Introduction
Gastric cancer is one of the most common types of
cancer globally, with a particularly high incidence in Northeast
Asian countries, including China, Japan and Korea (1). The traditional therapeutic strategies
for gastric cancer have failed to effectively treat the condition
(2). In previous years, targeted
therapies against the molecular variants specific to the
carcinogenesis of various types of human cancer have been
developed. However, the development of targeted therapies for
gastric cancer lags behind other types of cancer. Trastuzumab, a
monoclonal antibody against human epidermal growth factor receptor
2 (HER2) widely used for the treatment of breast cancer, has been
approved for the treatment of patients with gastric cancer
expressing HER2 based on the encouraging results of Trastuzumab for
the treatment of gastric cancer (3). Gefitinib, an epidermal growth factor
receptor-tyrosine kinase inhibitor (EGFR-TKI) successfully used for
the treatment of non-small cell lung cancer with an EGFR mutation
in exon 19 or 21, appears to be effective for certain patients with
gastric cancer (4). Furthermore,
EGFR mutations in exon 21 have been identified in patients with
gastric cancer by a Portuguese research group and in our previous
study (5,6). These findings indicate that certain
patients with gastric cancer would benefit from TKIs. However, only
a specific subpopulation of patients are able to benefit from
treatment with trastuzumab or EGFR-TKIs.
Cetuximab, an immunoglobulin G1 chimeric monoclonal
antibody directed against EGFR, exhibits anticancer effects by
binding to the extracellular domain of EGFR with a higher affinity
than the natural ligands of the receptor, inhibiting its
dimerization and phosphorylation, and then blocking the downstream
signaling transduction pathways, mainly KRAS/BRAF/mitogen activated
protein kinase and phosphoinositide 3-kinase (PI3K)/AKT/mammalian
target of rapamycin (7). Cetuximab
was initially approved for the treatment of patients with
colorectal cancer (CRC), particularly those with EGFR-positive
tissue samples. Notably, it was reported that over half of all
patients with gastric cancer expressed EGFR, suggesting that
certain patients with gastric cancer may benefit from cetuximab
treatment (8). Furthermore,
cetuximab, either alone or in combination with various
chemotherapeutic drugs, has been evaluated in clinical trials for
the treatment of patients with gastric cancer (9–11).
However, the majority of patients, including those with
EGFR-positive tissue samples were not sensitive to cetuximab. The
previous results appear to be conflicting. Therefore, it is
hypothesized that certain factors may impair the sensitivity of
patients with gastric cancer to cetuximab. Currently, cetuximab has
been well demonstrated to be effective in patients with CRC without
KRAS mutations as mutant KRAS with an elevated kinase activity
would result in a continuously activated state, leading to a
continuous and self-sufficient signaling transduction pathway, thus
rendering cetuximab ineffective (12). Furthermore, it has been clearly
demonstrated that mutations of other downstream effectors of EGFR,
such as BRAF and PIK3CA, are also able to render cetuximab
ineffective (13,14). Therefore, the evaluation of KRAS,
BRAF and PIK3CA mutations in gastric cancer may aid the selection
of patients eligible for treatment with cetuximab.
In previous years, KRAS mutations have been reported
in gastric cancer with a frequency, ranging between 0 and 21%
(15–18). However, the exact frequency of
mutations in the KRAS gene and their association with
clinicopatho-logical features of the patients remains to be
elucidated. Although previous studies had considered BRAF mutations
to be rare in gastric cancer, further studies are required to
confirm this hypothesis as no more than ten studies have
investigated the status of the BRAF mutation to date, to the best
of our knowledge (18–25). In contrast to KRAS and BRAF
mutations, a mutation in PIK3CA has been identified in gastric
cancer with a relatively higher frequency between 4.3 and 25.0%
(18,26–28).
However, no definitive conclusion can be drawn regarding whether
the PIK3CA mutation coexists with the KRAS mutation in gastric
cancer.
Therefore, the previously mentioned data revealed
that mutations in KRAS, BRAF and PIK3CA may be involved in gastric
cancer. In addition, these variants were usually evaluated by
different groups from different countries and data regarding
comprehensive analysis of KRAS, BRAF and PIK3CA mutations in
gastric cancer were limited. The aim of the present study was to
evaluate concomitantly the status of KRAS, BRAF and PIK3CA
mutations in 156 gastric cancer samples in order to aid the
selection of patients eligible for treatment with cetuximab.
Patients and methods
Study population
A total of 156 gastric cancer samples [53
fresh-frozen and 103 formalin-fixed, paraffin-embedded (FFPE)] were
collected from patients who underwent surgical resection at The
First Affiliated Hospital of Dalian Medical University and The
Third People’s Hospital of Dalian (Dalian, China). The present
study was initiated with the approval of the ethics committee of
The Second Affiliated Hospital of Dalian Medical University.
Informed consent was obtained from patients prior to the collection
of the samples. None of patients in the present study had received
preoperative radio-or chemotherapy. The clinicopathological
features of all patients were verified by two experienced
pathologists (Table I).
 | Table IAssociations between gene mutations
and clinicopathological features of patients. |
Table I
Associations between gene mutations
and clinicopathological features of patients.
| Clinicopathological
feature | No. | KRAS
| PIK3CA
| KRAS/PIK3CA
|
|---|
| Mutation | P-value | Mutation | P-value | Mutation | P-value |
|---|
| Gender | | | 1 | | 1 | | 0.956 |
| Male | 115 | 5 | | 4 | | 9 | |
| Female | 41 | 2 | | 2 | | 4 | |
| Age (years) | | | 0.439 | | 1 | | 0.385 |
| ≤65 | 78 | 2 | | 3 | | 5 | |
| >65 | 78 | 5 | | 3 | | 8 | |
| Lymph node
metastasis | | | 0.131 | | 0.359 | | 0.036 |
| Present | 116 | 3 | | 3 | | 6 | |
| Absent | 40 | 4 | | 3 | | 7 | |
| Differentiation
grade | | | 0.742a | | 0.53a | | 0.317a |
| Well | 5 | 1 | | 1 | | 2 | |
| Moderate | 42 | 2 | | 2 | | 4 | |
| Poor | 92 | 4 | | 3 | | 7 | |
| MAC | 17 | 0 | | 0 | | 0 | |
| Depth of
invasion | | | 0.433 | | 1 | | 0.296 |
| T1 | 4 | 1 | | 0 | | 1 | |
| T2–T4 | 152 | 6 | | 6 | | 12 | |
DNA extraction
The DNA extraction of the 53 fresh-frozen samples
has been previously completed by our group (17). Therefore, in the present study, DNA
was only extracted from the 103 FFPF samples. An ~5-μm thick single
section from each sample was selected for hematoxylin and eosin
(H&E) staining. An experienced pathologist analyzed the H&E
sections and marked the area containing at least 70% tumor cells.
Subsequently, according to the marked area in the H&E section,
the tumor tissue was dissected from eight sections of each FFPE
sample and then genomic DNA was extracted using an FFPE DNA
extraction kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according
to the manufacturer’s instructions.
Mutation analysis of KRAS (exon 2), BRAF
(exon 15) and PIK3CA (exon 9 and 20)
The status of the KRAS (exon 2) mutation in the 53
fresh-frozen samples has been previously analyzed and reported by
Liu et al (17). In the
present study, the mutation status of KRAS (exon 2) in 103 FFPE
tumor samples, and BRAF (exon 15) and PIK3CA (exon 9 and 20) in 156
tumor samples was screened using polymerase chain reaction-high
resolution melting (PCR-HRM) analysis. PCR primers for KRAS, BRAF
and PIK3CA are shown in Table II.
The final PCR reaction mixture (10 μl) contained: i) KRAS exon 2:
1X PCR buffer, 200 μM dNTPs, 2.5 mM MgCl2, 1 μM each
primer, 0.25 units HotStart Taq (Takara Bio Inc., Dalian, China), 5
ng genomic DNA and 1X LC Green Plus (Biofire Diagnostics, Salt Lake
City, UT, USA); ii) BRAF exon 15 and PIK3CA exon 9: 1X PCR Buffer,
200 μM dNTPs, 2 mM MgCl2, 1 μM each primer, 0.25 units
HotStart Taq, 5 ng genomic DNA and 1X LC Green Plus; iii) PIK3CA
exon 20: 1X PCR Buffer, 200 μM dNTPs, 2.5 mM MgCl2, 0.5
μM each primer, 0.25 units HotStart Taq, 5 ng genomic DNA and 1X LC
Green Plus. The PCR amplification conditions were as follows: i)
KRAS exon 2: 95 °C for 10 min; 45 cycles of 95°C for 30 sec, 54°C
for 10 sec and 72°C for 1 min; ii) BRAF exon 15 and PIK3CA exon 20:
95°C for 10 min; 45 cycles of 95°C for 30 sec, 56°C for 10 sec and
72°C for 30 sec; and iii) PIK3CA exon 9: 95°C for 10 min; 45 cycles
of 95°C for 30 sec, 60°C for 10 sec and 72°C for 30 sec.
 | Table IIPrimers for mutation analysis using
polymerase chain reaction-high resolution melting. |
Table II
Primers for mutation analysis using
polymerase chain reaction-high resolution melting.
| Primer | Sequence (from 5′
to 3′) |
|---|
| KRAS exon 2 | Forward:
AGGCCTGCTGAAAATGACTG |
| Reverse:
TCAAAGAATGGTCCTGCACC |
| BRAF exon 15 | Forward:
CTCTTCATAATGCTTGCTCTGATAGG |
| Reverse:
TAGTAACTCAGCAGCATCTCAGG |
| PIK3CA exon 9 | Forward:
CTAGCTAGAGACAATGAATTAAGGGAAA |
| Reverse:
CATTTTAGCACTTACCTGTGACTCCA |
| PIK3CA exon 20 | Forward:
TGAGCAAGAGGCTTTGGAGT |
| Reverse:
TCATTTTCTCAGTTATCTTTTCAGTTCAAT |
DNA sequencing
The PCR product was purified using the GeneJET™ gel
extraction kit (Thermo Fisher Scientific, Rockford, IL, USA)
according to the manufacturer’s instructions and then sequenced on
an ABI Prism 3730 sequence detection system (Takara Bio, Inc.). A
173 bp amplicon was generated by sequencing primers of KRAS exon 2
following the same method as the PCR-HRM primers above. A 124 bp
amplicon was generated by sequencing primers of PIK3CA exon 9,
forward: 5′-GTAACAGACTAGCTAGAG-3′ and reverse:
5′-CTGTGACTCCATAGAAAATC-3′. A 158 bp amplicon was generated by
sequencing primers of PIK3CA exon 20, forward:
5′-GAATGCCAGAACTACAATC-3′ and reverse:
5′-TGTGTGGAAGATCCAATC-3′.
The sequencing conditions for KRAS and PIK3CA were
as follows: 95°C for 10 min; 45 cycles of 95°C for 30 sec, 54°C for
10 sec and 72°C for 1 min.
Statistical analysis
All statistical analyses were conducted using the
χ2 test and Fisher’s exact test with SPSS 13.0
statistical software (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
A total of 156 gastric cancer samples were screened
for mutations in KRAS (exon 2), BRAF (exon 15) and PIK3CA (exon 9
and 20). As a whole, 8.3% (13/156) of the analyzed samples harbored
a mutation in either KRAS or PIK3CA. No mutations in the BRAF gene
were identified. Notably, the frequency of mutations in either KRAS
or PIK3CA was significantly higher in patients without lymph node
metastasis than those with (17.5 vs. 5.2%, P=0.036). No significant
association was observed between mutations in either KRAS or PIK3CA
and other clinicopathological features (Table I).
KRAS mutations
Mutations in the KRAS gene were identified in 7 out
of 156 gastric cancer samples (4.5%). In addition to the five
mutations (two G13D, one G12V and two G12D) previously described by
Liu et al (17), a further
two mutations were found to occur at codon 12, leading to the
substitutions of glycine for aspartic acid (G12D) and alanine
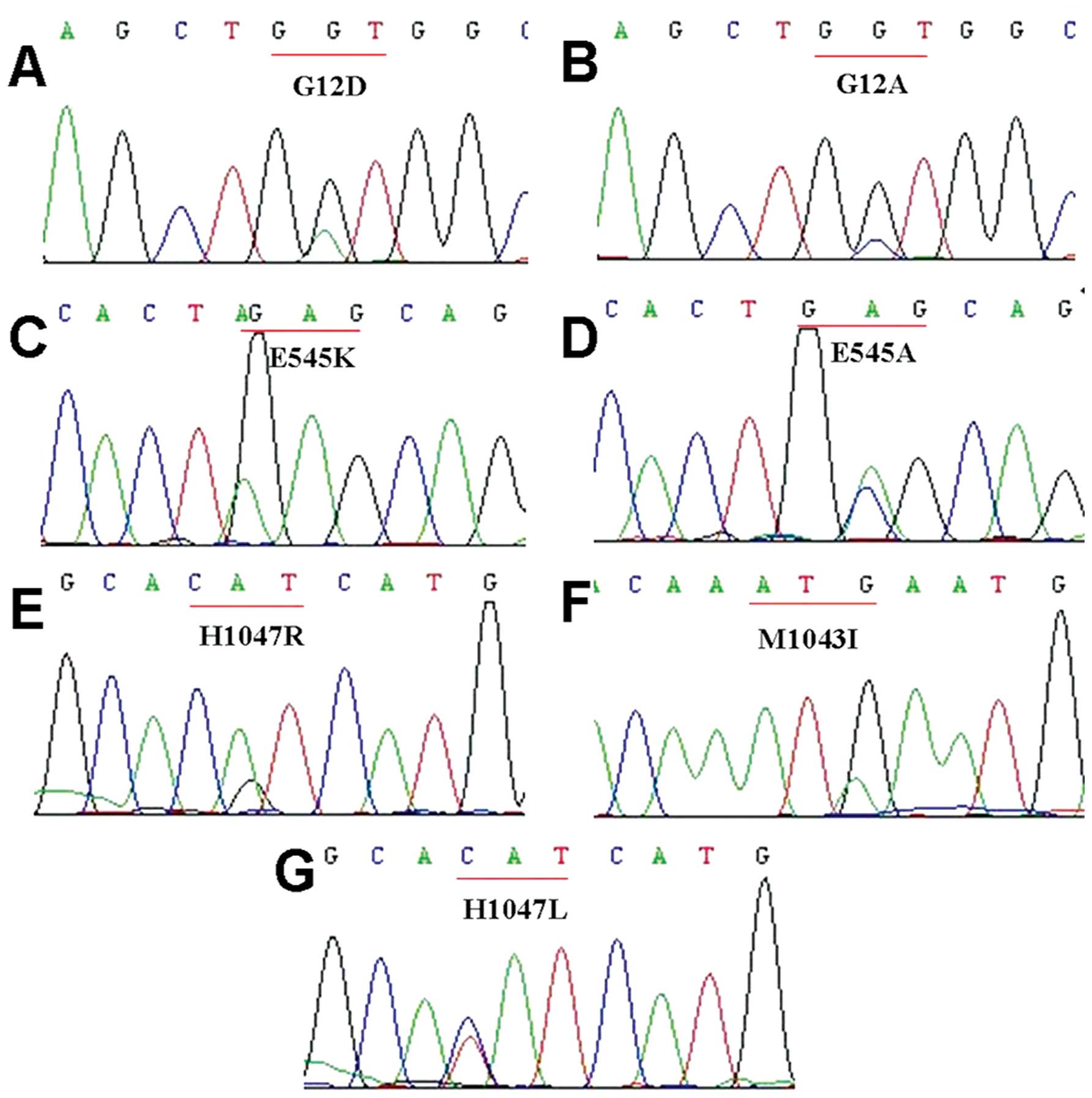
(G12A), respectively (Fig. 1). In
addition, the two mutations occurred exclusively in female patients
aged over 65 years whose tumors were histo-logically classified
into poorly-differentiated adenocarcinoma without lymph node
metastasis. The mutation types and the clinicopathological features
of the patients are summarized in Table III. The results demonstrated that
no significant association was found between mutations in the KRAS
gene and the clinicopathological features of the patients (Table I).
 | Table IIIMutation types and
clinicopathological features of the patients. |
Table III
Mutation types and
clinicopathological features of the patients.
| Patient no. | KRAS | PIK3CA
| Gender | Age (years) | Differentiation
grade | Lymph node
metastasis |
|---|
| Exon 9 | Exon 20 |
|---|
| 1 | G13D | | | Male | 61 | Well | Absent |
| 9 | G12D | | | Male | 72 | Moderate | Present |
| 27 | G12V | | | Male | 64 | Poor | Present |
| 29 | G12D | | | Male | 70 | Poor | Present |
| 49 | G13D | | | Male | 73 | Moderate | Absent |
| 68 | G12D | | | Female | 69 | Poor | Absent |
| 152 | G12A | | | Female | 75 | Poor | Absent |
| 39 | | | H1047R | Female | 81 | Moderate | Absent |
| 45 | | E545K | | Male | 65 | Moderate | Present |
| 70 | | | H1047R | Female | 73 | Well | Absent |
| 83 | | | M1043I | Male | 45 | Poor | Absent |
| 108 | | E545K | | Male | 61 | Poor | Present |
| 122 | | E545A | H1047L | Male | 67 | Poor | Present |
BRAF mutations
Mutations in the BRAF gene were not detected in the
present study, which is consistent with the results of previous
studies.
PIK3CA mutations
Mutations in the PIK3CA gene were identified in 6
out of 156 gastric cancer samples (3.8%), including three mutations
in exon 9 (two E545K and one E545A) and four mutations in exon 20
(two H1047R, one M1043I and one H1047L) (Fig. 1). Furthermore, two different types
of PIK3CA mutations, namely E545A in exon 9 and H1047L in exon 20,
occurred simultaneously in one patient with gastric cancer. By
contrast, mutations in the PIK3CA gene were found to occur in a
mutually exclusive manner with mutations in the KRAS gene. The
mutation types and the clinicopathological features of the patients
are summarized in Table III. In
addition, no significant association was found between mutations in
the PIK3CA gene and the clinicopathological features of the
patients (Table I).
Discussion
In the previous decade, molecular targeted therapy
has been an important area of investigation for the treatment of
human cancer. However, contemporary targeted therapy for gastric
cancer is less than satisfactory, with few encouraging therapeutic
effects. In addition to trastuzumab, which has been approved for
the treatment of patients with gastric cancer, gefitinib appears to
be useful clinically. It is worth noting that trastuzumab and
gefitinib are applicable to only a small faction of patients with
gastric cancer. Cetuximab, which was initially implemented for the
treatment of patients with CRC, is currently undergoing clinical
trials for the treatment of patients with gastric cancer. However,
clinical outcomes from certain trials have been unsuccessful
(9,10). In CRC, cetuximab has been well
demonstrated to be effective in patients without mutations in the
KRAS gene (12). However, certain
patients without mutations in the KRAS gene do not respond to
cetuximab in clinical practice, suggesting that other molecular
variants may be associated with the inefficacy of cetuximab.
Various studies have demonstrated that patients with CRC carrying
mutations of other downstream effectors of EGFR, such as BRAF and
PIK3CA, also exhibit resistance to treatment with cetuximab
(13,14). Therefore, a previous study
suggested that patients with CRC without KRAS, BRAF and PIK3CA
mutations are more likely to respond to cetux-imab (29). Based on the therapeutic success of
cetuximab in CRC, it was hypothesized that it is necessary to
evaluate the status of KRAS, BRAF and PIK3CA mutations in gastric
cancer for selecting patients eligible for treatment with
cetuximab.
KRAS, a molecular switch of intracellular signaling
transduction pathways, is essential in transferring extracellular
growth signals into the nucleus. Mutant KRAS protein exhibits
elevated kinase activity and is able to constitutively activate
downstream signaling transduction pathways independent of stimuli
from activated EGFRs. A review conducted by Kiaris and Spandidos
(30) demonstrated that a mutation
in the KRAS gene was associated with numerous types of human
cancer, including pancreatic cancer, CRC and lung cancer (30). To date, a wide variety of studies
have investigated the status of mutations in the KRAS gene in
gastric adenocarcinoma with the frequency ranging between 0 and 21%
(15–18). The frequency of mutations in the
KRAS gene in different studies has varied markedly. In order to
improve understanding of the status of mutations in the KRAS gene
in gastric cancer, a large international multi-center study,
including 712 patients with gastric cancer was conducted in 2013
(31). In the study, which is the
largest to date, the overall frequency of mutations in the KRAS
gene was 4% and KRAS mutation was not associated with
clinicopatho-logical features, including ethnicity, gender and
stage of tumor differentiation. The discrepancy between previous
studies and the multi-center study may be predominantly due to the
small sample size. Thus far, the majority of the studies included
≤100 patients with gastric cancer, rendering the interpretation of
any conclusion difficult. In our previous study (17), the frequency of mutations in the
KRAS gene was 9.6%. Furthermore, the patients with KRAS mutations
were exclusively males although no significant difference was
found, due to the small sample size. However, in the current study
with a larger sample size, it was identified that the frequency of
mutations in the KRAS gene was 4.5%. In addition, the patients with
mutations in the KRAS gene were not only males but also females. No
significant association was identified between KRAS mutation and
the clinicopathological features of the patient, including gender,
age, differentiation grade and depth of invasion. Thus, mutations
in the KRAS gene may occur in all patients with gastric cancer,
rendering its evaluation necessary for the identification of
patients suitable for cetuximab therapy.
BRAF, a member of the RAF kinase family, is an
essential downstream effector of KRAS. BRAF is commonly activated
by somatic mutation and mutated BRAF is able to constitutively
activate downstream signaling transduction pathways regardless of
stimulus from EGFR or KRAS. To date, BRAF mutations have been
identified in numerous types of human cancer, including melanoma,
papillary thyroid cancer and CRC (32–34).
However, data regarding the BRAF mutation in gastric cancer are
limited. To date, a small number of studies involving 943 patients
with gastric cancer have investigated the status of the BRAF
mutation and found that the average frequency of mutations in the
BRAF gene was only 2.5% (ranging between 0 and 11%) (18–25).
Therefore, it is not noteworthy that no mutations in the BRAF gene
were found in the present study. Mutations in the BRAF gene were
absent or occurred only occasionally in Chinese patients with
gastric cancer. Therefore, it is not necessary to evaluate the
status of the BRAF mutation in order to select patients eligible
for treatment with cetuximab.
PI3K, another downstream effector of EGFR, usually
interacts with KRAS in the regulation of cellular functions.
PIK3CA, encoding the catalytic subunit of PI3K, was initially
demonstrated to be mutated in human cancer by Samuels et al
(27). Subsequently, PIK3CA
mutations have been identified in various types of human cancer,
including breast cancer, CRC, liver cancer and ovarian cancer
(28,35,36).
In gastric cancer, the frequency of PIK3CA mutations varied between
4.3 and 25% (18,26–28).
In the present study, the frequency of PIK3CA mutations was 3.8%,
slightly lower than that observed in previous studies. The
discrepancy among these studies may be due to a number of reasons,
including sample size, mutation detection method used and the
ethnicity of the patients. In CRC, it has been reported that PIK3CA
mutations usually coexist with KRAS mutations (28,37).
Notably, the PIK3CA mutation was also observed in a concomitant
manner with KRAS mutation in gastric cancer (26,28).
However, in the present study, it was identified that PIK3CA and
KRAS mutations were mutually exclusive. Therefore, a definitive
conclusion regarding the association of the PIK3CA mutation with
the KRAS mutation in gastric cancer requires confirmation in
further studies. By contrast, it was identified that mutations in
either KRAS or PIK3CA were more likely to occur in patients without
lymph node metastasis, which indicated that mutations of downstream
effectors of EGFR occurred earlier in gastric carcinogenesis. In
any case, KRAS and PIK3CA mutations should be evaluated prior to
patients receiving cetuximab.
In conclusion, in the present study ~8.3% of Chinese
patients with gastric cancer harbored a mutation in either KRAS or
PIK3CA, suggesting that these patients would not benefit from
cetuximab. Therefore, in the future, KRAS and PIK3CA, but not BRAF
mutations should be evaluated in order to select patients eligible
for treatment with cetuximab.
Acknowledgments
The authors would like to thank the doctors of the
Second Affiliated Hospital of Dalian Medical University for the
collection of the samples. This study was supported by the National
Natural Science Foundation of China (grant no. 81071805).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Janunger KG, Hafström L, Nygren P and
Glimelius B; SBU-group. Swedish Council of Technology Assessment in
Health Care: A systematic overview of chemotherapy effects in
gastric cancer. Acta Oncol. 40:309–326. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bang YJ, Van Cutsem E, Feyereislova A, et
al: Trastuzumab in combination with chemotherapy versus
chemotherapy alone for treatment of HER2-positive advanced gastric
or gastro-oesophageal junction cancer (ToGA): a phase 3,
open-label, randomised controlled trial. Lancet. 376:687–697. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rojo F, Tabernero J, Albanell J, et al:
Pharmacodynamic studies of gefitinib in tumor biopsy specimens from
patients with advanced gastric carcinoma. J Clin Oncol.
24:4309–4316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moutinho C, Mateus AR, Milanezi F,
Carneiro F, Seruca R and Suriano G: Epidermal growth factor
receptor structural alterations in gastric cancer. BMC Cancer.
8:102008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Z, Liu L, Li M, et al: Epidermal
growth factor receptor mutation in gastric cancer. Pathology.
43:234–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ciardiello F and Tortora G: EGFR
antagonists in cancer treatment. N Engl J Med. 358:1160–1174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rivera F, Vega-Villegas ME and López-Brea
MF: Cetuximab, its clinical use and future perspectives. Anticancer
Drugs. 19:99–113. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan JA, Blaszkowsky LS, Enzinger PC, et
al: A multicenter phase II trial of single-agent cetuximab in
advanced esophageal and gastric adenocarcinoma. Ann Oncol.
22:1367–1373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schoennemann KR, Bjerregaard JK, Hansen
TP, et al: Biweekly cetuximab and irinotecan as second-line therapy
in patients with gastro-esophageal cancer previously treated with
platinum. Gastric Cancer. 14:219–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi M, Ji J, Wu J, et al: Cetuximab
combined with FOLFOX4 as the first-line treatment for advanced
gastric cancer: report of 25 cases from a single institution.
Hepatogastroenterology. 59:1054–1058. 2012.PubMed/NCBI
|
|
12
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
et al: K-ras mutations and benefit from cetuximab in advanced
colorectal cancer. N Engl J Med. 359:1757–1765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Roock W, Claes B, Bernasconi D, et al:
Effects of KRAS, BRAF, NRAS and PIK3CA mutations on the efficacy of
cetuximab plus chemotherapy in chemotherapy-refractory metastatic
colorectal cancer: a retrospective consortium analysis. Lancet
Oncol. 11:753–762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sartore-Bianchi A, Di Nicolantonio F,
Nichelatti M, et al: Multi-determinants analysis of molecular
alterations for predicting clinical benefit to EGFR-targeted
monoclonal antibodies in colorectal cancer. PLoS One. 4:e72872009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Victor T, Du Toit R, Jordaan AM, Bester AJ
and van Helden PD: No evidence for point mutations in codons 12, 13
and 61 of the ras gene in a high-incidence area for esophageal and
gastric cancers. Cancer Res. 50:4911–4914. 1990.PubMed/NCBI
|
|
16
|
Hongyo T, Buzard GS, Palli D, et al:
Mutations of the K-ras and p53 genes in gastric adenocarcinomas
from a high-incidence region around Florence, Italy. Cancer Res.
55:2665–2672. 1995.PubMed/NCBI
|
|
17
|
Liu ZM, Liu LN, Li M, Zhang QP, Cheng SH
and Lu S: Mutation detection of KRAS by high-resolution melting
analysis in Chinese gastric cancer. Oncol Rep. 22:515–520.
2009.PubMed/NCBI
|
|
18
|
Corso G, Velho S, Paredes J, et al:
Oncogenic mutations in gastric cancer with microsatellite
instability. Eur J Cancer. 47:443–451. 2011. View Article : Google Scholar
|
|
19
|
Lee SH, Lee JW, Soung YH, et al: BRAF and
KRAS mutations in stomach cancer. Oncogene. 22:6942–6945. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim IJ, Park JH, Kang HC, et al:
Mutational analysis of BRAF and K-ras in gastric cancers: absence
of BRAF mutations in gastric cancers. Hum Genet. 114:118–120. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oliveira C, Pinto M, Duval A, et al: BRAF
mutations characterize colon but not gastric cancer with mismatch
repair deficiency. Oncogene. 22:9192–9196. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu M, Semba S, Oue N, Ikehara N, Yasui W
and Yokozaki H: BRAF/K-ras mutation, microsatellite instability and
promoter hypermethylation of hMLH1/MGMT in human gastric
carcinomas. Gastric Cancer. 7:246–253. 2004. View Article : Google Scholar
|
|
23
|
Zhao W, Chan TL, Chu KM, Chan AS, Stratton
MR, Yuen ST and Leung SY: Mutations of BRAF and KRAS in gastric
cancer and their association with microsatellite instability. Int J
Cancer. 108:167–169. 2004. View Article : Google Scholar
|
|
24
|
Sasao S, Hiyama T, Tanaka S, Yoshihara M,
Yasui W and Chayama K: Clinicopathologic and genetic
characteristics of gastric cancer in young male and female
patients. Oncol Rep. 16:11–15. 2006.PubMed/NCBI
|
|
25
|
Stella G, Rojas Llimpe F, Barone C, et al:
KRAS and BRAF mutational status as response biomarkers to cetuximab
combination therapy in advanced gastric cancer patients. ASCO Meet
Abstr. 27:e155032009.
|
|
26
|
Li VS, Wong CW, Chan TL, et al: Mutations
of PIK3CA in gastric adenocarcinoma. BMC Cancer. 5:292005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Samuels Y, Wang Z, Bardelli A, et al: High
frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Velho S, Oliveira C, Ferreira A, et al:
The prevalence of PIK3CA mutations in gastric and colon cancer. Eur
J Cancer. 41:1649–1654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bardelli A and Siena S: Molecular
mechanisms of resistance to cetuximab and panitumumab in colorectal
cancer. J Clin Oncol. 28:1254–1261. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kiaris H and Spandidos D: Mutations of ras
genes in human tumors (review). Int J Oncol. 7:413–421.
1995.PubMed/NCBI
|
|
31
|
van Grieken NC, Aoyama T, Chambers PA, et
al: KRAS and BRAF mutations are rare and related to DNA mismatch
repair deficiency in gastric cancer from the East and the West:
Results from a large international multicentre study. Br J Cancer.
108:1495–1501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Davies H, Bignell GR, Cox C, et al:
Mutations of the BRAF gene in human cancer. Nature. 417:949–954.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee SH, Ahn BK, Baek SU and Chang HK: BRAF
mutation in multiple primary cancer with colorectal cancer and
stomach cancer. Gastroenterol Rep (Oxf). 1:70–74. 2013. View Article : Google Scholar
|
|
34
|
Henke LE, Perkins SM, Pfeifer JD, Ma C,
Chen Y, DeWees T and Grigsby PW: BRAF V600E mutational status in
pediatric thyroid cancer. Pediatr Blood Cancer. 61:1168–1172. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Campbell IG, Russell SE, Choong DY, et al:
Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer
Res. 64:7678–7681. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JW, Soung YH, Kim SY, et al: PIK3CA
gene is frequently mutated in breast carcinomas and hepatocellular
carcinomas. Oncogene. 24:1477–1480. 2005. View Article : Google Scholar
|
|
37
|
Simi L, Pratesi N, Vignoli M, et al:
High-resolution melting analysis for rapid detection of KRAS, BRAF
and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol.
130:247–253. 2008. View Article : Google Scholar : PubMed/NCBI
|