Introduction
Abdominal aortic aneurysm (AAA), the most common
form of aortic aneurysm, refers to a localized dilatation of the
abdominal aorta exceeding the normal aortic diameter by >50%
(1). The most severe complication
of AAA is AAA rupture, which is the tenth primary cause of
mortality for American Caucasian males between 65–74 years old
(2). As AAA rupture is fatal
within minutes, in the majority of cases, the patients do not reach
hospital in time to receive treatment. Therefore, the mortality
rate of AAA rupture is ~90% (3).
Thus, there is an urgent requirement for the
development of effective therapies for AAA, and elucidating the
etiology of AAA is important in this development. Chronic tobacco
smoking has been identified as the most important risk factor for
AAA (4,5). In addition to smoking, inherited
susceptibility to AAA has also been identified as a critical
causative factor accounting for this disease (6). The molecular mechanism of AAA has
been widely investigated. In particular, protease inhibitors, such
as metalloproteinase inhibitor-2 and plasminogen activator
inhibitor-1 have been reported to be involved in the development of
AAA from original atherosclerotic plaques (7). β-arrestin-2, a scaffolding protein,
has been suggested to promote AAA formation induced by angiotensin
II in mice in a previous study (8). Additional studies suggested that
Sortilin-1 and microRNA-29b were also involved in the pathogenesis
of AAA (9,10).
Thus, previous studies have made progress in
understanding the pathogenesis of AAA; however, the underlying
mechanisms of AAA remain to be fully elucidated. The current study
aimed to clarify the molecular mechanism of AAA using
bioinformatics techniques. Differentially expressed genes (DEGs)
were identified through analyzing the whole genome gene expression
profiles of 14 AAA samples. Gene ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analyses were conducted
for DEGs, and a protein-protein interaction (PPI) network was
constructed followed by analysis of clusters from the PPI network.
Additionally, important transcription factors (TFs) that regulated
DEGs were investigated. The observations of the current study may
provide novel insights into the pathogenesis of this disease and
aid in the development of future therapeutic approaches to treat
AAA.
Materials and methods
Microarray data
In order to investigate the molecular mechanism of
AAA, the gene expression profile of GSE47472 was obtained from the
National Center of Biotechnology Information (NCBI) Gene Expression
Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). This was used with
the Illumina HumanHT-12 V4.0 Expression BeadChip kit (Illumina,
Inc., San Diego, CA, USA), which had a total of 22 gene chips,
consisting of 14 AAA samples and 8 control aortic samples.
Data preprocessing and DEG analysis
The probe-level data in CEL files was converted into
expression measures using the Log2 transformation following data
preprocessing with the median method using the preprocessCore
package (11). In total, there
were 39,426 probes for 17,393 genes. When there were multiple
probes corresponding to one given gene, expression values of these
probes were averaged.
DEGs between AAA specimens and controls were
identified using the multiple linear regression package limma
(12) in Bioconductor (13), followed by multiple testing
correction using the Bayesian inference method. |logFC| >1 and
false discovery rate (FDR)<0.05 were set as the strict
cutoffs.
GO and KEGG enrichment analysis for
DEGs
For functional annotation of DEGs, the Database for
Annotation, Visualization and Integrated Discovery (DAVID)
(14) was utilized to perform GO
(15) and KEGG (16) analyses for the identified DEGs.
P<0.05 was set as the strict threshold.
PPI network construction
In order to investigate the interactions among
proteins encoded by DEGs and their associations with diseases, the
Search Tool for the Retrieval of Interacting Genes online tool
(17) was applied to construct a
PPI network, which was visualized using Cytoscape 3.0.0 software
(http://www.cytoscape.org/) (18). Hub proteins were identified by
analyzing the node degree. The proteins in the network were defined
as the ‘nodes’ and a pair of interacted proteins were linked by an
edge. The ‘degree’ of a node represents the number of interactions
that node has. The nodes with high degrees were considered as hub
nodes.
Analysis of network clustering
Network clustering was performed using ClusterONE
(19) in Cytoscape software with
P<0.01 as a cutoff. The DAVID online tool was applied to perform
GO and KEGG pathway analysis for network clusters with P<0.05
set as the threshold.
Detection of upstream regulatory
elements
To investigate TFs that modulated the up- and
downregulated DEGs, the upstream regulatory elements for TFs, which
were also termed transcription factor binding sites (TFBS) were
identified using the Whole-Genome rVISTA online tool (http://genome.lbl.gov/cgi-bin/WGRVistaInputCommon.pl).
The length parameter for the gene promoter region was set at 1,000
base pairs upstream of the transcriptional start site with
P<0.0001 as the strict cutoff.
Results
DEG screening
In total, 346 DEGs were selected between 14 AAA and
8 control samples. Among these DEGs, 61% (212) were upregulated,
while 39% (134) were downregulated.
GO and KEGG enrichment analysis
GO functional analysis was performed for up- and
downregulated DEGs. As presented in Table I, up- and downregulated DEGs were
enriched in 6 and 4 GO function terms, respectively. Cellular
sodium ion homeostasis and regulation of RNA metabolic process were
identified to be the most significant GO function terms for the up-
and downregulated DEGs, respectively. Using P<0.05 as the
threshold for significance, no DEG was observed to be significantly
enriched in any sub-pathway of KEGG.
 | Table IGO enrichment analysis of DEGs. |
Table I
GO enrichment analysis of DEGs.
A, Upregulated genes
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| GO:0006883 | Cellular sodium ion
homeostasis | C7, NEDD4L | 0.046429183 | 52.31338009 |
| GO:0003924 | GTPase activity | SEPT5, GBP5, RAB11B,
ERAS, MX1, GNG5 | 0.044676591 | 45.15145139 |
| GO:0005886 | Plasma membrane | TGOLN2, SEPT5, OR2J2,
PCDHGA5, LGR5, TAAR9, OR9A4, S1PR1, DYNLL1, RAET1G, CEACAM6, SV2B,
ERAS, GNG4, SV2C, GNG5, HTR1F, GJD4, ZP1, GBP5, PCDHB5, CACNG8,
PIK3C2A, SLC34A1, MPP5, MRGPRF, IFNAR1, SLC26A3, GRASP, TM4SF5, C7,
GPRC5D, OR1L8, OR2T3, ABI1, CDH8, GORASP1, RAB11B, KCNE1, SERPINC1,
PCSK9, OR2T8, SPRN, IL2RB, DLGAP2, TMEM47, OR51A2, OR8B12,
EMR3 | 0.007482427 | 9.0256518 |
| GO:0016023 | Cytoplasmic membrane
bounded vesicle | SEPT5, TGOLN2,
ANGPTL6, CACNG8, PIK3C2A, DLD, RAB11B, SV2B, VGF, SV2C,
ARHGDIB | 0.032382066 | 33.93883034 |
| GO:0030136 | Clathrin-coated
vesicle | SEPT5, TGOLN2,
PIK3C2A, SV2B, SV2C | 0.034758195 | 35.95313043 |
| GO:0031988 | Membrane bounded
vesicle | SEPT5, TGOLN2,
ANGPTL6, CACNG8, PIK3C2A, DLD, RAB11B, SV2B, VGF, SV2C,
ARHGDIB | 0.039021284 | 39.42595138 |
B, Downregulated
genes
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| GO:0051252 | Regulation of RNA
metabolic process | ZFP36, ZNF584, HSFX2,
ETV7, HOXA13, RFX7, SIX3, ZNF230, PA2G4, FOXF1, NR2F6, HOXA10,
ZNF462, PER3, CARM1, TCF3, NCOR1, ZNF257 | 0.046908154 | 52.01470738 |
| GO:0006350 | Transcription | ZNF584, HSFX2, ETV7,
SNAPC2, HOXA13, ZNF507, PPP1R10, ZNF230, PA2G4, NCOA5, FOXF1,
NR2F6, HOXA10, ZNF462, PER3, CARM1, TCF3, ZNF575, NCOR1,
ZNF257 | 0.049224445 | 53.76642617 |
| GO:0043565 | Sequence-specific DNA
binding | HSFX2, ETV7,
HOXA13, FOXF1, SIX3, HOXA10, NR2F6, TCF3, NCOR1 | 0.035160922 | 37.25923349 |
| GO:0003677 | DNA binding | H1F0, ZFP36,
ZNF584, HSFX2, ETV7, SNAPC2, HOXA13, ZNF507, RFX7, SIX3, PPP1R10,
ZNF230, XPA, PA2G4, FOXF1, NR2F6, HOXA10, ZNF462, TCF3, ZNF575,
NCOR1, ZNF257 | 0.046429791 | 46.16041287 |
PPI network construction
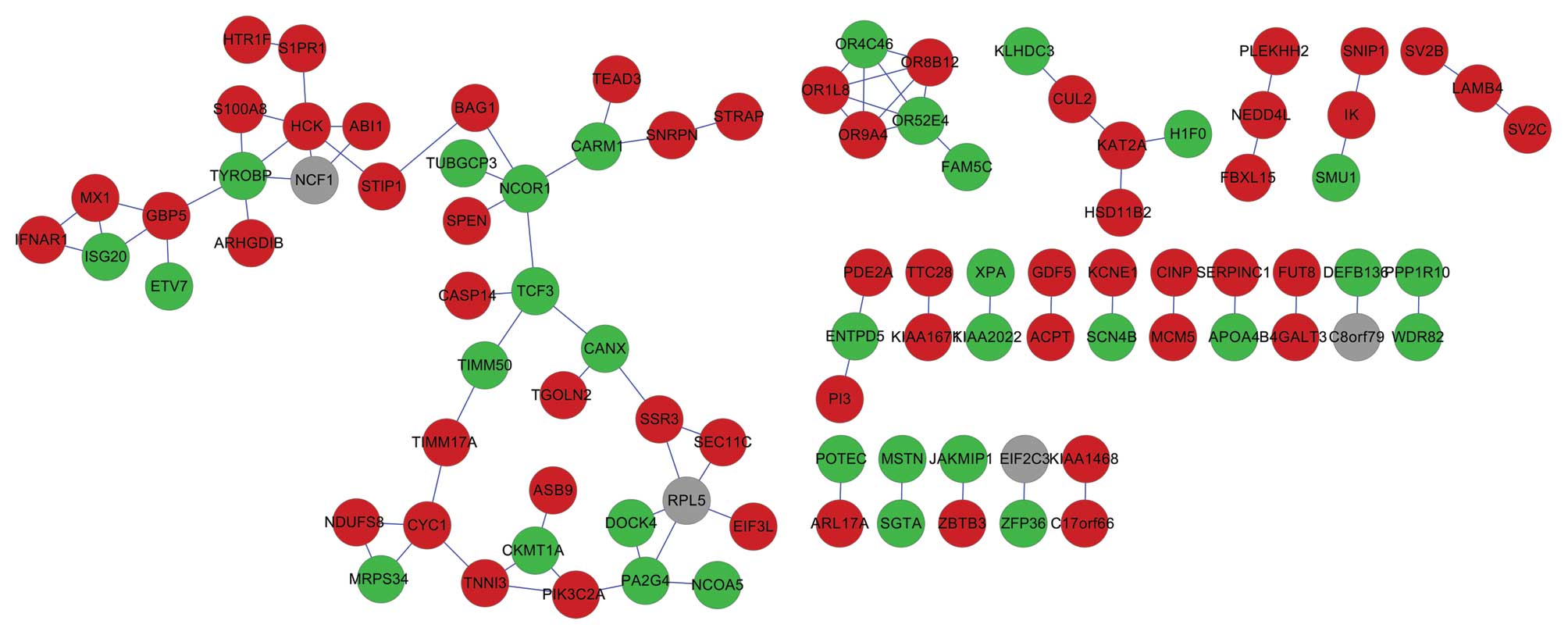
As demonstrated in Fig
1, 88 interactions were identified among proteins in the PPI
network. Proteins with a degree >4 consisted of tyrosine-protein
kinase (HCK), ribosomal protein L5 (RPL5), olfactory receptor 52E4
(OR52E4), TYRO protein tyrosine kinase-binding protein (TYROBP) and
nuclear receptor co-repressor 1 (NCOR1).
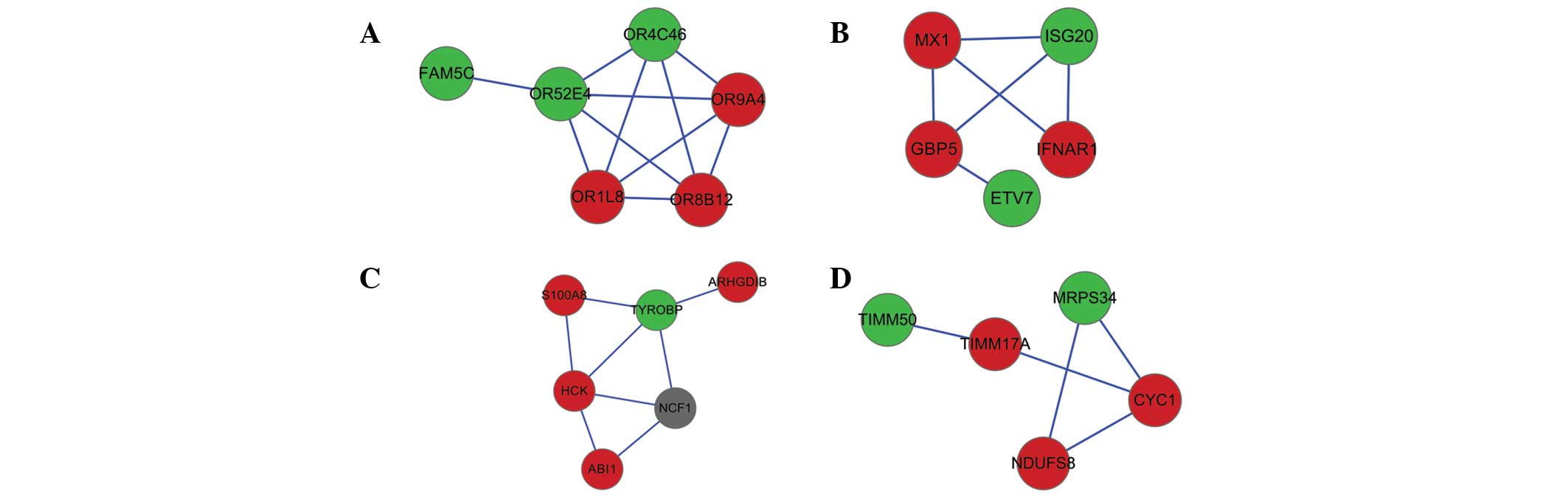
Network cluster analysis
Four network clusters were identified from the PPI
network (Fig. 2). Among the four
clusters, cluster 1 included OR52E4, and cluster 3 included HCK and
TYROBP. The results of the GO enrichment analysis for the 4
clusters are presented in Table
II. Cluster 1 was identified to be most significantly enriched
in the plasma membrane and sensory perception of smell. A number of
olfactory receptor genes were included in cluster 1. The most
significant GO terms for cluster 2 and 3 were response to virus and
defense response, respectively. Interferon-induced GTP-binding
protein (MX1), interferon-α/β receptor (IFNAR) and
interferon-stimulated gene 20 kDa protein (ISG20) were enriched in
cluster 2. In addition, cluster 4 was enriched in
mitochondria-associated functions. As presented in Table III, for cluster 1, the olfactory
transduction subpathway was the most significantly enriched KEGG
subpathway. DEGs including NADH dehydrogenase (ubiquinone)
iron-sulfur protein 8 (NDUFS8) and cytochrome c1 (CYC1) in cluster
4 were observed to be enriched in the oxidative phosphorylation
subpathway.
 | Table IIGO enrichment analysis for four
clusters. |
Table II
GO enrichment analysis for four
clusters.
A, Cluster 1
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| GO:0005886 | Plasma
membrane | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
2.91×10−2 | 10.65718378 |
| GO:0007608 | Sensory perception
of smell | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
4.95×10−6 | 0.00285105 |
| GO:0007606 | Sensory perception
of chemical stimulus | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
7.48×10−6 | 0.004306871 |
| GO:0007600 | Sensory
perception | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
6.08×10−5 | 0.034971424 |
| GO:0050890 | Cognition | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
9.59×10−5 | 0.055165167 |
| GO:0007186 | G-protein coupled
receptor protein signaling pathway | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
2.21×10−4 | 0.126911001 |
| GO:0050877 | Neurological system
process | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
2.96×10−4 | 0.170141842 |
| GO:0007166 | Cell surface
receptor linked signal transduction | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 | 0.001573052 | 0.901913236 |
| GO:0004984 | Olfactory receptor
activity | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
1.20×10−6 |
1.20×10−4 |
B, Cluster 2
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| GO:0009615 | Response to
virus | MX1, IFNAR1,
ISG20 |
3.82×10−4 | 0.354852933 |
C, Cluster 3
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| GO:0006952 | Defense
response | S100A8, NCF1, HCK,
TYROBP |
8.73×10−4 | 0.795749167 |
| GO:0006968 | Cellular defense
response | NCF1, TYROBP |
2.23×10−2 | 18.6793053 |
| GO:0005739 | Mitochondrion | MRPS34, TIMM17A,
NDUFS8, CYC1, TIMM50 |
5.20×10−5 | 0.044320277 |
| GO:0005743 | Mitochondrial inner
membrane | TIMM17A, NDUFS8,
CYC1, TIMM50 |
5.34×10−5 | 0.045471679 |
| GO:0019866 | Organelle inner
membrane | TIMM17A, NDUFS8,
CYC1, TIMM50 |
6.63×10−5 | 0.056473574 |
D, Cluster 4
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| GO:0031966 | Mitochondrial
membrane | TIMM17A, NDUFS8,
CYC1, TIMM50 |
1.14×10−4 | 0.096745997 |
| GO:0005740 | Mitochondrial
envelope | TIMM17A, NDUFS8,
CYC1, TIMM50 |
1.37×10−4 | 0.116223644 |
| GO:0044429 | Mitochondrial
part | TIMM17A, NDUFS8,
CYC1, TIMM50 |
3.88×10−4 | 0.329680741 |
| GO:0031967 | Organelle
envelope | TIMM17A, NDUFS8,
CYC1, TIMM50 |
4.38×10−4 | 0.372446101 |
| GO:0031975 | Envelope | TIMM17A, NDUFS8,
CYC1, TIMM50 |
4.42×10−4 | 0.376016181 |
| GO:0044455 | Mitochondrial
membrane part | TIMM17A, NDUFS8,
TIMM50 |
5.62×10−4 | 0.47771995 |
| GO:0031090 | Organelle
membrane | TIMM17A, NDUFS8,
CYC1, TIMM50 |
2.4×10−3 | 1.987624717 |
| GO:0005744 | Mitochondrial inner
membrane presequence translocase complex | TIMM17A,
TIMM50 |
3.7×10−3 | 3.15011107 |
| GO:0070469 | Respiratory
chain | NDUFS8, CYC1 |
2.3×10−2 | 18.17151041 |
| GO:0007005 | Mitochondrion
organization | TIMM17A, NDUFS8,
TIMM50 |
3.08×10−4 | 0.286135591 |
| GO:0022900 | Electron transport
chain | NDUFS8, CYC1 |
2.5×10−2 | 21.04388668 |
 | Table IIIKEGG subpathway enrichment analysis
for 4 clusters. |
Table III
KEGG subpathway enrichment analysis
for 4 clusters.
A, Cluster 1
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| hsa04740 | Olfactory
transduction | OR9A4, OR1L8,
OR8B12, OR52E4, OR4C46 |
3.04×10−5 | 0.003040948 |
B, Cluster 4
|
|---|
| ID | Name | Gene symbol | P-value | FDR |
|---|
| hsa05012 | Parkinson’s
disease | NDUFS8, CYC1 | 0.025172075 | 10.49506004 |
| hsa00190 | Oxidative
phosphorylation | NDUFS8, CYC1 | 0.025565388 | 10.6520097 |
| hsa05010 | Alzheimer’s
disease | NDUFS8, CYC1 | 0.032055064 | 13.21121831 |
| hsa05016 | Huntington’s
disease | NDUFS8, CYC1 | 0.03539823 | 14.50736559 |
Identification of TFBS and TFs
For the upregulated DEGs, the important TFs
identified included early growth response protein 1 (EGR-1) and
Myc. In the downregulated DEGs however, activating transcription
factor 5 (ATF5), specificity protein (SP)1:SP3, E2F transcription
factor 4 (E2F4) and TFII-I were the critical TFs identified.
Discussion
In the current study, a total of 212 upregulated
DEGs and 134 downregulated DEGs were identified between AAA samples
and control samples. HCK, OR52E4 and TYROBP were identified as hub
nodes in the PPI network. Four clusters from the PPI network were
validated in terms of GO functions and KEGG pathways individually.
In addition, important TFs were identified for up- and
downregulated DEGs.
Of the four clusters from the PPI network, cluster 1
was identified to be enriched in sensory perception of smell and
the olfactory transduction subpathway. Consistently, OR52E4, which
was part of cluster 1, is a member of the family of olfactory
receptor proteins (20). It has
been reported that tobacco exposure is able to impair olfactory
function in a dose-dependent manner (21), thus given that tobacco smoking is
an important risk factor for AAA, it is not surprising that
olfactory receptor genes, including OR52E4, were enriched in
cluster 1.
According to the result of GO function analysis
cluster 2 was enriched in response to viruses and MX1, IFNAR and
ISG20 were all included in cluster 2. Among these three DEGs, MX1
and IFNAR were observed to be upregulated, while ISG20 was
downregulated. Previous studies have demonstrated that MX1, IFNAR
and ISG20 are associated with the immune response to viruses
(22–24). Accordingly, immune and inflammatory
responses have been identified to be critical in AAA formation
(25). These previous studies
further confirm the role of the immune response in the pathogenesis
of AAA.
HCK is an enzyme encoded by the HCK gene and is a
member of the Src family of tyrosine kinases. HCK has been
demonstrated to be involved in the migration and degranulation of
neutrophils (26). TYROBP is an
adaptor protein encoded by the TYROBP gene, which has been
previously observed to be abnormally expressed in AAA (27). In agreement with this, the current
study demonstrated that TYROBP and HCK were enriched in cluster
3.
Various studies have established that oxidative
stress may promote inflammation in the pathogenesis of AAA
(28–30). In addition, mitochondrial-dependent
apoptosis is reported to promote AAA formation in rodent
experimental models (31). In
accordance with this, the present study demonstrated that cluster 4
was enriched in mitochondria-associated functions and the oxidative
phosphorylation subpathway. A variety of genes were enriched in
cluster 4, including NDUFS8 and CYC1. NDUFS8 is encoded by the
NDUFS8 gene and is a subunit of mitochondrial NADH (32). Consistently, NDUFS8 has been
previously observed to be involved in oxidative phosphorylation
(33). CYC1 is a heme protein
encoded by the CYC1 gene, and cytochrome c is a critical
component of the electron transport chain in mitochondria (34). These studies are in agreement with
the results of the current study, further suggesting the importance
of mitochondria and oxidative phosphorylation in AAA.
The current study demonstrated that EGR-1 and Myc
were important TFs modulating upregulated DEGs, while ATF5 and
SP1:SP3 were critical TFs for downregulated DEGs. EGR-1 has been
identified to be an important TF and a tumor suppressor gene in
addition (35). EGR-1 has been
observed to be involved in thrombus formation and the inflammatory
pathogenesis of AAA (36). The Myc
protein is a member of the Myc family of transcription factors and
has been demonstrated to be involved in regulating cell apoptosis
and proliferation (37). The
current study hypothesized that Myc may promote cell proliferation
in AAA. In addition, SP1 is a protein encoded by the SP1 gene and
belongs to the SP/Krüppel-like factor family of transcription
factors (38). SP1 has been
observed to modulate inflammation associated with AAA by increasing
the expression levels of cyclooxy-genase-2 (39). ATF5, encoded by ATF5 gene, belongs
to the ATF/cyclic adenosine monophosphate response-element binding
(CREB) protein family (40). CREB
has been reported to serve a role in modulating the apoptosis and
proliferation of vascular smooth muscle cells (41,42).
In conclusion, the current study indicated critical
roles of the olfactory transduction, mitochondria and oxidative
phosphorylation subpathways, and suggested the importance of the
immune response in the pathogenesis of AAA. In addition, crucial
TFs, including EGR-1, Myc, ATF5 and SP1:SP3, were identified and
were suggested as potential treatment targets for this disease.
Thus the present study aided in the further investigation of the
pathogenesis of AAA. Further experimental studies are required in
order to validate the results of the current study.
References
|
1
|
Schermerhorn M: A 66-year-old man with an
abdominal aortic aneurysm: Review of screening and treatment. JAMA.
302:2015–2022. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Upchurch GR Jr and Schaub TA: Abdominal
aortic aneurysm. Am Fam Physician. 73:1198–1204. 2006.PubMed/NCBI
|
|
3
|
Shimazaki Y and Ueda H: Abdominal Aortic
Aneurysm. Interdisciplinary Concepts in Cardiovascular Health
Volume III: Cardiovascular events. Wakabayashi I and Groschne K:
Springer; Cham, Switzerland: pp. 161–179. 2014, View Article : Google Scholar
|
|
4
|
Sode BF, Nordestgaard BG, Grønbæk M and
Dahl M: Tobacco smoking and aortic aneurysm: Two population-based
studies. Int J Cardiol. 167:2271–2277. 2013. View Article : Google Scholar
|
|
5
|
Kent KC, Zwolak RM, Egorova NN, Riles TS,
Manganaro A, Moskowitz AJ, Gelijns AC and Greco G: Analysis of risk
factors for abdominal aortic aneurysm in a cohort of more than 3
million individuals. J Vasc Surg. 52:539–548. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kuivaniemi H, Shibamura H, Arthur C, et
al: Familial abdominal aortic aneurysms: Collection of 233
multiplex families. J Vasc Surg. 37:340–345. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Defawe OD, Colige A, Lambert CA, et al:
TIMP-2 and PAI-1 mRNA levels are lower in aneurysmal as compared to
athero-occlusive abdominal aortas. Cardiovasc Res. 60:205–213.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Trivedi DB, Loftin CD, Clark J, Myers P,
DeGraff LM, Cheng J, Zeldin DC and Langenbach R: β-Arrestin-2
deficiency attenuates abdominal aortic aneurysm formation in mice.
Circ Res. 112:1219–1229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jones GT, Bown MJ, Gretarsdottir S, et al:
A sequence variant associated with sortilin-1 (SORT1) on 1p13.3 is
independently associated with abdominal aortic aneurysm. Hum Mol
Genet. 22:2941–2947. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maegdefessel L, Azuma J, Toh R, et al:
Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm
development. J Clin Invest. 122:497–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bolstad BM: preprocessCore: A collection
of pre-processing functions. R package, version 1.28.0. 2013,
Available at: http://bioconductor.org/packages/release/bioc/html/preprocessCore.html.
|
|
12
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gentleman RC, Carey VJ, Bates DM, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashburner M, Ball CA, Blake JA, et al Gene
ontology: Tool for the unification of biology. The Gene Ontology
Consortium. Nat Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arakawa K, Kono N, Yamada Y, Mori H and
Tomita M: KEGG-based pathway visualization tool for complex omics
data. In Silico Biol. 5:419–423. 2005.PubMed/NCBI
|
|
17
|
Franceschini A, Szklarczyk D, Frankild S,
et al: STRING v9.1: Protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res. 41(Database
issue): D808–D815. 2013. View Article : Google Scholar :
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nepusz T, Yu H and Paccanaro A: Detecting
overlapping protein complexes in protein-protein interaction
networks. Nat Methods. 9:471–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malnic B, Godfrey PA and Buck LB: The
human olfactory receptor gene family. Proc Natl Acad Sci USA.
101:2584–2589. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Katotomichelakis M, Balatsouras D,
Tripsianis G, Davris S, Maroudias N, Danielides V and Simopoulos C:
The effect of smoking on the olfactory function. Rhinology.
45:273–280. 2007.PubMed/NCBI
|
|
22
|
Haller O, Staeheli P and Kochs G:
Interferon-induced Mx proteins in antiviral host defense.
Biochimie. 89:812–818. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Theofilopoulos AN, Baccala R, Beutler B
and Kono DH: Type I interferons (alpha/beta) in immunity and
autoimmunity. Annu Rev Immunol. 23:307–336. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schoggins JW and Rice CM:
Interferon-stimulated genes and their antiviral effector functions.
Curr Opin Virol. 1:519–525. 2011. View Article : Google Scholar
|
|
25
|
Kwon YS and Kim DK: Pathogenesis of
abdominal aortic aneurysm. J Korean Soc Vasc Surg. 21:78–83.
2005.In Korean.
|
|
26
|
Stanglmaier M, Warmuth M, Kleinlein I,
Reis S and Hallek M: The interaction of the Bcr-Abl tyrosine kinase
with the Src kinase Hck is mediated by multiple binding domains.
Leukemia. 17:283–289. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lillvis JH, Kyo Y, Tromp G, et al:
Analysis of positional candidate genes in the AAA1 susceptibility
locus for abdominal aortic aneurysms on chromosome 19. BMC Med
Genet. 12:142011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McCormick ML, Gavrila D and Weintraub NL:
Role of oxidative stress in the pathogenesis of abdominal aortic
aneurysms. Arterioscler Thromb Vasc Biol. 27:461–469. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yajima N, Masuda M, Miyazaki M, Nakajima
N, Chien S and Shyy JY: Oxidative stress is involved in the
development of experimental abdominal aortic aneurysm: A study of
the transcription profile with complementary DNA microarray. J Vasc
Surg. 36:379–385. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kaneko H, Anzai T, Morisawa M, et al:
Resveratrol prevents the development of abdominal aortic aneurysm
through attenuation of inflammation, oxidative stress, and
neovascularization. Atherosclerosis. 217:350–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sinha I, Sinha-Hikim AP, Hannawa KK, Henke
PK, Eagleton MJ, Stanley JC and Upchurch GR Jr:
Mitochondrial-dependent apoptosis in experimental rodent abdominal
aortic aneurysms. Surgery. 138:806–811. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Procaccio V and Wallace DC: Late-onset
Leigh syndrome in a patient with mitochondrial complex I NDUFS8
mutations. Neurology. 62:1899–1901. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Janssen RJ, van den Heuvel LP and Smeitink
JA: Genetic defects in the oxidative phosphorylation (OXPHOS)
system. Expert Rev Mol Diagn. 4:143–156. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nord FF and Green DE: Electron transport
and oxidative phosphorylation. Adv Enzymol Relat Areas Mol Biol.
21:732006.
|
|
35
|
Gómez-Martín D, Díaz-Zamudio M,
Galindo-Campos M and Alcocer-Varela J: Early growth response
transcription factors and the modulation of immune response:
Implications towards autoimmunity. Autoimmun Rev. 9:454–458. 2010.
View Article : Google Scholar
|
|
36
|
Shin IS, Kim JM, Kim KL, Jang SY, Jeon ES,
Choi SH, Kim DK, Suh W and Kim YW: Early growth response factor-1
is associated with intraluminal thrombus formation in human
abdominal aortic aneurysm. J Am Coll Cardiol. 53:792–799. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pelengaris S, Rudolph B and Littlewood T:
Action of Myc in vivo - proliferation and apoptosis. Curr Opin
Genet Dev. 10:100–105. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Safe S and Abdelrahim M: Sp transcription
factor family and its role in cancer. Eur J Cancer. 41:2438–2448.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu Q, Ji YS and Schmedtje JF Jr: Sp1
increases expression of cyclooxygenase-2 in hypoxic vascular
endothelium. Implications for the mechanisms of aortic aneurysm and
heart failure. J Biol Chem. 275:24583–24589. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vinson C, Myakishev M, Acharya A, Mir AA,
Moll JR and Bonovich M: Classification of human B-ZIP proteins
based on dimerization properties. Mol Cell Biol. 22:6321–6335.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tokunou T, Shibata R, Kai H, et al:
Apoptosis induced by inhibition of cyclic AMP response
element-binding protein in vascular smooth muscle cells.
Circulation. 108:1246–1252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ono H, Ichiki T, Fukuyama K, Iino N,
Masuda S, Egashira K and Takeshita A: cAMP-response element-binding
protein mediates tumor necrosis factor-alpha-induced vascular
smooth muscle cell migration. Arterioscler Thromb Vasc Biol.
24:1634–1639. 2004. View Article : Google Scholar : PubMed/NCBI
|