative and differentiative functions in gastric
and colon cancer (8,9). In accordance with previous studies
(6,8,9,20),
our results demonstrated that the low expression of QKI is due to
the high expression of miR-155.
In addition, we revealed that inhibiting the
function of miR-155 restored the expression of QKI. Furthermore,
both overexpression of QKI and inhibition of miR-155 have the same
effect on the cell proliferation and invasion abilities of colon
cancer cells. In order to confirm the effect of miR-155 on the role
of invasion abilities of colon cancer cells, we also analyzed the
expression of e-cadherin and lactase, two enterocyte markers of
colon epithelial cells. The expression levels of both markers were
increased by overexpression of QKI, whose effects are reached
uniformly through the inhibition of miR-155. The high expression of
β-catenin detected in colon cancer promotes the proliferation of
colon cancer cells and can be suppressed by miR-320a (21). In our results, the expression of
β-catenin was intensively suppressed by miR-155 inhibition and QKI
overexpression, while QKI was considered to be a suppressor of
β-catenin.
In conclusion, the present study has revealed that
miR-155 regulates the cell cycle and invasion ability of colon
cancer cells via the modulation of QKI expression. The study
provides novel therapeutic strategies for colon cancer therapy.
Acknowledgements
The authors would like to thank everyone who
assisted in this study.
References
|
1
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shibuya H, Iinuma H, Shimada R, Horiuchi A
and Watanabe T: Clinicopathological and prognostic value of
microRNA-21 and microRNA-155 in colorectal cancer. Oncology.
79:313–320. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iorio MV, Ferracin M, Liu C-G, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bian Y, Wang L, Lu H, et al:
Downregulation of tumor suppressor QKI in gastric cancer and its
implication in cancer prognosis. Biochem Biophys Res Commun.
422:187–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang G, Fu H, Zhang J, et al: RNA-binding
protein quaking, a critical regulator of colon epithelial
differentiation and a suppressor of colon cancer. Gastroenterology.
138:231–240. e1–e5. 2010. View Article : Google Scholar
|
|
10
|
Thomas M, Lieberman J and Lal A:
Desperately seeking microRNA targets. Nat Struct Mol Biol.
17:1169–1174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
van de Wetering M, Sancho E, Verweij C, et
al: The β-catenin/TCF-4 complex imposes a crypt progenitor
phenotype on colorectal cancer cells. Cell. 111:241–250. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Irby RB and Yeatman TJ: Increased Src
activity disrupts cadherin/catenin-mediated homotypic adhesion in
human colon cancer and transformed rodent cells. Cancer Res.
62:2669–2674. 2002.PubMed/NCBI
|
|
14
|
Zweibaum A, Laburthe M, Grasset E and
Louvard D: Use of cultured cell lines in studies of intestinal cell
differentiation and function. Comprehensive Physiology. Published
online. Wiley-Blackwell; pp. 223–255. 2011
|
|
15
|
Tili E, Croce CM and Michaille JJ:
miR-155: on the crosstalk between inflammation and cancer. Int Rev
Immunol. 28:264–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan LX, Huang XF, Shao Q, et al: MicroRNA
miR-21 overexpression in human breast cancer is associated with
advanced clinical stage, lymph node metastasis and patient poor
prognosis. RNA. 14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang S, Zhang HW, Lu MH, et al:
MicroRNA-155 functions as an OncomiR in breast cancer by targeting
the suppressor of cytokine signaling 1 gene. Cancer Res.
70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Donnem T, Eklo K, Berg T, et al:
Prognostic impact of MiR-155 in non-small cell lung cancer
evaluated by in situ hybridization. J Transl Med. 9:1–9. 2011.
View Article : Google Scholar
|
|
19
|
Marcucci G, Maharry KS, Metzeler KH, et
al: Clinical role of microRNAs in cytogenetically normal acute
myeloid leukemia: miR-155 upregulation independently identifies
high-risk patients. J Clin Oncol. 31:2086–2093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tili E, Michaille JJ, Wernicke D, et al:
Mutator activity induced by microRNA-155 (miR-155) links
inflammation and cancer. Proc Natl Acad Sci USA. 108:4908–4913.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun JY, Huang Y, Li JP, et al:
MicroRNA-320a suppresses human colon cancer cell proliferation by
directly targeting β-catenin. Biochem Biophys Res Commun.
420:787–792. 2012. View Article : Google Scholar : PubMed/NCBI
|