Introduction
Schizophrenia is a severe cognitive disorder that is
characterized by psychosis, disorganized speech and behavioral
symptoms. The observed lifetime prevalence estimates and morbidity
rate of this condition in the general population is 4.0 in 1,000
and 7.2 in 1,000 people, respectively (1,2). In
the etiology of schizophrenia, the disease development is
attributable to a combination of environmental and genetic risk
factors. Previous molecular genetic studies have identified
abnormalities in the structure of oligodendrocytes and myelin in
schizophrenic brains (3–8), factors that may contribute to the
disorganized behavioral symptoms in schizophrenia patients
(9). Despite the well-defined
factors involved in disease development, the exact molecular
mechanisms of schizophrenia remain unclear and require further
investigation.
Proteins involved in the dopamine or glutamate
signaling pathways, which regulate nerve growth and development,
have been hypothesized to be candidate molecules for indicating the
risk of schizophrenia. The human Ran-binding protein 9 (RANBP9;
NM_005493.2) is a member of the Ran-binding family of proteins and
interacts with multiple receptors, acting as a scaffold protein for
signal transduction processes (10). To date, although the function of
RANBP9 is poorly understood, it is considered to be an important
regulator of neurite growth through interactions with neural cell
adhesion molecules, including L1 (10). It has been suggested that RANBP9,
as a ligand for Rho-GEF, may be involved in regulating dendritic
spine and branch morphology (11).
In addition, RANBP9 has been demonstrated to interact with several
important proteins associated with schizophrenia, such as disrupted
in schizophrenia 1 (DISC1) and the dopamine D1 receptor in brain
tissue (12,13). DISC1 is hypothesized to be
associated with major mental illnesses, including schizophrenia,
autism and bipolar disorder, and is involved in the regulation of
neuronal axon and dendrite outgrowth (14,15).
The aberrant dopamine D1 receptor is linked to various
neuropsychiatric disorders such as substance abuse, schizophrenia
and Parkinson’s disease (16).
Despite insufficient knowledge with regard to the association
between RANBP9 and schizophrenia, RANBP9 is predicted to be
involved in the pathogenesis of the disease.
In the present study, a case-control association
analysis was conducted between genetic variations in RANBP9 and the
risk of schizophrenia and smooth pursuit eye movement (SPEM)
abnormalities in a Korean population.
Materials and methods
Subjects
Trained psychiatrists diagnosed schizophrenia based
on the criteria set forth by the Diagnostic and Statistical Manual
of Mental Disorders, fourth edition (17). Patients with additional,
complicating diagnoses of mental retardation, organic brain damage,
drug or alcohol abuse, neurological disorders, autoimmune disorders
or low comprehension skills were excluded from the study.
Schizophrenia patients (n=449) were recruited over 10 years from
Jinju Mental Hospital (Jinju, Korea), Soonyoung Hospital (Sacheon,
Korea), Hadong Wooridle Hospital (Gyeongsang Nam Do, Korea), Seoul
National University Hospital (Seoul, Korea) and Keyo Hospital
(Kyunggi-Do, Korea). Unrelated healthy controls (n=393) were
recruited from Hangang Sacred Heart Hospital and the Center for
Health Promotion at Seoul National University Hospital. Peripheral
blood leukocytes from 842 samples were collected in tubes
containing sodium EDTA and genomic DNA was subsequently obtained
using the QIAamp blood extraction kit (Qiagen, Valencia, CA, USA).
The study protocols were approved by the institutional review
boards of each hospital (Jinju Mental Hospital, Soonyoung Hospital,
Hadong Wooridle Hospital, Seoul National University Hospital and
Keyo Hospital) and the experiments were conducted with the
understanding and written informed consent from each patient.
Measurement of SPEM
In total, 125 schizophrenia patients, who were able
to understand the experimental procedure underwent an eye-tracking
task as described previously (18). The electrooculographic recordings
of the eye movements were used to quantify SPEM abnormality and
calculate the natural logarithmic values of the signal/noise (Ln
S/N) ratio.
Genetic variation selection and
genotyping in the RANBP9 gene
In total, five single nucleotide polymorphisms
(rs24023, rs442458, rs6940759, rs204226 and rs11759518) and an
insertion/deletion (rs16724) variation in RANBP9 were selected from
the International HapMap Database (http://hapmap.ncbi.nlm.nih.gov/). Commercially
available predesigned TaqMan probes and designed primers (Applied
Biosystems, Foster City, CA, USA) were used in genotyping (Table I). All polymorphisms satisfied the
following criteria: (i) A minimum call rate of 95% and (ii)
Hardy-Weinberg equilibrium (HWE) of P>0.05. Haplotypes were
inferred from the genotyped polymorphisms using the PHASE
algorithm, version. 2.0 (19) and
those with a frequency of >0.05 were included in the association
analysis.
 | Table IAssay IDs of human Ran-binding protein
9 variations. |
Table I
Assay IDs of human Ran-binding protein
9 variations.
| Loci | | Probe sequence or
Assay-by-design ID* | Method |
|---|
| rs24023 | | C__11661329_10 | TaqMan assay |
| rs442458 | | C____580920_10 | TaqMan assay |
| rs6940759 | Forward |
ACCAATGCAGCACAATGATAGGTTA | TaqMan assay by
design |
| Reverse |
CAGTAGCAGTAGCAAAGGAGAAGTA | |
| VIC® |
CTTATTCTGTGACTTTAAAG | |
| FAM |
CTTATTCTGTGATTTTAAAG | |
| rs204226 | | C___7615918_20 | TaqMan assay |
| rs16724 | Forward |
GCTATTCCCTCCACCTTTCCTT | TaqMan assay by
design |
| Reverse |
TTGAGGAAGTAAAGCTTGAGCTAAGATTT | |
| VIC® |
CCTGAGTAGGTAGGTCCTA | |
| FAM |
TAGCCTGAGTAGGTCCTA | |
| rs11759518 | | C___2974708_10 | TaqMan assay |
Statistical analysis
Lewontin’s D′ (|D′|) and linkage disequilibrium (LD)
coefficient r2 between all pairs of biallelic loci using
the Haploview algorithm were examined (20). The odds ratio (95% confidence
interval) and corresponding P-values were calculated using a
logistic model, controlling for age (continuous value) as a
covariate. P<0.05 was considered to indicated a statistically
significant difference between values. The Ln S/N ratio was
calculated from analysis of the power spectrum curves and the
results were used in logistic analysis of SPEM abnormality,
controlling for age as a covariate. Values are presented as the
mean ± standard deviation.
Results
Characteristics of study subjects
To elucidate whether there is a genetic association
between RANBP9 polymorphisms and schizophrenia, a case-control
association analysis with 449 schizophrenia patients and 393
healthy subjects was conducted. Table
II provides a summary of the clinical profiles in this study.
Although the mean age of the control subjects was slightly higher
than that of the experimental group, the range of ages was similar
in the two groups (Table II). The
percentage of males in both groups was also similar (males in case
vs. control groups, 55.7% vs. 56.5% respectively). Among the case
subjects, 125 schizophrenia patients were measured for Ln S/N
ratios of SPEM abnormality using an eye-tracking task. The patients
were categorized into ‘good’ and ‘poor’ performers, according to
their SPEM function, based on whether their Ln S/N ratio was above
or below 3.97. The Ln S/N ratios (mean ± standard deviation) of the
good and poor SPEM function groups were 4.32±0.30 and 3.23±0.65,
respectively. A significant difference between good and poor groups
(P<0.001) was observed (Table
II). Association analysis of SPEM function abnormality in
normal controls was not performed due to the low incidence of SPEM
impairment in healthy subjects (<10%) and association analysis
could not retain adequate statistical power due to the low number
of samples.
 | Table IISummary of clinical profiles. |
Table II
Summary of clinical profiles.
| Parameters | Schizophrenia
patients | Healthy controls |
|---|
| Subjects, n | 449 | 393 |
| Average age, years
(range) | 44.92 (23–76) | 54.62 (28–80) |
| Gender, n
(male/female) | 250/199 | 222/171 |
|
| |
| Poor (<3.97) | Good (≥3.97) | |
|
| |
| Subjects, n | 58 | 67 | |
| Average age,
years | 46.0 ± 9.18 | 44.2 ± 9.27 | |
| Ln S/N
ratio* | 3.23 ± 0.65 | 4.32 ± 0.30 | |
Genotyping of RANBP9 polymorphisms
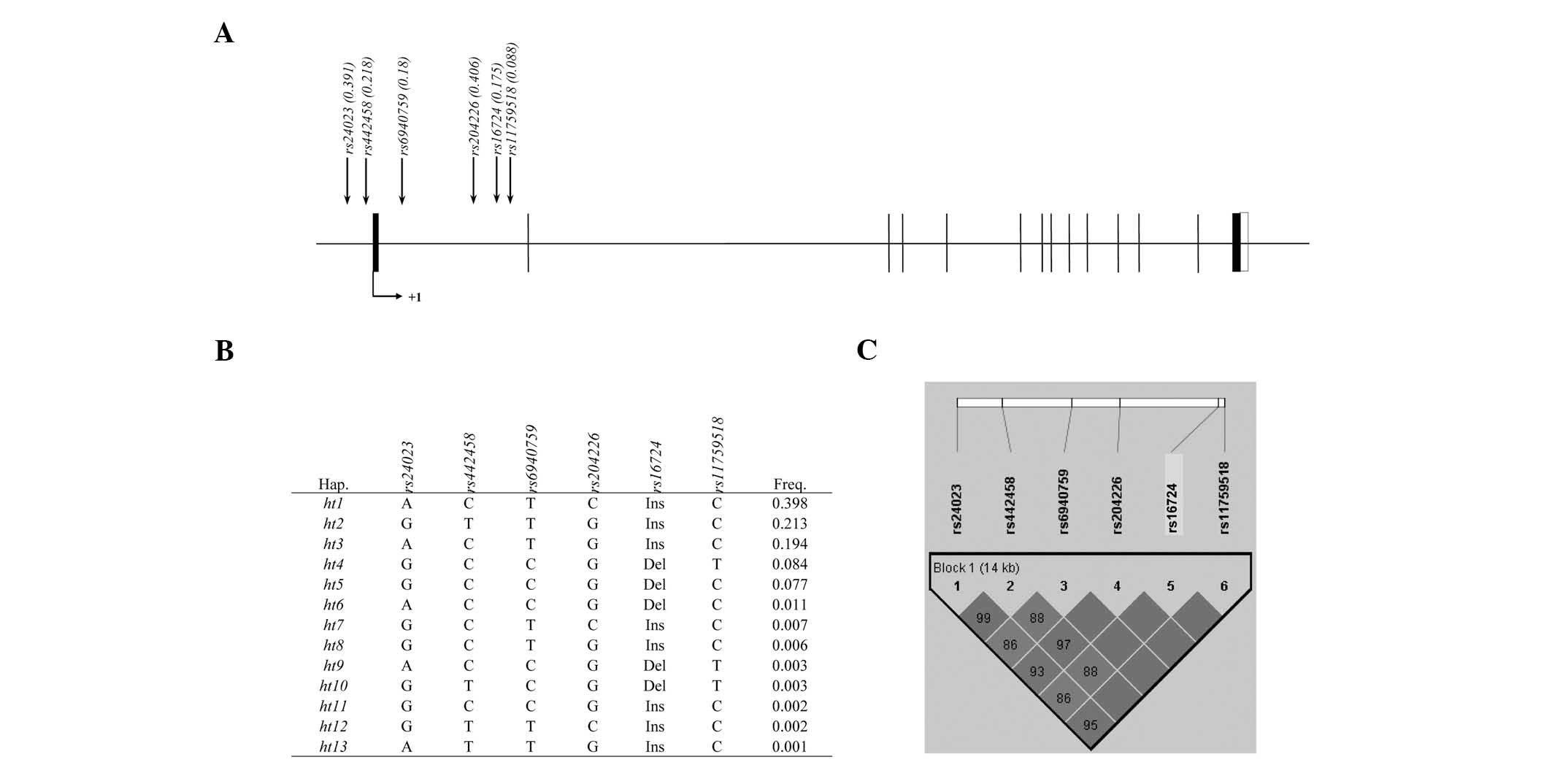
All RANBP9 polymorphisms were successfully genotyped
in a total of 842 subjects; two variations were localized in the
promoter (rs24023 and rs442458) and the other four variants in the
intron (rs6940759, rs204226, rs16724 and rs11759518) of the gene
(Fig. 1A). No significant
deviations from HWE were observed in any of the polymorphisms
(P>0.05). Based on the results of a pair-wise comparison of the
variations, a total of 13 haplotypes were inferred (Fig. 1B). Of these, five common haplotypes
with frequencies >0.05 were identified.
Association analysis of RANBP9
polymorphisms with schizophrenia and SPEM abnormality
Allele frequencies of six polymorphisms were
compared between schizophrenia patients and healthy controls using
logistic analysis, controlling for age as a covariate. However, no
associations between RANBP9 variations and schizophrenia were
identified (Table III). In the
analysis of SPEM abnormality, no significant association was
revealed following the logistic analysis of RANBP9 polymorphisms
and haplotypes (Table IV).
 | Table IIIAssociation analysis of RANBP9
variations with schizophrenia in a Korean population. |
Table III
Association analysis of RANBP9
variations with schizophrenia in a Korean population.
| MAF | | | |
|---|
|
| | | |
|---|
| Loci | Schizophrenia
(n=449) | Control
(n=393) | OR (95% CI) | P-value | Statistical
powera |
|---|
| rs24023 | 0.389 | 0.394 | 1.01
(0.81–1.26) | 0.94 | 95.04 |
| rs442458 | 0.219 | 0.217 | 1.08
(0.83–1.41) | 0.56 | 87.56 |
| rs6940759 | 0.179 | 0.18 | 0.98
(0.74–1.30) | 0.89 | 83.04 |
| rs204226 | 0.422 | 0.388 | 1.14
(0.91–1.42) | 0.25 | 94.97 |
| rs16724 | 0.174 | 0.177 | 0.96
(0.72–1.27) | 0.76 | 82.58 |
| rs11759518 | 0.089 | 0.087 | 1.04
(0.71–1.53) | 0.84 | 58.15 |
| ht1 | 0.417 | 0.377 | 1.16
(0.93–1.45) | 0.19 | 94.81 |
| ht2 | 0.213 | 0.213 | 1.06
(0.81–1.38) | 0.68 | 87.16 |
| ht3 | 0.177 | 0.213 | 0.77
(0.58–1.03) | 0.08 | 87.16 |
| ht4 | 0.084 | 0.083 | 1.04
(0.70–1.55) | 0.86 | 56.40 |
| ht5 | 0.073 | 0.081 | 0.89
(0.59–1.34) | 0.57 | 55.50 |
 | Table IVAssociation analysis of RAN binding
protein 9 variations with smooth pursuit eye movement
abnormality. |
Table IV
Association analysis of RAN binding
protein 9 variations with smooth pursuit eye movement
abnormality.
| MAF | | |
|---|
|
| | |
|---|
| Loci | Poor (<3.97,
n=58) | Good (≥3.97,
n=67) | OR (95% CI) | P-value |
|---|
| rs24023 | 0.439 | 0.368 | 1.28
(0.78–2.10) | 0.33 |
| rs442458 | 0.239 | 0.193 | 1.14
(0.62–2.08) | 0.67 |
| rs6940759 | 0.206 | 0.202 | 1.15
(0.65–2.04) | 0.62 |
| rs204226 | 0.425 | 0.421 | 0.99
(0.61–1.61) | 0.98 |
| rs16724 | 0.206 | 0.202 | 1.15
(0.65–2.04) | 0.62 |
| rs11759518 | 0.108 | 0.088 | 1.46
(0.60–3.54) | 0.41 |
| ht1 | 0.396 | 0.421 | 0.90
(0.56–1.46) | 0.67 |
| ht2 | 0.209 | 0.193 | 0.98
(0.52–1.84) | 0.95 |
| ht3 | 0.157 | 0.184 | 0.85
(0.44–1.62) | 0.62 |
| ht4 | 0.104 | 0.088 | 1.45
(0.59–3.54) | 0.41 |
| ht5 | 0.075 | 0.088 | 0.91
(0.34–2.41) | 0.84 |
Discussion
RANBP9, also known as RanBPM, was first reported in
2002 as a protein that interacts with the MET proto-oncogene
(21). In previous studies, RANBP9
was revealed to interact with other important molecules such as
amyloid precursor protein (APP) and protein kinase C (PKC) δ and γ
(16,22). The two kinases have been reported
to be associated with mental disorders. Missense mutations in APP
have been shown to be associated with cerebral hemorrhage,
multi-infarct dementia and Alzheimer’s disease, but not directly
with schizophrenia (23). However,
a recent study has reported the defective metabolism of APP in
older schizophrenia patients (age, >70), suggesting that APP may
be involved in schizophrenia etiology (24). PKC is known to phosphorylate the
dopamine D1 receptor and negatively regulate dopaminergic signaling
(12). A defect in dopamine
signaling has been associated with cognitive disorders such as
alcohol abuse and schizophrenia and dopamine D1 receptor antagonist
SKF-38393 treatment was revealed to improve the cognitive function
in animal models with cognitive deficits similar to schizophrenia
(25).
Despite the potentially critical role of RANBP9 in
mental disorders as previously demonstrated, relatively few studies
have been conducted investigating this gene. In a previous study,
RANBP9 was observed to be highly expressed in the neurons of the
cerebral cortex and the cerebral Purkinje cells in a similar
pattern to fragile X mental retardation protein (FMRP) (26). FMRP is known to cause fragile X
syndrome, a common form of hereditary mental retardation. The
authors reported that RANBP9 was closely associated with the
disorder, and was found to modulate the RNA binding properties of
FMRP. In another study, researchers revealed that the concentration
of RANBP9-N60, a processed form of RANBP9 almost identical to the
RANBP9-Delta1/N60 mutant, was increased in the brain cells of
Alzheimer’s patients (22). In the
study, the authors demonstrated that RANBP9-N60 increased the
amyloid β generation rate by more than five-fold and therefore
drove the amyloid cascade in Alzheimer’s disease. The authors
hypothesized that the proteolytic processing of RANBP9 may be an
ideal therapeutic target.
Although several reports have suggested a possible
association between RANBP9 and schizophrenia, the results presented
in the current study demonstrated that the genetic variants of
RANBP9 were not associated with schizophrenia and SPEM abnormality
in a Korean population. However, the present study is the first to
conduct association analyses between RANBP9 polymorphisms and
schizophrenia. To the best of our knowledge, although, an
association study of RANBP9 variants with cognitive disorders such
as schizophrenia has not been previously conducted, genetic
variants of RANBP1 were recently investigated by our group for
possible associations with schizophrenia and SPEM abnormality in a
Korean population (27). In the
study, two polymorphisms of RANBP1, rs2238798 and rs175162, were
genotyped for association analysis. The results indicated that,
while the two variations did not exhibit any association with
schizophrenia, the haplotype rs2238978G-rs175162T was significantly
associated with the risk of SPEM abnormality. According to these
results, although RANBP1 and RANBP9 have a similar role, their
effects on SPEM abnormality vary. While the exact functions of the
proteins are not fully understood, the most prominent difference
between the two is that RANBP1 binds specifically with
guanosine-5′-triphosphate (GTP)-charged RAN and increases GTP
hydrolysis (28). This difference
may account for the association between RANBP1 and SPEM
abnormality, however further studies would be required to confirm
this hypothesis.
To the best of our knowledge, while the present
study is the first to investigate the possible association of
RANBP9 variants with schizophrenia and SPEM abnormality, certain
limitations are apparent. Due to the rarity of the disease, the
number of patients in the study may not have been large enough to
generate adequate statistical power. Moreover, the number of
schizophrenia patients who were able to test for SPEM was also
small. Despite this, the method used to measure SPEM function is
used worldwide and is adequate for a genetic association analysis.
However, a larger number patient population may be required to
confirm the results of the present study. Furthermore, a functional
study comparing and contrasting the roles of RANBP1 and RANBP9 may
be crucial for deducing the causal variation in SPEM
abnormality.
In conclusion, association analyses between RANBP9
variants and the risk of schizophrenia were conducted, however no
significant association was identified. Logistic analyses were also
conducted to search for a possible association between RANBP9
variants and SPEM abnormality. However, no significant difference
was identified. As RANBP9 was only recently discovered, the exact
mechanism and function of the protein is not fully understood.
Therefore, despite the lack of positive results, the present study
may provide a meaningful insight into the role of RANBP9. Further
studies comparing a larger number of samples may be beneficial to
confirm the results of the present study, as well as elucidate the
precise differences between RANBP1 and RANBP9, which may be
critical in determining the exact cause of SPEM abnormality.
Acknowledgements
This study was supported by a grant from the Korea
Healthcare Technology R and D Project, Ministry of Health and
Welfare, Republic of Korea (grant no. A101023), The National
Research Foundation of Korea grant, funded by the Korean government
(grant no. 2011-0011935) and the Ministry of Education, Science and
Technology, Republic of Korea (grant no. 2009-0093822).
References
|
1
|
McGrath J, Saha S, Chant D and Welham J:
Schizophrenia: a concise overview of incidence, prevalence, and
mortality. Epidemiol Rev. 30:67–76. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torrey EF: Prevalence studies in
schizophrenia. Br J Psychiatry. 150:598–608. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Park HJ, Westin CF, Kubicki M, et al:
White matter hemisphere asymmetries in healthy subjects and in
schizophrenia: a diffusion tensor MRI study. Neuroimage.
23:213–223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hof PR, Haroutunian V, Friedrich VL Jr, et
al: Loss and altered spatial distribution of oligodendrocytes in
the superior frontal gyrus in schizophrenia. Biol Psychiatry.
53:1075–1085. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hulshoff Pol HE, Schnack HG, Mandl RC, et
al: Focal white matter density changes in schizophrenia: reduced
inter-hemispheric connectivity. Neuroimage. 21:27–35. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kubicki M, McCarley RW and Shenton ME:
Evidence for white matter abnormalities in schizophrenia. Curr Opin
Psychiatry. 18:121–134. 2005. View Article : Google Scholar
|
|
7
|
Uranova N, Orlovskaya D, Vikhreva O, et
al: Electron microscopy of oligodendroglia in severe mental
illness. Brain Res Bull. 55:597–610. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Uranova NA, Vostrikov VM, Orlovskaya DD
and Rachmanova VI: Oligodendroglial density in the prefrontal
cortex in schizophrenia and mood disorders: a study from the
Stanley Neuropathology Consortium. Schizophr Res. 67:269–275. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mitterauer B: The incoherence hypothesis
of schizophrenia: based on decomposed oligodendrocyte-axonic
relations. Med Hypotheses. 69:1299–1304. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murrin LC and Talbot JN: RanBPM, a
scaffolding protein in the immune and nervous systems. J
Neuroimmune Pharmacol. 2:290–295. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nakayama AY, Harms MB and Luo L: Small
GTPases Rac and Rho in the maintenance of dendritic spines and
branches in hippocampal pyramidal neurons. J Neurosci.
20:5329–5338. 2000.PubMed/NCBI
|
|
12
|
Rex EB, Rankin ML, Ariano MA and Sibley
DR: Ethanol regulation of D(1) dopamine receptor signaling is
mediated by protein kinase C in an isozyme-specific manner.
Neuropsychopharmacology. 33:2900–2911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kutay U, Hartmann E, Treichel N, et al:
Identification of two novel RanGTP-binding proteins belonging to
the importin beta superfamily. J Biol Chem. 275:40163–40168. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blackwood DH, Fordyce A, Walker MT, St
Clair DM, Porteous DJ and Muir WJ: Schizophrenia and affective
disorders--cosegregation with a translocation at chromosome 1q42
that directly disrupts brain-expressed genes: clinical and P300
findings in a family. Am J Hum Genet. 69:428–433. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Millar JK, Christie S, Anderson S, et al:
Genomic structure and localisation within a linkage hotspot of
Disrupted In Schizophrenia 1, a gene disrupted by a translocation
segregating with schizophrenia. Mol Psychiatry. 6:173–178. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rex EB, Rankin ML, Yang Y, et al:
Identification of RanBP 9/10 as interacting partners for protein
kinase C (PKC) gamma/delta and the D1 dopamine receptor: regulation
of PKC-mediated receptor phosphorylation. Mol Pharmacol. 78:69–80.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
American Psychiatric Association.
Diagnostic and Statistical Manual of Mental Disorders. 4th edition.
American Psychiatric Association; Washington, D.C., WA: 1994
|
|
18
|
Park BL, Shin HD, Cheong HS, et al:
Association analysis of COMT polymorphisms with schizophrenia and
smooth pursuit eye movement abnormality. J Hum Genet. 54:709–712.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stephens M, Smith NJ and Donnelly P: A new
statistical method for haplotype reconstruction from population
data. Am J Hum Genet. 68:978–989. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar
|
|
21
|
Wang D, Li Z, Messing EM and Wu G:
Activation of Ras/Erk pathway by a novel MET-interacting protein
RanBPM. J Biol Chem. 277:36216–36222. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lakshmana MK, Hayes CD, Bennett SP, et al:
Role of RanBP9 on amyloidogenic processing of APP and synaptic
protein levels in the mouse brain. FASEB J. 26:2072–2083. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Morris S, Leung J, Sharp C, Blackwood D,
Muir W and St Clair D: Screening schizophrenic patients for
mutations in the amyloid precursor protein gene. Psychiatr Genet.
4:23–27. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Albertini V, Benussi L, Paterlini A, et
al: Distinct cerebrospinal fluid amyloid-beta peptide signatures in
cognitive decline associated with Alzheimer’s disease and
schizophrenia. Electrophoresis. 33:3738–3744. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McLean SL, Idris NF, Woolley ML and Neill
JC: D(1)-like receptor activation improves PCP-induced cognitive
deficits in animal models: Implications for mechanisms of improved
cognitive function in schizophrenia. Eur Neuropsychopharmacol.
19:440–450. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Menon RP, Gibson TJ and Pastore A: The C
terminus of fragile X mental retardation protein interacts with the
multi-domain Ran-binding protein in the microtubule-organising
centre. J Mol Biol. 343:43–53. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheong HS, Park BL, Kim EM, et al:
Association of RANBP1 haplotype with smooth pursuit eye movement
abnormality. Am J Med Genet B Neuropsychiatr Genet. 156B:67–71.
2011. View Article : Google Scholar
|
|
28
|
Kehlenbach RH, Dickmanns A, Kehlenbach A,
Guan T and Gerace L: A role for RanBP1 in the release of CRM1 from
the nuclear pore complex in a terminal step of nuclear export. J
Cell Biol. 145:645–657. 1999. View Article : Google Scholar : PubMed/NCBI
|