Introduction
Breast cancer is one of the most common types of
cancer, accounting for 13.7% of cancer-related mortalities in 2008
according to the World Cancer Research cancer report (1). Several factors have a key role in
breast carcinogenesis, including ovarian steroids, estrogen and
progesterone. Breast tumors are usually screened for hormone
receptors, for example estrogen receptor (ER), progesterone
receptor (PR) and overexpression of HER2, and based on the
expression levels, patients are directed to endocrine or
chemotherapy. It has been determined that the failure of cancer
treatment is due to the persistence of cancer stem cells (CSCs)
that evade the treatment regimen and are responsible for minimal
residual disease (2). These cells
have been found to possess characteristics usually associated with
stem cells, such as self-renewal, and exhibit a high in vivo
tumorigenicity, differentiation potential multi-drug and apoptosis
resistance (3). Cells that exclude
Hoechst 33342 dye are referred to as side population (SP) cells.
These cells share characteristics with CSCs; specifically they are
enriched for tumor initiating capacity, express stem-like genes and
are resistant to chemotherapeutic drugs. Furthermore, SP cells also
overexpress the adenosine triphosphatase binding cassette (ABC)
transporters ABCB1 [multi-drug resistance transporter 1 (MDR1)],
ABCC1 and ABCG2 (BCRP1), which contribute to multi-drug resistance
and express stem cell surface markers. SP cells have been
identified in several solid tumors and cancer cell lines based on
Hoechst 33342 dye efflux by fluorescence activated cell sorting
(FACS) (4–6). SP cells have been isolated from
malignant and non-malignant tissues and they have been shown to
exhibit stem cell characteristics and drug resistance (7–9). It
has been reported that the presence of SP cells in mammary gland
tissue of breast cancer accounts for 2.0–3.0% of total epithelial
cells in mice (8) and 2.0–5.0% in
humans (10). These SP cells have
overexpression of ABC transporters, including ABCG2 and ABCB1
(MDR1) (11), and a high
expression level of stem cell surface markers, which contribute to
multi-drug resistance, high proliferation and survival rate,
respectively (7). Hence, the
sorting and characterization of SP cells would assist in
elucidating the mechanisms of oncogenesis and drug resistance
(8) and aid in the design of novel
therapeutic strategies that may selectively target CSCs.
Consequently, the present study is designed to isolate SP cells in
the MDU-22 breast cancer cell line and analyze their expression
levels of ABC transporters and stem cell surface markers.
Materials and methods
Cell line and cell culture
Samples from breast cancer tissues were obtained
from a range of patients at the time of surgery. Written informed
consent was obtained from all participants. All the patients were
recruited from the Jiangxi Provincial Tumor Hospital (Jiangxi,
China). Ethical approval was obtained from the Jiangxi Provincial
Tumor Hospital. The patients details were as follows: Age range,
33–43 years; region, ductal mammary gland; grade, 4 (recurrent
type). The tumor samples were washed immediately and tissues were
broken down using collagenase (Sigma-Aldrich, Shanghai, China). The
cells were continuously cultured in Dulbecco’s modified Eagle’s
medium (DMEM; Sigma-Aldrich) and a cell line, MDU-22, was
established. The cell line was cultured in DMEM with 10% fetal
bovine serum (Life Technologies, Shanghai, China), supplemented
with antibiotics (penicillin and streptomycin; Sigma-Aldrich) and
maintained in T-75 flasks at 37°C in a humidified 5% CO2
and 95% air atmosphere. Once confluent, cells were removed from the
culture flask using Trypsin-EDTA (0.25%, 53 mM EDTA) washed, cells
suspended in 10% DMEM and centrifuged at 6,200 × g for 6 min. Cells
were resuspended in 10% DMEM. Cell count was measured using a
hemocytometer (Bio-Rad, Inc., Shanghai, China).
Labeling with Hoechst 33342
All reagents mentioned in this paragraph were
obtained from Sigma-Aldrich, with the exception of those already
specified. The cells were divided into two groups, control cells +
Hoechst 33342 (n=4) and verapamil treated cells + verapamil +
Hoechst 33342 (n=4). Cells in staining medium (~106
cells/ml of 10% DMEM) were labeled with Hoechst 33342 stock
(Sigma-Aldrich) and bis-benzimide (5 μl/ml), either with dye
alone or in combination with verapamil (0.8 μl/ml). The
cells were mixed and placed in water bath at 37°C for exactly 90
min. Subsequently, the cells were spun down (2,000 rpm for 10 min
at 4°C) and resuspended in 500 μl of Hank’s buffered salt
solution containing 10 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid. Finally, the
cells were counter stained with propidium iodide (PI; 2
μg/ml sample) at 4°C. The cells were filtered through a 50
μm nylon mesh (BD Biosciences, Franklin Lakes, NJ, USA) into
labeled FACS tubes in order to remove cell clumps. Separate tubes
with fresh medium (10% DMEM) were kept for sterile sorting of SP
cells and main population cells. The cells were sorted using a
FACSAria II flow cytom eter (BD Biosciences, Mountain View, CA,
USA). A wavelength of 355 nm was used to excite the Hoechst 33342
dye, and its dual-wavelength fluorescence was analyzed (blue, 450
nm; red, 675 nm).
In vitro proliferation activity
assay
The sorted SP and non-SP cells were seeded into a
96-well plate at a density of 2×106 cells/well and
cultured in a CO2 incubator. Each group was set up in
triplicate. Cell proliferation activity was measured every day for
7 days. Each well was supplemented with CCK-8 solution (10
μl) and incubated in CO2 incubator for 2–3 h. The
optical density (OD) was determined at a wavelength of 450 nm.
These data were used to calculate cell growth graphs based on the
mean value of OD450 and standard deviation values for
each well.
Cell resistance assay
The obtained SP and non-SP cells were cultured in
96-well plates at a concentration of 1×103 cells/plate.
After 24 h, 5-fluorouracil (5-FU) was added to all cultures to a
final concentration of 10 μg/ml. The plates were placed in a
hatch box for 48 h. Each well was supplemented with CCK-8 (10
μl) solution and the plates were incubated for 3 h. The mean
value of OD450 obtained was represented as a graph. Cell
resistance in the two groups was calculated using the following
formula: Cell resistance (%) = (experimental group OD450
value/control group OD450 value) × 100.
Immunocytochemistry
The sorted SP cells and non-SP cells were seeded
into 35 mm culture plates (~100 μl), maintained in an
incubator for 3 h and supplemented with 1 ml DMEM (10%). Following
overnight incubation, the cells were rinsed with phosphate-buffered
saline (PBS; Life Technologies) and fixed in 4% paraformaldehyde
(Life Technologies) in 1X PBS, for 5 min at 4°C. After washing with
1X PBS, cells were blocked with 1% bovine serum albumin in
Tris-buffered saline (BSA-TBS; Life Technologies) with RNase (10
μl per 1,000 μl of 3% BSA-TBS; Life Technologies).
Following a 1-h incubation at room temperature the cells were
rinsed with PBS, and FAK primary mouse polyclonal antibody (Life
Technologies) in 1% BSA-TBS was added (dilution, 1:100; 2/200
μl) prior to incubation overnight at 4°C. Once washed with
1X PBS, the cells were incubated with Rabbit Anti-Mouse IgG (H+L)
Superclonal™ Secondary antibody (dilution, 1:100 in 1% BSA-TBS), at
room temperature for 1 h. The cells underwent a further PBS wash
and PI was added (1/200 μl of PBS). The cells were viewed
under a confocal laser scanning microscope (Leica TCS; Leica
Microsystems, Wetzlar, Germany). Image analysis and figures were
prepared using Adobe Photoshop CS4 (Adobe Systems, Inc., San Jose,
CA, USA).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
PCR was carried out as previously described
(13). Total RNA was isolated from
the SP and non-SP cells using the Ambion RNAqueous®
Micro kit (Applied Biosystems, Warrington, UK). cDNA was
synthesized using the Bioline cDNA synthesis kit (Bioline, London,
UK). qPCR was performed using 2–3 μl cDNA and 2X TaqMan Gene
Expression Master mix (Applied Biosystems) in 30 μl reaction
volumes. GAPDH was used as a reference. The primer sequences were
as follows (14): forward, 5′-AGC
TGC AAG GAA AGA TCC AA-3′ and reverse, 5′-TCC AGA CAC ACC ACG GAT
AA-3′ for ABCG2; forward, 5′-ATC CTG GGG GTT CTA TTT GG-3′ and
reverse, 5′-CTC CAG GTT GCC TCT CAC TC-3′ for OCT-4; forward,
5′-CTG CCA AAT GTT TGG TGA TG-3′ and reverse, 5′-ACG CGT TGT GAT
CTC CTT CT-3′, for epithelial cell adhesion molecule (EpCAM); and
forward, 5′-ATG TCG TGG AGT CTA CTG GC-3′ and reverse, 5′-TGA CCT
TGC CCA CAG CCT TG-3′ for GAPDH. The amplified products were
separated by electrophoresis on ethidium bromide-stained 1.2%
agarose gels. Band intensity was measured by ImageJ (National
Institutes of Health, Bethesda, MD, USA) from two independent
experiments.
Statistical analysis
A one-way analysis of variance test was performed to
determine any differences between the treatment groups. Student’s
t-tests were performed to compare the effect of different
treatments between the SP and non-SP populations.
*P<0.05 and **P<0.01 were considered to
indicate a statistically significant difference.
Results
Analysis of SP Cells FACS using Hoechst
33342
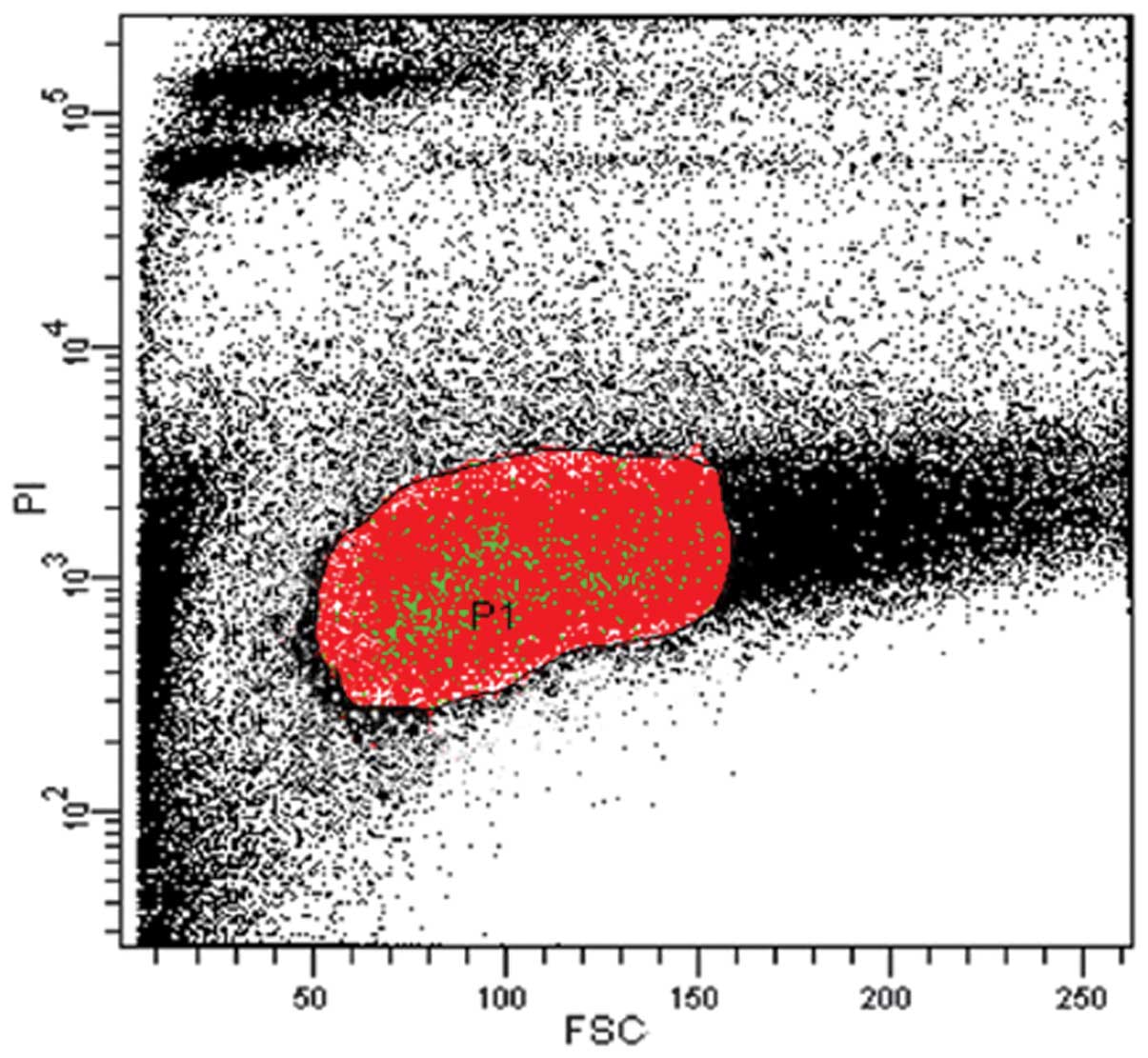
The live cell population (P1 gated region) was
selected against PI as the P1 gated population. PI is used to
exclude the dead cells from the sample (Fig. 1). SP cells were sorted out from the
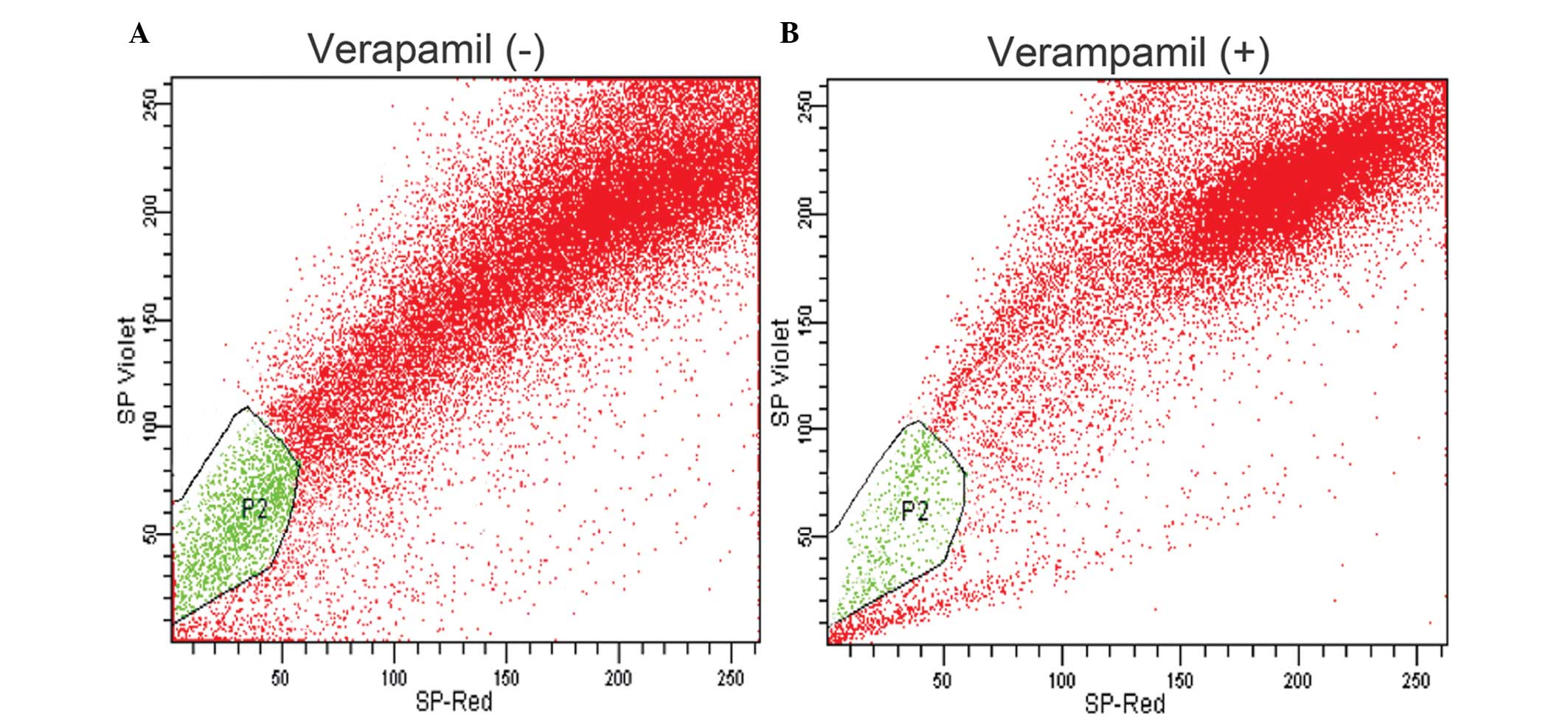
gated live cell population (P1) using Hoechst 33342, which is a DNA
binding dye. Hoechst 33342 emits light at wavelengths of ~450 nm
(SP-Violet) and 675 nm (SP-Red) following UV excitation, and a set
of cells were observed that displayed low blue and red
fluorescence. This distinct cell population (P2) found towards the
SP-violet region of the dot plot of the FACS profile are the
so-called SP cells (Fig. 2A). The
exclusion of Hoechst 33342 by SP cells is an active process
involving MDR1, a member of the ABC transporter transmembrane
proteins (Fig. 2A). The number of
cells collected was ~62 × 103, which is ~56% of the
initial cell count. Of the 62 × 103 cells (P1) analyzed,
Hoechst dye was effluxed by 3.8% of cells in the P2 gated region
(Fig. 2A). Verapamil-treated cells
of the same cell line were sorted out with the same Hoechst 33342
efflux. The resulting P1 cells obtained were ~48,800 in number,
which was ~44% of the initial cell count. Following treatment with
verapamil, the percentage of SP cells (P2 gated) was reduced from
3.8 to 0.6% (Fig 2B). Verapamil is
a MDR1 transporter protein inhibitor, which blocks the drug efflux
action by the cells. Hence, the sorted SP cells are highly
resistant to drug uptake, which may be due to the over expression
of ABC transporters. Therefore, these cells were further analyzed
for the expression of ABC transporters and stem cell surface
markers.
Analysis of ABCG2 expression and stem
cell surface markers in SP cells
It has been previously reported that ABCG2 is
expressed by malignant breast tissue, and it was observed that
ABCG2 expression increases with higher tumor grades (15,6).
Therefore, to examine and compare the expression level of the
ABCB2 gene and other stem cell genes, including OCT-4
and EpCAM, between SP and non-SP cells, the extracted RNA
was analyzed using RT-qPCR. The ABCB2, EpCAM and
OCT-4 levels were higher in SP cells compared to non-SP
cells (Fig. 3A). The
quantification graph clearly shows that the levels of these genes
in SP cells are significantly higher (Fig. 3B). GAPDH was used as a reference
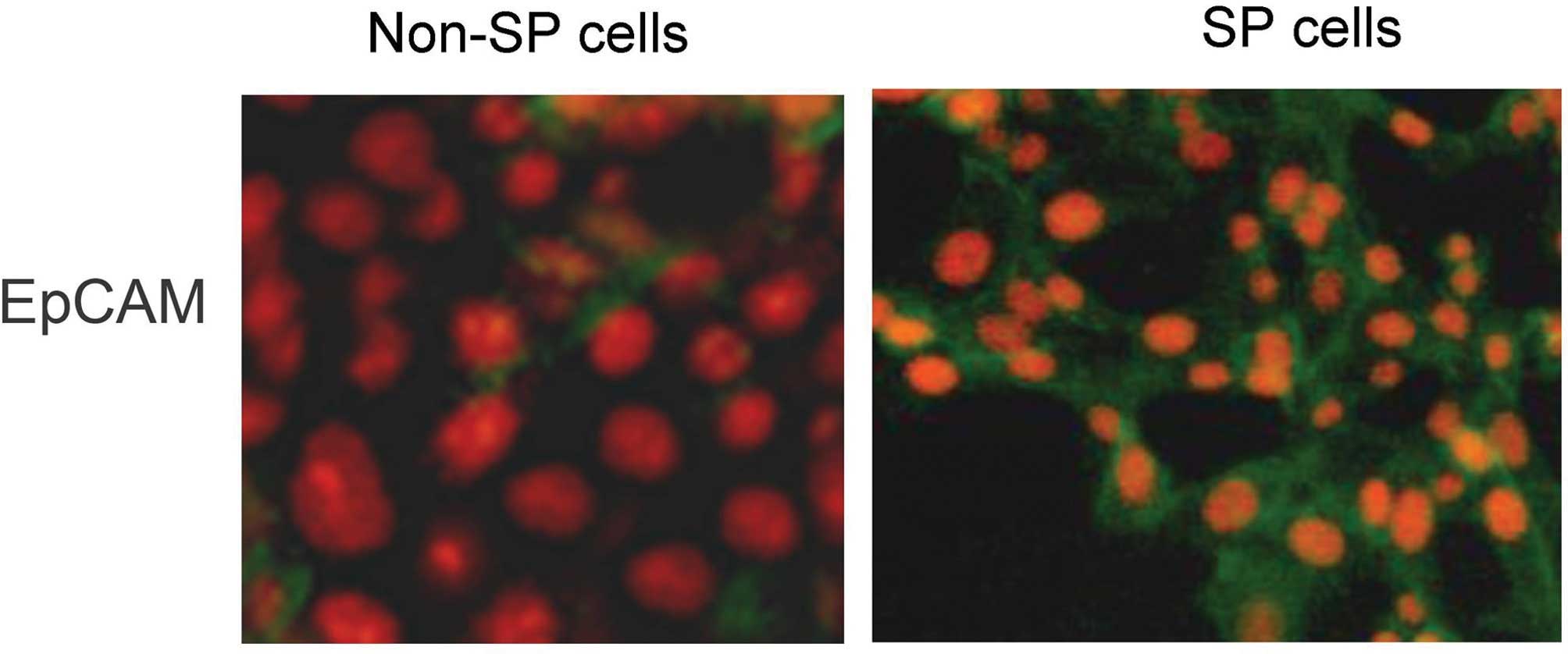
gene. Notably, fluorescence microscopic analysis revealed that,
compared with the non-SP cells, the SP cells displayed a greater
positive expression of cell surface proteins such as EpCAM
(Fig. 4) which further confirms
the RT-qPCR results. These results suggest that a high expression
level of ABCG2 and OCT-4 in SP cells may act as a
crucial factor in drug resistance and the massive proliferation of
cancer cells.
Characterization of sorted SP cells
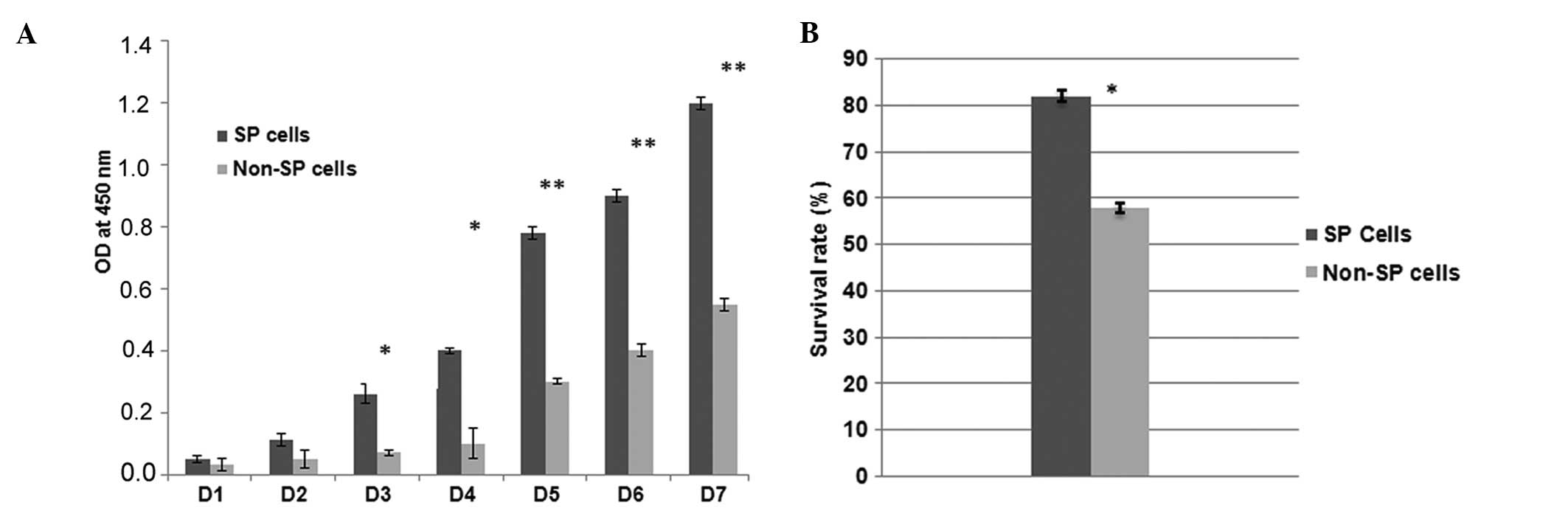
FACS sorted SP and non-SP cells were further
subjected to in vitro cell proliferation and cell survival
assays. The isolated breast carcinoma SP cells underwent rapid cell
proliferation starting from the third day and becoming more
confluent on the eighth day (data not shown) (Fig. 5A). However, the growth rates of the
non-SP cells were significantly lower when compared with those of
the SP cells (Fig. 5A). The
measured growth rate of SP cells at 450 nm was significantly higher
than non-SP cells (*P<0.05; **P<0.01).
Following the in vitro proliferation assay, the SP cells
were further subjected to a drug resistance assay. The survival
rate of the SP cells (83%) following exposure to 10 μg/ml
5-FU were significantly higher (Fig.
5B) compared with that of the non-SP cells (58%). Hence these
data suggest that the sorted SP cells have high proliferation rate
and an increased resistance to chemotherapeutic agent.
Discussion
CSCs have the ability to resist chemotherapeutic
agents via increased expression levels of efflux pumps, and they
have an enhanced DNA repair activity. The theory of cancer stem
cells hypothesizes that treatment failure and minimal residual
disease are caused by the presence of small population of CSCs
(7,8), which are responsible for tumor
growth, metastasis and tumor relapse. Hence the eradication of CSCs
is an important goal for providing effective cancer treatment and
long term disease-free survival.
SP cells have been observed in several types of
solid tumors as well as a number of breast cancer cell lines
(7,9). SP cells were first identified in
primary human breast cancer cells, and they demonstrated the
expression of HER2 signaling which is involved in drug
efflux via ABC transporters (16).
In the present study, we demonstrated that SP cells was identified
and isolated from breast carcinoma cell line (MDU-22). SP cells
were sorted out based on Hoechst 33342 dye exclusion by FACS. The
exclusion of Hoechst 33342 by SP cells is due to highly induced
ABCG2 (MDR1), a member of the ABC transporter transmembrane
proteins that are involved in multi-drug resistance (Fig. 2A). The results of the current study
demonstrated that, following treatment with verapamil (a MDR1
protein inhibitor), the percentage of SP cells was reduced from 3.8
to 0.6% (Fig. 2B). This is a
confirmatory test for the presence of MDR1 protein in MDU-22 cells.
Furthermore, the results of the RT-qPCR showed that the expression
levels of genes such as ABCG2, OCT-4 and EpCAM
are significantly higher in SP cells than those in the non-SP
population (Fig. 3). A previous
study has shown that the ABCG2 protein is over expressed in the
MCF-7 cell line, and that these cells were more drug resistant
compared with the MCF-7 non-SP cells (6). Therefore, the results of the current
study fit well with the recent study by Britton et al
(6), 2012 and with other previous
studies (9,17) where a differential increase in the
expression levels of mRNA encoding ABCG2 was observed in the
SP population when compared with that of the non-SP cells.
Similarly, the OCT-4 gene, a member of the POU family of
transcription factors, was shown to be involved in the
proliferation potential (18) and
survival of CSCs partly through the OCT-4/Tcl1/Akt1 pathway
(19). Hence the results of the
current study indicate that the overexpression of ABCG2 may act as
a suitable marker for the identification of SP cells in breast
cancers. Subsequently, the results revealed that SP cells displayed
a greater positive expression of cell surface markers such as
epithelial cell adhesion molecule (EpCAM), a CSC marker (Fig. 4). In line with the present results,
CD44+ cells in the head and neck squamous cell
carcinomas of mice were proven to be enriched with tumorigenic CSCs
able to propagate tumor formation, whereas CD44− cells
were not able to form tumors (5).
Furthermore, the in vitro proliferation and
drug resistance assays revealed that MDU-22 SP cells have an
increased proliferation capacity and are highly resistant to 5-FU,
hence they have increased survival rate compared with that of the
non-SP cells, which are very sensitive (Fig. 4). It has been previously reported
that MDA-MB-231 cell lines have a significantly increased survival
rate when treated with doxorubicin, methotrexate and 5-FU, compared
with untreated cells (20). Taken
together, the results of the present study indicate that the sorted
SP cells show elevated expression levels of ABCG2,
OCT-4 and EpCAM, hence, the interactions of these
genes may be collectively involved in the drug resistance and
enhanced survival rate of SP cells. However, the signaling pathways
and cascade of events involved in expression of these genes in CSCs
remains speculative. However, the identification and
characterization of SP cells provides a strategy to design novel
therapeutic targets with the eventual goal of eliminating residual
disease and preventing tumor recurrence.
References
|
1
|
World Cancer Report: International Agency
for Research on Cancer. 2008, Retrieved 2011-02-26. (cancer
statistics often exclude non-melanoma skin cancers such as basal
cell carcinoma which though very common are rarely fatal).
|
|
2
|
Ramachandran C and Melnick SJ: Multidrug
resistance in human tumors - molecular diagnosis and clinical
significance. Mol Diagn. 4:81–94. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gil J, Stembalska A, Pesz KA and Sasiadek
MM: Cancer stem cells: the theory and perspectives in cancer
therapy. J Appl Genet. 49:193–199. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Robey RW, Shukla S, Finley EM, et al:
Inhibition of P-glycoprotein (ABCB1)- and multidrug
resistance-associated protein 1 (ABCC1)-mediated transport by the
orally administered inhibitor, CBT-1((R)). Biochem Pharmacol.
75:1302–1312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yanamoto S, Kawasaki G, Yoshitomi I,
Iwamoto T, Hirata K and Mizuno A: Clinicopathologic significance of
EpCAM expression in squamous cell carcinoma of the tongue and its
possibility as a potential target for tongue cancer gene therapy.
Oral Oncol. 43:869–877. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Britton KM, Eyre R, Harvey IJ, et al:
Breast cancer, side population cells and ABCG2 expression. Cancer
Lett. 323:97–105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hirschmann-Jax C, Foster AE, Wulf GG, et
al: A distinct ‘side population’ of cells with high drug efflux
capacity in human tumor cells. Proc Natl Acad USA. 101:14228–14233.
2004. View Article : Google Scholar
|
|
8
|
Kondo T, Setoguchi T and Taga T:
Persistence of a small subpopulation of cancer stem-like cells in
the C6 glioma cell line. Proc Natl Acad Sci USA. 101:781–786. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Patrawala L, Calhoun T,
Schneider-Broussard R, Zhou J, Claypool K and Tang DG: Side
population is enriched in tumorigenic, stem-like cancer cells,
whereas ABCG2+ and ABCG2− cancer cells are
similarly tumorigenic. Cancer Res. 65:6207–6219. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Welm BE, Tepera SB, Venezia T, et al:
Sca-1(pos) cells in the mouse mammary gland represent an enriched
progenitor cell population. Dev Biol. 245:42–56. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clarke R, Spence K, Anderson E, Howell A,
Okano H and Potten C: A putative human breast stem cell population
is enriched for steroid receptor-positive cells. Dev Biol.
277:443–456. 2005. View Article : Google Scholar
|
|
12
|
Jonker JW, Freeman J, Bolscher E, et al:
Contribution of the ABC transporters Bcrp1 and Mdr1a/1b to the side
population phenotype in mammary gland and bone marrow of mice. Stem
Cells. 23:1059–1065. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yanamoto S1, Kawasaki G, Yamada S,
Yoshitomi I, et al: Isolation and characterization of cancer
stem-like side population cells in human oral cancer cells. Oral
Oncol. 47:855–860. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Britton KM1, Eyre R, Harvey IJ,
Stemke-Hale K, et al: Breast cancer, side population cells and
ABCG2 expression. Cancer Lett. 323:97–105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Faneyte IF, Kristel PM, Maliepaard M,
Scheffer GL, Scheper RJ, Schellens JH and van de Vijver MJ:
Expression of the breast cancer resistance protein n breast cancer.
Clin Cancer Res. 8:1068–1074. 2002.PubMed/NCBI
|
|
16
|
Nakanishi T, Chumsri S, Khakpour N, et al:
Side-population cells in luminal-type breast cancer have
tumour-initiating cell properties, and are regulated by HER2
expression and signaling. Br J Cancer. 102:815–826. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Engelmann K, Shen K and Finn OJ: MCF7 side
population cells with characteristics of cancer stem/progenitor
cells express the tumor antigen MUC1. Cancer Res. 68:2419–2426.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Campbell PA, Perez-Iratxeta C,
Andrade-Navarro MA and Rudnicki MA: Oct4 targets regulatory nodes
to modulate stem cell function. PLoS One. 2:e5532007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu T, Liu S, Breiter DR, Wang F, Tang Y
and Sun S: Octamer 4 small interfering RNA results in cancer stem
cell-like cell apoptosis. Cancer Res. 68:6533–6540. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Steiniger SC1, Coppinger JA, Krüger JA,
Yates J III and Janda KD: Quantitative mass spectrometry identifies
drug targets in cancer stem cell-containing side population. Stem
Cells. 26:3037–3046. 2008. View Article : Google Scholar : PubMed/NCBI
|