Introduction
Lung cancer comprises two types: Small cell lung
cancer and non-small cell lung cancer, and is one of the most
commonly diagnosed malignancies worldwide (1,2).
Previous studies have demonstrated that the development of lung
cancer arises from a dysregulation of numerous oncogenes and tumor
suppressors, including p53, phosphatase and tensin homolog, as well
as β-catenin (3,4). However, the underlying regulatory
mechanisms remain poorly understood.
Roles of the Forkhead transcription factor family
(FOXO) have previously been reported in cell proliferation,
differentiation, apoptosis and metabolic pathways (5). There are four members of the FOXO
family in humans: FOXO1, −3, −4 and −6. The role of FOXO1 in tumor
cell proliferation has been well documented (6). FOXO1 overexpression has been shown to
result in cell-cycle arrest through upregulation of p27 and p21 and
downregulation of cyclin D1 (5).
Furthermore, FOXO3 is also considered a tumor suppressor in
numerous types of cancer, including neuroblastoma (7), as well as colon (8) and thyroid cancer (9). Previous studies have demonstrated
that FOXO6 may regulate memory consolidation and synaptic function
(10). Additionally, FOXO6 have
been identified to promote gluconeogenesis and integrate insulin
signaling with microsomal triglyceride transfer protein for the
regulation of very low-density lipoprotein production in the liver
(11,12). However, the functions of FOXO6 in
tumorigenesis remain unclear. In the present study, the expression
levels of FOXO6 in lung cancer tissue were determined by
quantitative PCR and western blot analysis. Additionally, USP7, a
ubiquitin-specific protease, was shown to promote p53 protein
stabilization (13). Therefore,
the role and mechanism of FOXO6 in the regulation of USP7
expression was further investigated.
Materials and methods
Tissue samples
A total of 30 paired primary lung cancer tissue and
adjacent normal tissue samples were obtained from patients at the
Department of Thoracic Tumor Surgery 2 (Xinxiang Central Hospital,
Xinxiang, China). All patients provided informed consent. The
present study was approved by the Institutional Review Board of
Xinxiang Central Hospital. The study was approved by the ethics
committee of Xinxiang Central Hospital (Xinxiang, China). Written
informed consent was obtained from the patients or their
families.
Cell culture and reagents
The A549 human lung cancer cells were provided by
The Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China). The cells were cultured in Dulbecco’s
modified Eagle’s medium (Sigma-Aldrich, St. Louis, MO, USA)
supplemented with 10% fetal calf serum (Invitrogen Life
Technologies, Shanghai, China), 100 IU/ml penicillin (Invitrogen
Life Technologies) and 100 mg/ml streptomycin (Invitrogen Life
Technologies). The cell cultures were maintained at 37°C in a
humidified atmosphere containing 5% CO2. Tumor necrosis
factor (TNF α (5 ng/ml), interleukin (IL)-1β (2 ng/ml) and IL-6 (10
ng/ml) were obtained from Sigma-Aldrich. Cells were seeded into
6-well plates and treated with these cytokines or the vehicle
control (PBS). Following either 24 or 36-h incubation, the cells
were harvested for RNA or protein extraction.
Promoter construction and luciferase
assays
The 5′-regulatory sequence of the human
ubiquitin-specific-processing protease 7 (USP7) promoter was cloned
by polymerase chain reaction (PCR) and inserted into a pGL3 vector
(Promega Corp., Madison, WI, USA) using Taq enzymes (Invitrogen
Life Technologies. Site-directed mutagenesis was conducted using a
PCR kit (Toyobo Co., Ltd., Osaka, Japan) with the following primer
(mutation sites in bold): 5′-CCATGCTTTGGGGAACGACTACA-3′. MDM2 and
YY1 expression levels were measured using quantitative PCR. All the
transient transfection experiments were performed on cells seeded
into 24-well plates at 70–80% confluence, using
Lipofectamine® 2000 (Invitrogen Life Technologies),
according to the manufacturer’s instructions.. Luciferase
activities were normalized against pRL-TK activity (Promega
Corporation) using the Dual Luciferase Reporter Assay system
(Promega Corp.).
Small interfering (si)RNA, RNA extraction
and quantitative PCR (qPCR)
Non-targeting siRNA (5′-UCUAUACGGUCUACUACGG-3′) and
FOXO6 siRNA (5′-CAUGACUUAGCAUACGAAGUAC-3′) oligonucleotides were
obtained from Genepharma Co., Ltd. (Shanghai, China). Tissues were
homogenized by liquid nitrogen grinding and total RNA was extracted
using TRIzol® reagent (Invitrogen Life Technologies).
First-strand complementary DNA (cDNA) synthesis was performed for
each RNA sample using the Promega Reverse Transcription system
(Promega Corp.). Random primers were used to prime cDNA synthesis.
qPCR was performed using SYBR Green Premix Ex Taq (Roche
Diagnostics, Basel, Switzerland) on a Light Cycler 480 (Roche
Diagnostics). The relative quantification for each target gene was
corrected to GAPDH mRNA values. The following primers were used:
FOXO6 forward, 5′-GGCGGCGCTCGTATACC-3′ and reverse,
5′-TACACAGGGCCGGCCG-3′; USP7 forward, 5′-GGCTCCTGGCATTAGGTCA-3′ and
reverse, 5′-CTGGCTAATTGTGCTGATGT-3′); p53 forward,
5′-TGGAGCCCGTGAATTAGAGC-3′ and reverse,
5′-TCTGGTTCATCTTAGAAACCAC-3′; p27 forward,
5′-ACCTTAACCAGTGCTCCTA-3′ and reverse, 5′-CCCAGCCTTGAAACAATCC-3′;
p21 forward, 5′-ACTCTTCGTGCAAGGGCG-3′ and reverse,
5′-GCTCAAGAAAGTGCTGATCCC-3′; and GAPDH forward,
5′-CATGTACGCCATATCCAGGC-3′ and reverse, 5′-CTCCATTGATGTCACGCGGAT-3′
(Biosune Co., Shanghai, China). PCR conditions included an initial
holding period at 95°C for 5 min, followed by a two-step PCR
program comprising 94°C for 5 sec and 60°C for 30 sec for 40
cycles. Relative quantification analysis of the gene expression
results were analyzed using the 2−ΔΔCt method.
Western blot analysis
Extracts from the cells or tissues were prepared
using lysis buffer containing 50 mM Tris-HCl (pH 6.8), 100 mM 2-ME,
2% w/v SDS and 10% glycerol. Proteins in the supernatants were
quantified using a Bicinchoninic Acid Protein Assay kit (Pierce
Biotechnology, Inc., Rockford, IL, USA). Equal amounts of protein
(50 μg) were separated by 10% SDS-PAGE. Proteins
electrophoretically separated on denaturing gels were transferred
to polyvinylidine difluoride membranes (Millipore, Bedford, MA,
USA). Then membranes were blocked using 10% non-fat milk and washed
by PBST solution (Beyotime Company, Nantong, China). Anti-GAPDH
antibody was purchased from Santa Cruz Biotechnology, Inc.,
(Dallas, TX, USA). Rabbit polyclonal anti-FOXO6 (ab48730), rabbit
monoclonal anti-p53 (EPR17343), rabbit polyclonal anti-p21
(ab7960), rabbit monoclonal anti-p27 (Y236), rabbit polyclonal
anti-growth arrest and DNA damage-inducible 45 (GADD45; ab105060),
rabbit monoclonal anti-signal transducer and activator of
transcription 3 (Stat3; EPR787Y), rabbit polyclonal anti-FOXO1
(ab70382), mouse monoclonal anti-E2F transcription factor 1 (E2F1;
8G9), rabbit monoclonal anti-β-catenin (E247) and rabbit polyclonal
anti-retinoblastoma (Rb; ab6075) antibodies were all purchased from
Abcam (Cambridge, MA, USA). All the antibodies were used at 1:2,000
dilution and incubated overnight at 4°C. The proteins were
visualized using an enhanced chemiluminescence detection kit (GE
Healthcare Life Sciences, Little Chalfont, UK).
Bromodeoxyuridine (BrdU) assay
A cell proliferation enzyme-linked immunosorbent
assay (BrdU kit; Beyotime Company) was used to measure the
incorporation of BrdU during DNA synthesis, according to the
manufacturer’s instructions. Following transfection for 24 h, BrdU
was added and the cells were incubated for an additional 4 h at
37°C.
Chromatin immunoprecipitation (ChIP)
assays
The A549 cells were fixed with 1% formaldehyde,
followed by numerous intervals of sonication. The
immunoprecipitated DNA fragments were quantified by PCR. The
following primer sequences used were: Regions −600 to −400 forward,
5′-CAGCTTCGACGTCAC-3′ and reverse, 5′-GACTGCAACATTGCACT-3′; and
regions −3,000 to −2,800 forward, 5′-CATTAGCATCAGATCCATTA-3′ and
reverse, 5′-CAGCATCAGGACTTACGCA-3′.
Transcription element search system
(TESS) analysis
TESS software http://www.cbil.upenn.edu/cgi-bin/tess was used to
automatically identify potential FOXO6 motifs.
Statistical analysis
Values are expressed as the mean ± standard error of
the mean. Statistical analyses were performed using GraphPad
software version 5.0 (La Jolla, CA, USA). Statistical differences
were determined by Student’s t test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Reduced expression of FOXO6 in lung
cancer tissue
FOXO6 mRNA expression levels were initially measured
by qPCR in 30 paired lung cancer and adjacent normal tissue
samples. FOXO6 mRNA expression levels were significantly
downregulated in the lung cancer samples, as compared with those in
the adjacent normal tissue (Fig.
1A). This finding was further confirmed by western blot
analysis (Fig. 1B).
Identification of FOXO6 as a downstream
target of inflammation
The present study aimed to determine potential
mechanisms underlying the downregulation of FOXO6. Previous studies
have shown that inflammatory response-mediated nuclear factor
(NF)-κB/p65 activation is critical for the development of lung
carcinoma (14). To investigate
whether NF-κB/p65 was able to regulate FOXO6, A549 cells were
treated with various pro-inflammatory cytokines. Treatment with
TNFα, IL-1β and IL-6 suppressed FOXO6 mRNA and protein expression
levels (Fig. 2A and B).
Furthermore, FOXO6 expression was also inhibited by p65
overexpression in A549 cells (Fig. 2C
and D).
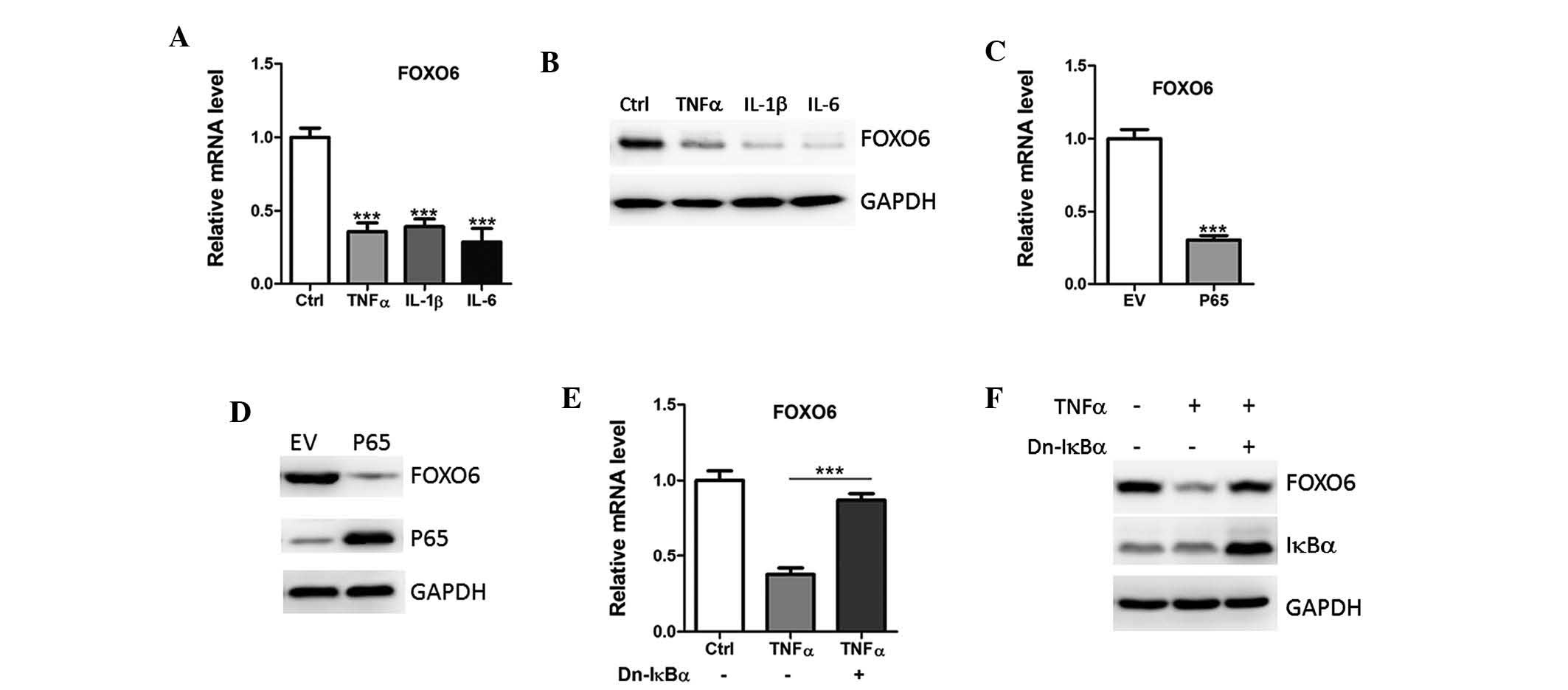 | Figure 2NF-κB activation downregulates FOXO6
expression. (A) mRNA and (B) protein expression levels of FOXO6
were analyzed in A549 human lung cancer cells treated with Ctrl or
TNFα (5 ng/ml), IL-1β (2 ng/ml) or IL-6 (10 ng/ml) by qPCR and
western blotting, respectively. (C) mRNA and (D) protein expression
levels of FOXO6 were analyzed in A549 cells transfected with EV or
p65 by qPCR and western blotting, respectively. (E) mRNA and (F)
protein expression levels of FOXO6 in A549 cells transfected with
dominant-negative (Dn) IkBα, in the absence or presence of TNFα (5
ng/ml).FOXO6, forkhead box O6; TNFα, tumor necrosis factor-α; IL,
interleukin; EV, empty vector; Ctrl, vehicle control; qPCR,
quantitative polymerase chain reaction. |
To further analyze whether downregulation of FOXO6
was NF-κB/p65-dependent, the A549 cells were transfected with
dominant negative IkBα, which inhibits the translocation of p65
from the cytoplasm to the nucleus (15). As predicted, overexpression of IkBα
reversed the inhibition of FOXO6 by TNFα (Fig. 2E and F). Therefore, it is
hypothesized that the downregulation of FOXO6 may partly be due to
the hyperinflammatory status of the lung cancer tissue.
FOXO6 inhibits lung cancer cell
proliferation
To determine the functions of FOXO6, A549 cells were
transfected with empty vector or FOXO6 lentiviruses (Fig. 3A). Overexpression of FOXO6 reduced
the cell proliferation of A549 cells (Fig. 3B), which was further confirmed by a
bromodeoxyuridine analysis (Fig.
3C).
In addition, endogenous FOXO6 expression in A549
cells was deleted by siRNA targeting FOXO6 (Fig. 3D). Ablation of FOXO6 increased the
number of A549 cells and enhanced the cell proliferative abilities
(Fig. 3E and F).
FOXO6 overexpression induces p53 protein
accumulation in lung cancer cells
To investigate the molecular mechanisms of FOXO6 on
cell proliferation, the expression levels of numerous oncogenes and
tumor suppressors were determined. None of the transcription
factors were altered at the transcriptional level in response to
FOXO6 overexpression (Fig. 4A).
However, western blot analysis demonstrated that p53 protein
expression levels were increased in the A549 cells overexpressing
FOXO6 (Fig. 4B). In addition,
downstream targets of p53, including p21, p27 and GADD45 (16), were also upregulated by FOXO6
overexpression (Fig. 4C and D).
Furthermore, FOXO6 deficiency resulted in reduced levels of p53,
p21, p27 and GADD45 (Fig. 4E).
These results indicated that FOXO6 may modulate p53 protein
expression at the post-transcriptional or translational level.
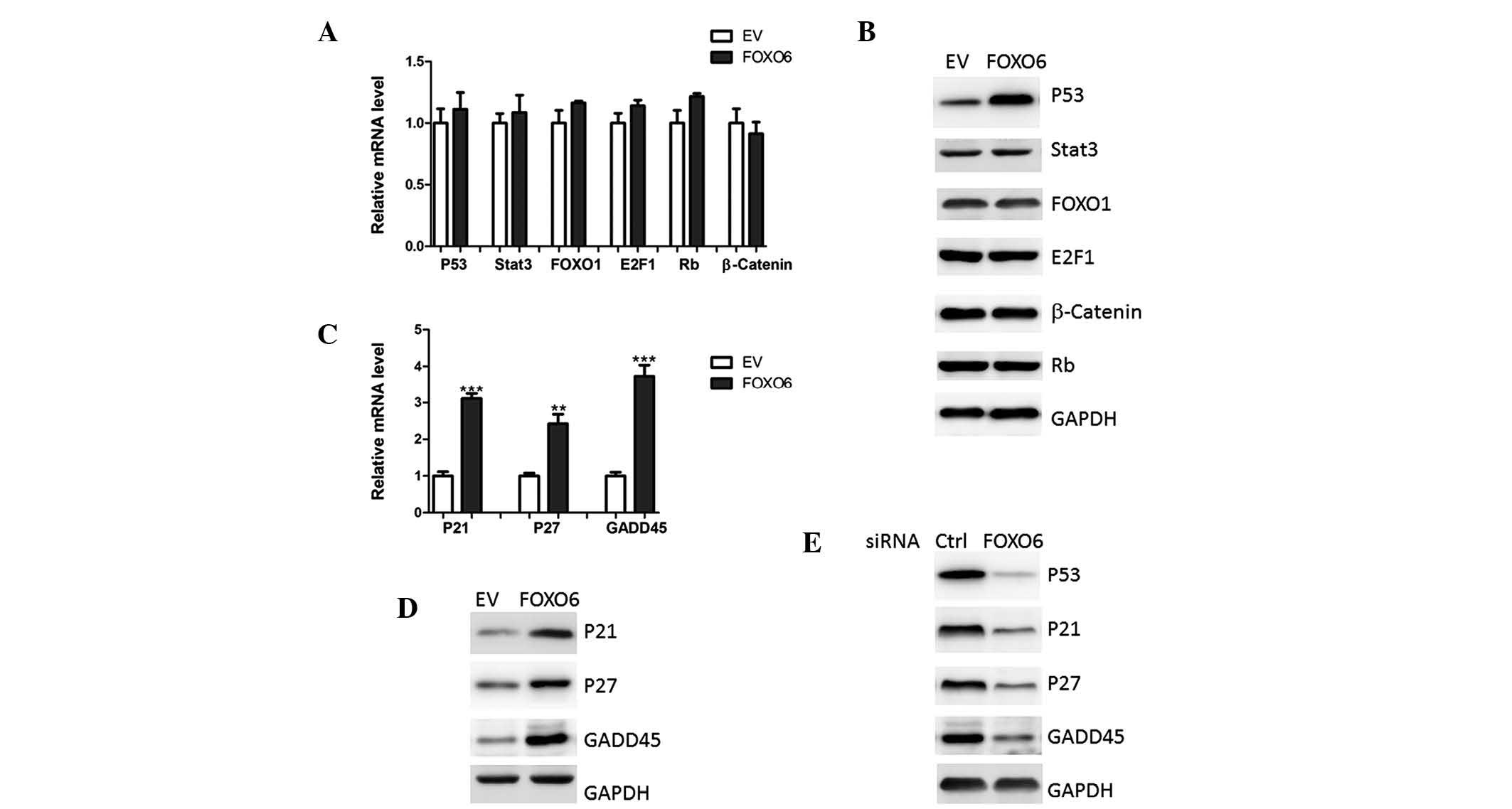 | Figure 4FOXO6 upregulates p53 protein
expression levels. (A) mRNA and (B) protein expression levels of
p53, Stat3, FOXO1, E2F1, Rb and β-catenin were analyzed in A549
human lung cancer cells transfected with EV or FOXO6 by qPCR and
western blotting, respectively. (C) mRNA and (D) protein expression
levels of p21, p27 and GADD45 were analyzed in A549 cells
transfected with EV or FOXO6, by qPCR and western blotting,
respectively. (E) Protein expression levels of p53, p21, p27 and
GADD45 were analyzed by western blotting in A549 cells transfected
with FOXO6 or Ctrl. FOXO1/O6, forkhead box O1/O6; Stat3, signal
transducer and activator of transcription 3; E2F1, E2F
transcription factor 1; Rb, retinoblastoma; GADD45, growth arrest
and DNA damage-inducible 45; siRNA, small interfering RNA; qPCR,
quantitative polymerase chain reaction; EV, empty vector; Ctrl,
scramble siRNA. |
FOXO6 modulates p53 protein expression
through upregulation of USP7
It has been well-established that p53 protein
abundance is regulated by various proteins, including USP7, mouse
double minute 2 homolog (MDM2) and Yin Yang 1 (YY1) (17–19).
In the present study, USP7 expression levels were markedly
increased in the A549 cells with FOXO6 overexpression (Fig. 5A and B), whereas MDM2 and YY1
expression remained unaltered (data not shown). Conversely,
knockdown of FOXO6 expression inhibited USP7 expression (Fig. 5C and D).
 | Figure 5FOXO6 transcriptionally regulates USP7
expression. (A) mRNA and (B) protein expression levels of USP7 were
analyzed in A549 cells transfected with EV or FOXO6, by qPCR and
western blotting, respectively. (C) mRNA and (D) protein expression
levels of USP7 were analyzed in A549 cells transfected with FOXO6
or Ctrl by qPCR and western blotting, respectively. (E) Human USP7
promoter constructs containing a potential FOXO6 motif (−512 bp to
−503 bp). Point mutations were induced in the FOXO6 motif
(TTTGTTTAAC to TTTGGGGAAC). The transcription start site was set as
+1 bp. (F) Promoter region from −750 to +120 bp (Wt-Luc and
Mut-Luc) was cloned and co-transfected with FOXO6 expression
plasmids in A549 cells. Cells were lysed and luciferase activity
was determined 36 hours after transfection. (G) Chromatin
immunoprecipitation assays showing the recruitment of FOXO6 onto
the USP7 promoter. The promoter region from −3,000 to −2,800 bp was
set as a negative control. PCR was performed to quantify the
binding. FOXO6, forkhead box O6; USP7,
ubiquitin-specific-processing protease 7; siRNA, small interfering
RNA; qPCR, quantitative polymerase chain reaction; EV, empty
vector; Ctrl, scramble siRNA; WT, wild-typ; Mut, mutated. |
The present study aimed to investigate the
regulatory pathway involving FOXO6 and USP7. A potential FOXO6
motif (TTTGTTTAAC) was identified at −512 bp to −503 bp using TESS
software (http://www.cbil.upenn.edu/cgi-bin/tess) (Fig. 5E). A luciferase reporter was
generated containing the binding site from position −750 bp to +120
bp. USP7 promoter activity was markedly elevated by FOXO6
overexpression (Fig. 5F). However,
mutation of the potential binding site completely abrogated the
function of FOXO6 (Fig. 5F). ChIP
assays also confirmed that FOXO6 protein could bind with the
proximal promoter region in A549 cells, but not with the distal
region (Fig. 5G). These results
suggested that FOXO6 regulated USP7 expression at the
transcriptional level.
Discussion
The present study established FOXO6 as a novel tumor
suppressor in lung cancer. The results of the present study
demonstrated that FOXO6 was downregulated, at least partly, by
inflammatory response-mediated NF-κB/p65 activation in lung cancer
tissue. Since cytokines and growth factors secreted by tumor cells
are viewed as causative factors in constitutive NF-κB/p65
activation, these results provide an alternative role for NF-κB/P65
in tumor cells; however, the regulatory mechanisms still require
further study.
The present study showed that FOXO6 was able to
upregulate USP7 expression through binding to the proximal region
of its promoter. USP7 is a member of the ubiquitin-specific
protease family, and was initially considered a binding partner of
the herpes simplex virus protein Vmw110/ICP0 (20). Subsequent studies have indicated
that USP7 is able to stabilize p53 protein, through a complex
mechanism targeting both p53 and its E3 ubiquitin ligase (13). Ubiquitination-mediated modification
of p53 protein may regulate its degradation, transcriptional
activity and subcellular localization (21). MDM2 is an oncogenic E3 ligase for
p53, which promotes p53 degradation and prevents its activation in
the presence of genotoxic stress (22). Furthermore, upregulation of MDM2
has previously been observed in numerous types of human cancer,
including lung and breast cancer, as well as osteosarcoma (23,24).
Therefore, USP7-mediated regulation of p53 protein stability may be
a potential therapeutic target for the treatment of human
cancer.
In conclusion, the results of the present study
indicated an important role for FOXO6 in controlling lung cancer
development. Of note, Qinyu et al (25) demonstrated that FOXO6 promoted
gastric cancer progression through trans-activation of C-myc
expression. These results suggested that the roles of FOXO6 may be
cell or tissue-specific. Therefore, the precise roles of FOXO6 in
cancer biology require further study, using other models, such as
tissue-specific FOXO6 knockout mice.
References
|
1
|
Hutchinson L: Screening: Improved model
for lung cancer detection. Nat Rev Clin Oncol. 10:1832013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Keith RL and Miller YE: Lung cancer
chemoprevention: current status and future prospects. Nat Rev Clin
Oncol. 10:334–343. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mallakin A, Sugiyama T, Taneja P, et al:
Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer.
Cancer Cell. 12:381–394. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Damsky WE, Curley DP, Santhanakrishnan M,
et al: β-catenin signaling controls metastasis in Braf-activated
Pten-deficient melanomas. Cancer Cell. 20:741–754. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eijkelenboom A and Burgering BM: FOXOs:
signalling integrators for homeostasis maintenance. Nat Rev Mol
Cell Biol. 14:83–97. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Santo EE, Stroeken P, Sluis PV, Koster J,
Versteeg R and Westerhout EM: FOXO3a is a major target of
inactivation by PI3K/AKT signaling in aggressive neuroblastoma.
Cancer Res. 73:2189–2198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Savkovic SD: Decreased FOXO3 within
advanced human colon cancer: implications of tumour suppressor
function. Br J Cancer. 109:297–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong ZY, Lee HJ, Shin DY, Kim SK, Seo M
and Lee EJ: Inhibition of Akt/FOXO3a signaling by constitutively
active FOXO3a suppresses growth of follicular thyroid cancer cell
lines. Cancer Lett. 314:34–40. 2012. View Article : Google Scholar
|
|
10
|
Salih DA, Rashid AJ, Colas D, et al: FoxO6
regulates memory consolidation and synaptic function. Genes Dev.
26:2780–2801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim DH, Perdomo G, Zhang T, et al: FoxO6
integrates insulin signaling with gluconeogenesis in the liver.
Diabetes. 60:2763–2774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim DH, Zhang T, Lee S, et al: FoxO6
integrates insulin signaling with MTP for regulating VLDL
production in the liver. Endocrinology. 155:1255–1267. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li M, Chen D, Shiloh A, et al:
Deubiquitination of p53 by HAUSP is an important pathway for p53
stabilization. Nature. 416:648–653. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hopewell EL, Zhao W, Fulp WJ, et al: Lung
tumor NF-κB signaling promotes T cell-mediated immune surveillance.
J Clin Invest. 123:2509–2522. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang G, Li J, Purkayastha S, et al:
Hypothalamic programming of systemic ageing involving IKK-β, NF-κB
and GnRH. Nature. 497:211–216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saha A, Kuzuhara T, Echigo N, Fujii A,
Suganuma M and Fujiki H: Apoptosis of human lung cancer cells by
curcumin mediated through up-regulation of “growth arrest and DNA
damage inducible genes 45 and 153”. Biol Pharm Bull. 33:1291–1299.
2010. View Article : Google Scholar
|
|
17
|
Dar A, Shibata E and Dutta A:
Deubiquitination of Tip60 by USP7 determines the activity of the
p53-dependent apoptotic pathway. Mol Cell Biol. 33:3309–3320. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
de Rozieres S, Maya R, Oren M and Lozano
G: The loss of mdm2 induces p53-mediated apoptosis. Oncogene.
19:1691–1697. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sui G, Affar el B, Shi Y, et al: Yin Yang
1 is a negative regulator of p53. Cell. 117:859–872. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Holowaty MN, Zeghouf M, Wu H, et al:
Protein profiling with Epstein-Barr nuclear antigen-1 reveals an
interaction with the herpesvirus-associated ubiquitin-specific
protease HAUSP/USP7. J Biol Chem. 278:29987–29994. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li M, Brooks CL, Kon N and Gu W: A dynamic
role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 13:879–886. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang S, Jensen JP, Ludwig RL, Vousden KH
and Weissman AM: Mdm2 is a RING finger-dependent ubiquitin protein
ligase for itself and p53. J Biol Chem. 275:8945–8951. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iwakuma T and Lozano G: MDM2, an
introduction. Mol Cancer Res. 1:993–1000. 2003.
|
|
24
|
Jung CH, Kim J, Park JK, et al: Mdm2
increases cellular invasiveness by binding to and stabilizing the
Slug mRNA. Cancer Lett. 335:270–277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qinyu L, Long C, Zhen-dong D, et al: FOXO6
promotes gastric cancer cell tumorigenicity via upregulation of
C-myc. FEBS Lett. 587:2105–2111. 2013. View Article : Google Scholar : PubMed/NCBI
|



















