Introduction
Glucocorticoid (GCs) steroid hormones are used in
the treatment of diseases, including rheumatoid arthritis, asthma
and various ocular diseases. It has been widely reported that
prolonged treatment with GCs can lead to the formation of posterior
subcapsular cataracts (1–3). Although numerous attempts have been
made to increase understanding of this, the mechanism underlying
GC-induced cataract formation remains to be elucidated (4,5).
GCs have important roles in numerous biological
processes, including regulation of anti-inflammatory activity and
immunosuppressive action (6,7). GCs
exert their effects through binding to GC receptors (GR), which
modulate the expression of target genes (8,9).
Alternatively, GCs have been proposed to act on the lens indirectly
through mechanisms involving oxidative stress and depletion of
glutathione (10,11). Global gene profiling was performed
to analyze novel GC-induced changes in the gene expression of human
lens epithelial cells (LECs) (12). Following this study, pathway
analysis was performed in immortalized and primary human LECs and
the results demonstrated that GC treatment of LECs activated the GR
to modulate the expression of mitogen-activated protein kinase and
phosphatidylinositol-3-kinase/AKT regulators (13).
To improve the understanding of the mechanism
involved in the formation of cataracts, GC’s induction of vascular
barrier function requires elucidation. GCs combine with a
cytoplasmic receptor that alters gene expression in two ways. One
way is dependent on the receptor binding directly to DNA and acts
as a transcription factor (positively or negatively). The other is
dependent on its binding to and interfering with other
transcription factors (14).
Transcription factor p54 is essential for GC-mediated expression of
occludin, claudin-5 and vascular barrier induction, and the p54/PSF
heterodimer may contribute to normal blood-retinal barrier
induction in vivo (15).
Thus, it is necessary to elucidate the transcription factors that
are activated in response to GCs.
The present study aimed to identify differentially
expressed genes (DEGs) and their common transcription factors in
order to gain a novel insight into the mechanism of action of GCs
in LECs.
Materials and methods
Affymetrix microarray data
The transcription profile of GSE3040 was obtained
from the gene expression omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database, which is
based on the GPL96 [HG-U133A] Affymetrix Human Genome U133A Array
(Affymetrix Inc., Santa Clara, CA, USA). There were 12 samples of
human LECs treated with vehicle or dexamethasone (Dex) at 4 and 16
h. At each time period, there were six samples, of which three
samples were treated with vehicle (control group) and three samples
were treated with Dex (Dex group). Freshly isolated human LECs were
obtained from capsulorhexis specimens following surgery, these were
the original cells used in the GEO (12).
Data preprocessing and DEG analysis
The GSE3040 datasets were converted into expression
values and pre-processing, including background correction and
quartile data normalization were performed using the robust
multiarray average algorithm (16)
with default parameters in the R language affy package (http://www.bioconductor.org/) (17,18).
The linear models for microarray analysis (Limma) package in the R
language (www.bioconductor.org/packages/release/bioc/html/limma.html)
(19) were used to identify DEGs
by performing Student’s t-test on the samples. A fold change value
>1 and P<0.05 were selected as the cut-off criteria.
Hierarchical cluster analysis of
DEGs
Gene hierarchical cluster analysis of DEGs was
performed using the Pearson correlation coefficient algorithm
(20) in cluster 3.0 (21).
Functional enrichment analysis of common
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.abcc.Ncifcrf.gov/) (22), a high-throughput and integrated
data-mining environment, analyzes gene lists derived from
high-throughput genomic experiments. After the common DEGs were
selected, DAVID was used to identify over-represented gene ontology
(GO; http://www.geneontology.org/) categories
in biological processes based on the hypergeometric distribution.
The GO terms with a value of P<0.05 were selected as
significantly enriched DEGs.
Transcription factors and binding site
analysis
A transcription factor is a protein, which binds to
specific DNA sequences. The TRANSFAC database comprising
information about transcription factors, target genes and binding
sites has been developed (23).
The TRANSFAC database was used to screen transcription factors and
binding sites on DEGs in response to GCs.
Results
DEG analysis
The publicly available microarray dataset, GSE3040,
was obtained from the GEO database. Student’s t-test was used to
identify genes specifically differentially expressed at 4 and 16 h
with the cut-off criteria of P<0.05 and fold change >1. The
results revealed that 696 and 949 genes at 4 and 16 h,
respectively, exhibited significant differential expression.
Hierarchical cluster analysis of DEGs
between Veh and Dex samples at two time periods
As indicated using hierarchical cluster analysis,
the expression levels of DEGs were markedly increased in Veh
samples compared with that of the Dex group, at 4 and 16 h
(Fig. 1).
Set comparison of DEGs between two time
periods
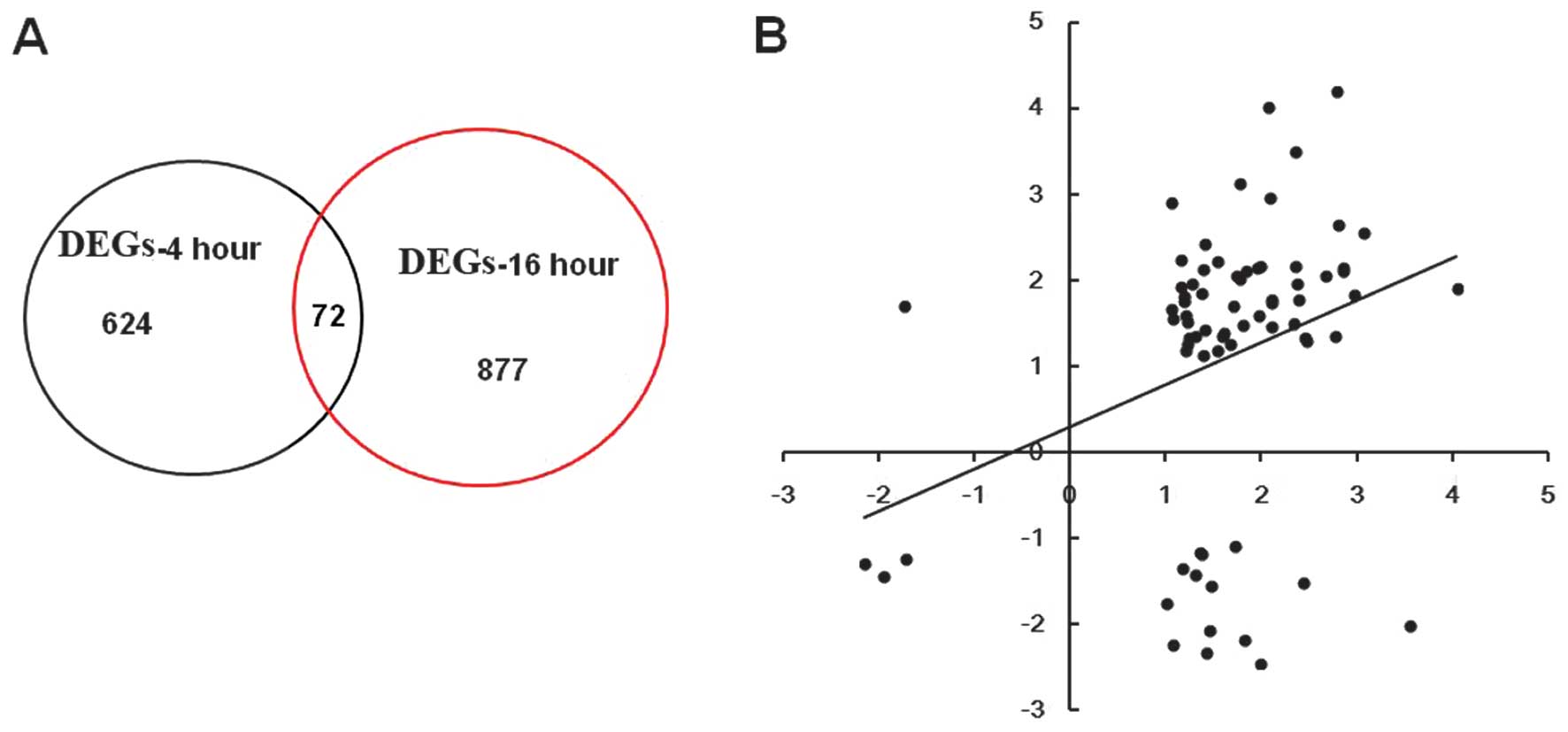
DEGs set at 4 and 16 h were compared and presented
as a Venn diagram (Fig. 2A). There
were 72 common DEGs. The expression folds of 72 DEGs at 4 and 16 h
are shown in Fig. 2B. The results
revealed that the gene expression trend at 4 h was the same as that
at 16 h.
GO enrichment analysis
To gain further insight into the function of genes
in our interaction network, the online biological classification
tool DAVID was used. A total of 13 significant GO function
enrichment nodes were obtained and the distributions of genes is
shown in Fig. 3. As Table I demonstrates, Chemokine (C-C
motif) ligand 2 (CCL2), dual-specificity phosphatase-1 (DUSP1) and
FAS were associated with the function of response to GC stimulus
(P=0.0496906). Expression of CCL2 was downregulated, and DUSP1 and
FAS were upregulated at 4 and 16 h (P<0.05).
 | Table IEnriched Gene Ontology terms of the
common differentially expressed genes at 4 h and 16 h
(P<0.05). |
Table I
Enriched Gene Ontology terms of the
common differentially expressed genes at 4 h and 16 h
(P<0.05).
| Term | Function | Count | P-value | Genes |
|---|
| GO:0048545 | Response to steroid
hormone stimulus | 7 | 0.000240124 | KCNMA1, CCL2, DUSP1,
LEPR, ESR1, FAS, CD24 |
| GO:0009725 | Response to hormone
stimulus | 9 | 0.000255726 | KCNMA1, CCL2, DUSP1,
LEPR, ESR1, FOXC2, FAS, CD24, STAT1 |
| GO:0009719 | Response to
endogenous stimulus | 9 | 0.000494549 | KCNMA1, CCL2, DUSP1,
LEPR, ESR1, FOXC2, FAS, CD24, STAT1 |
| GO:0042981 | Regulation of
apoptosis | 12 | 0.000955092 | KCNMA1, PRUNE2, CCL2,
DUSP1, MCL1, SOS2, ESR1, FOXC2, FAS, CD24, STAT1, ANGPTL4 |
| GO:0043067 | Regulation of
programmed cell death | 12 | 0.001035716 | KCNMA1, PRUNE2, CCL2,
DUSP1, MCL1, SOS2, ESR1, FOXC2, FAS, CD24, STAT1, ANGPTL4 |
| GO:0010941 | Regulation of cell
death | 12 | 0.001067374 | KCNMA1, PRUNE2, CCL2,
DUSP1, MCL1, SOS2, ESR1, FOXC2, FAS, CD24, STAT1, ANGPTL4 |
| GO:0043627 | Response to estrogen
stimulus | 5 | 0.001353113 | KCNMA1, DUSP1, LEPR,
ESR1, CD24 |
| GO:0010033 | Response to organic
substance | 10 | 0.005300596 | KCNMA1, CCL2, DUSP1,
MCL1, LEPR, ESR1, FOXC2, FAS, CD24, STAT1 |
| GO:0031960 | Response to
corticosteroid stimulus | 4 | 0.006933831 | KCNMA1, CCL2, DUSP1,
FAS |
| GO:0043065 | Positive regulation
of apoptosis | 7 | 0.013655682 | KCNMA1, PRUNE2,
DUSP1, SOS2, FAS, CD24, STAT1 |
| GO:0043068 | Positive regulation
of programmed cell death | 7 | 0.01409138 | KCNMA1, PRUNE2,
DUSP1, SOS2, FAS, CD24, STAT1 |
| GO:0010942 | Positive regulation
of cell death | 7 | 0.01438722 | KCNMA1, PRUNE2,
DUSP1, SOS2, FAS, CD24, STAT1 |
| GO:0051384 | Response to
glucocorticoid stimulus | 3 | 0.049690594 | CCL2, DUSP1,
FAS |
Transcription factor analysis
Using the TRANSFAS database, the transcription
factors and binding sites, which were associated with the three GC
response genes, CCL2, DUSP1 and FAS, were assessed. As Fig. 4 demonstrates, c-Jun binds to the
promoter regulatory regions of these three genes and was the common
transcription factor (Fig. 4).
Discussion
GCs have been used in clinical treatment for
decades; however, prolonged GC treatment may lead to the formation
of cataracts (24). In the current
study, using DNA microarray analysis, the gene expression profiles
of human LECs treated with Dex or vehicle were analyzed. A total of
13 significant GO functions were identified and CCL2, DUSP1 and FAS
genes were associated with a response to GC stimulus. The
transcription factor that binds to CCL2, DUSP1 and FAS were also
analyzed. The results demonstrated that c-Jun was a common
transcription factor between these genes.
CCL2 is also known as monocyte chemotactic protein-1
and is secreted by endothelial cells, fibroblasts and monocytes
(25). It has been reported that
CCL2 expression and macrophage accumulation were inhibited by
treatment with Dex in cholesterol-fed rabbits (26). GRs may bind specifically to CCL2
mRNA and the inflammatory response of the GR was mediated by
regulation of CCL2 mRNA stability (27). CCL2 was detected in the sample
obtained from patients following cataract surgery (28). DUSP1 is a member of the
threonine-tyrosine dual-specificity phosphatases (29). Increased expression of GILZ mRNA
and DUSP1 mRNA and protein was observed in immortalized and donor
immortalized primary LECs (13).
The induction of DUSP1 is dependent on the GR and typically occurs
within ≤1 h (30). The FAS
receptor is an important cell surface receptor protein of the tumor
necrosis factor receptor family (31). Yang et al (32) reported that FAS ligand expression
was inhibited by retinoic acid and GCs.
In the present study, c-Jun was observed to bind the
promoter regulatory regions of CCL2, DUSP1 and FAS. The c-Jun gene
encodes a basic region-leucine zipper transcription factor
implicated in numerous cellular processes. C-Jun regulates gene
expression and cell function by being involved in the formation of
a variety of dimeric complexes, which exhibit high affinity
sequence specific DNA-binding activity (33). It has been reported that c-Jun
attenuated MG132-induced activation of activator protein-1 and
expression of CCL2 (34). The
Hepatitis C virus core protein expression activated MAP kinase
phosphatase, increased DUSP1 expression and increased cell
proliferation, which was accompanied by an activation of c-Jun
(35). The expression of
dominant-negative c-Jun in melanoma cells efficiently increased Fas
expression (36). The present
results demonstrated that c-Jun may be the critical transcription
factor, which affected gene expression in LECs in response to
GCs.
In conclusion, the gene expression profiles of LECs
following GC treatment were analyzed using bioinformatics analysis
and it was found that CCL2, DUSP1 and FAS are involved in the
response to GC stimulus. The transcription factor c-Jun, when bound
to CCL2, DUSP1 and FAS, may affect their expression. CCL2, DUSP1,
FAS and transcription factor c-Jun may be used as specific
therapeutic molecular targets in order to treat cataracts induced
by GCs. However, further studies are required to confirm the
present results.
Acknowledgments
The authors would like to thank the Laboratory of
Medical Genetics, Harbin Medical University, Harbin, China for the
technical assistance that was required. The authors would also to
thank Fenghe (Shanghai) Information Technology Co., Ltd. Their
input and assistance provided a valuable added dimension to the
present study.
References
|
1
|
Black RL, Oglesby RB, von Sallmann L and
Bunim JJ: Posterior subcapsular cataracts induced by
corticosteroids in patients with rheumatoid arthritis. JAMA.
174:166–171. 1960. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Williamson J, Paterson RW, McGavin DD,
Jasani MK, Boyle JA and Doig WM: Posterior subcapsular cataracts
and glaucoma associated with long-term oral corticosteroid therapy.
In patients with rheumatoid arthritis and related conditions. Br J
Ophthalmol. 53:361–372. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Urban RC Jr and Cotlier E:
Corticosteroid-induced cataracts. Surv Ophthalmol. 31:102–110.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jobling AI and Augusteyn RC: What causes
steroid cataracts? A review of steroid-induced posterior
subcapsular cataracts. Clin Exp Optom. 85:61–75. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Auphan N, DiDonato JA, Rosette C, Helmberg
A and Karin M: Immunosuppression by glucocorticoids: inhibition of
NF-κB activity through induction of IκB synthesis. Science.
270:286–290. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ehrchen J, Steinmüller L, Barczyk K, et
al: Glucocorticoids induce differentiation of a specifically
activated, anti-inflammatory subtype of human monocytes. Blood.
109:1265–1274. 2007. View Article : Google Scholar
|
|
7
|
Reichardt HM, Tronche F, Berger S,
Kellendonk C and Schütz G: New insights into glucocorticoid and
mineralocorticoid signaling: lessons from gene targeting. Adv
Pharmacol. 47:1–21. 2000. View Article : Google Scholar
|
|
8
|
Aranda A and Pascual A: Nuclear hormone
receptors and gene expression. Physiol Rev. 81:1269–1304.
2001.PubMed/NCBI
|
|
9
|
Webster JC and Cidlowski JA: Mechanisms of
glucocorticoid-receptor-mediated repression of gene expression.
Trends Endocrinol Metab. 10:396–402. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nishigori H, Lee JW, Yamauchi Y and
Iwatsuru M: The alteration of lipid peroxide in
glucocorticoid-induced cataract of developing chick embryos and the
effect of ascorbic acid. Curr Eye Res. 5:37–40. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lou MF, Dickerson JE Jr, Garadi R and York
BM Jr: Glutathione depletion in the lens of galactosemic and
diabetic rats. Exp Eye Res. 46:517–530. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gupta V, Galante A, Soteropoulos P, Guo S
and Wagner BJ: Global gene profiling reveals novel glucocorticoid
induced changes in gene expression of human lens epithelial cells.
Mol Vis. 11:1018–1040. 2005.PubMed/NCBI
|
|
13
|
Gupta V, Awasthi N and Wagner BJ: Specific
activation of the glucocorticoid receptor and modulation of signal
transduction pathways in human lens epithelial cells. Invest
Ophthalmol Vis Sci. 48:1724–1734. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saklatvala J: Glucocorticoids: do we know
how they work? Arthritis Res. 4:146–150. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Keil JM, Liu X and Antonetti DA:
Glucocorticoid induction of occludin expression and endothelial
barrier requires transcription factor p54 NONO. Invest Ophthalmol
Vis Sci. 54:4007–4015. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Best CJ, Gillespie JW, Yi Y, et al:
Molecular alterations in primary prostate cancer after androgen
ablation therapy. Clin Cancer Res. 11:6823–6834. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Team RC: R: A language and environment for
statistical computing. Vienna, Austria: R Foundation for
Statistical Computing; pp. 1–1731. 2008
|
|
18
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy - analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reimers M and Carey VJ: Bioconductor: an
open source framework for bioinformatics and computational biology.
Methods Enzymol. 411:119–134. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sarle WS: Algorithms for clustering data.
Technometrics. 32:227–229. 1990. View Article : Google Scholar
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matys V, Fricke E, Geffers R, et al:
TRANSFAC: transcriptional regulation, from patterns to profiles.
Nucleic Acids Res. 31:374–378. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cumming RG, Mitchell P and Leeder SR: Use
of inhaled corticosteroids and the risk of cataracts. N Engl J Med.
337:8–14. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gu L, Tseng SC and Rollins BJ: Monocyte
chemoattractant protein-1. Chem Immunol. 72:7–29. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Poon M, Gertz SD, Fallon JT, et al:
Dexamethasone inhibits macrophage accumulation after balloon
arterial injury in cholesterol fed rabbits. Atherosclerosis.
155:371–380. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dhawan L, Liu B, Blaxall BC and Taubman
MB: A novel role for the glucocorticoid receptor in the regulation
of monocyte chemoattractant protein-1 mRNA stability. J Biol Chem.
282:10146–10152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Janciauskiene S, Westin K, Grip O and
Krakau T: Detection of Alzheimer peptides and chemokines in the
aqueous humor. Eur J Ophthalmol. 21:104–111. 2011. View Article : Google Scholar
|
|
29
|
Boutros T, Chevet E and Metrakos P:
Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase
regulation: roles in cell growth, death, and cancer. Pharmacol Rev.
60:261–310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Abraham SM, Lawrence T, Kleiman A, et al:
Antiinflammatory effects of dexamethasone are partly dependent on
induction of dual specificity phosphatase 1. J Exp Med.
203:1883–1889. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagata S: Fas and Fas ligand: a death
factor and its receptor. Adv Immunol. 57:129–144. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang Y, Merćep M, Ware CF and Ashwell JD:
Fas and activation-induced Fas ligand mediate apoptosis of T cell
hybridomas: inhibition of Fas ligand expression by retinoic acid
and glucocorticoids. J Exp Med. 181:1673–1682. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wisdom R, Johnson RS and Moore C: c-Jun
regulates cell cycle progression and apoptosis by distinct
mechanisms. EMBO J. 18:188–197. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakayama K, Furusu A, Xu Q, Konta T and
Kitamura M: Unexpected transcriptional induction of monocyte
chemoattractant protein 1 by proteasome inhibition: involvement of
the c-Jun N-terminal kinase-activator protein 1 pathway. J Immunol.
167:1145–1150. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Erhardt A, Hassan M, Heintges T and
Häussinger D: Hepatitis C virus core protein induces cell
proliferation and activates ERK, JNK, and p38 MAP kinases together
with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line.
Virology. 292:272–284. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Herdegen T, Claret FX, Kallunki T, et al:
Lasting N-terminal phosphorylation of c-Jun and activation of c-Jun
N-terminal kinases after neuronal injury. J Neurosci. 18:5124–5135.
1998.PubMed/NCBI
|