Introduction
Primary hepatocellular carcinoma (HCC), which occurs
in the liver parenchymal cells or the epithelial cells of the
intrahepatic bile duct, is one of the most clinically common and
damaging types of malignant cancer. The high rate of malignancy
that is associated with HCC is associated with reduced survival
rates in patients (1). In
particular, the 5-year survival rate for HCC is ~3–5%, which
corresponds to the third highest mortality rate of all types of
cancer worldwide (2,3). HCC is primarily caused by hepatitis B
virus (HBV) infection. Previous in-depth and extensive
investigations of HBV-induced HCC have been performed to identify
the specific pathogenic mechanisms by which HBV can cause HCC;
however, these mechanisms remain to be fully elucidated (4–6). In
our preliminary study, microarray screening was used to identify
genes with differences in expression between cancer tissues and
paracancerous tissues in patients with HCC and histories of chronic
HBV infection (7). From this
screen, kinesin family member 4A (KIF4A) was found to be highly
expressed in the tumor tissues relative to the paracancerous
tissues. The KIF4A gene maps to Xq13.1 in the human genome and
encodes a 140-kDa protein, which is composed of 1,232 amino acids
(8–10). KIF4A, which is located
predominantly in the cytoplasm and nuclei of cells, is a motor
protein that has been closely associated with the intracellular
movement of organelles, mitosis and meiosis, the growth and
development of tissues and organs, neuronal development and signal
transduction (11–13). The present study aimed to evaluate
the regulatory effects of HBV on the expression of KIF4A and to
examine the molecular mechanisms underlying these effects.
Materials and methods
Materials
Samples of paracancerous and cancerous tissue were
obtained from three male patients (45, 59 and 64 years old) with
HCC and histories of chronic HBV infection, who underwent surgery
in Zhongnan Hospital at Wuhan University (Wuhan, China). The HepG2
human liver cancer cell line was provided by the China Center for
Type Culture Collection at Wuhan University. The pKIF4A-Luc
plasmid, containing the luciferase gene driven by the KIF4A gene
promoter, and the pHBV1.3 plasmid, which produces an infectious
clone of HBV, were constructed in The State Key Laboratory of
Virology, College of Life Sciences, Wuhan University (Wuhan,
China). TRIzol, a RNA extraction reagent and Lipofectamine 2000, a
transfection reagent, were purchased from Invitrogen Life
Technologies (Carlsbad, CA, USA). M-MLV reverse tran-scriptase (200
U/µl) and luciferin, a luciferase substrate, were purchased
from Promega Corporation (Madison, WI, USA). A luminometer was
purchased from Bio-Rad Laboratories, Inc. (TD-20/20; Hercules, CA,
USA). The polyclonal rabbit anti-human anti-KIF4A antibody (1:600;
ABU010) was purchased from Sigma-Aldrich (St. Louis, CA, USA), and
the horseradish peroxide (HRP)-conjugated goat anti-rabbit
secondary antibody (1:5,000; ab6721) was purchased from Abcam
(Cambridge, UK). The study was approved by the Ethics Committee of
Renmin Hospital of Wuhan University (Wuhan, China) and informed
consent was obtained from the patients.
Cell culture and transfection
The HepG2 cells were cultured in RPMI-1640 medium
(Gibco Life Technologies, Carlsbad, CA, USA) containing 10% fetal
bovine serum (Invitrogen Life Technologies) and two antibiotics
(100 U/ml penicillin and 100 µg/ml streptomycin; Gibco Life
Technologies) in an incubator at 37°C and 5% CO2. HepG2
cells were also co-transfected with an empty vector, pBlue ks and
pKIF4A Luc. The transfection solutions were created by diluting 0.6
µg pHBV1.3 or pBlue ks and 0.2 µg pKIF4A Luc and 2
µl Lipofectamine 2000 transfection reagent (Invitrogen Life
Technologies) into 50 µl serum and antibiotic free RPMI-1640
(Gibco Life Technologies) culture medium. Following incubation of
these solutions at room temperature for 20 min, the appropriate
transfection solutions were transferred to the 24-well plates, and
the cells were placed in a 5% CO2 incubator for
continued culture. Prior to transfection, the HepG2 cells were
seeded onto 24- (density, ~ 1 × 105 cells/well) or
6-well (density, ~5×105 cells/well) plates. Cell
transfection was performed as follows: Transfection solutions were
created by diluting 0.8 µg plasmid DNA and 2 µl
Lipofectamine 2000 transfection reagent into 50 µl serum-
and antibiotic-free RPMI-1640 culture medium (24-well plate), or by
diluting 4 µg plasmid DNA and 6 µl Lipofectamine 2000
transfection reagent into 100 µl serum- and antibiotic-free
RPMI-1640 culture medium (6-well plate). Following incubation of
these solutions at room temperature for 20 min, the appropriate
transfection solutions were transferred to the cell culture plates,
and the cells were placed in a 5% CO2 incubator for
continued culture.
Luciferase assay
The HepG2 cells were cultured for 48 h at 37°C after
transfection. The culture medium was then removed, and the cells
were washed with phosphate-buffered saline (PBS; O'BioLab, Beijing,
China). Lysis buffer (Beijing BLKW Biotech Co., Ltd., Beijing,
China) was added to lyse the cells (100 µl/well). Following
complete lysis of the cells, 50 µl of the cell lysate was
mixed with 50 µl luciferin and a luminometer was used for
determination of the optical density. Each experiment was repeated
three times.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The total RNA was first extracted from the tissue
samples or the HepG2 cells using TRIzol, followed by treatment with
DNase I (5 U/µl; Shanghai Haoran Biotechnology Co., Ltd.,
Shanghai, China). Subsequently, reverse transcriptase from the
GoldScript One-Step RT-PCR kit (Invitrogen Life Technologies) was
used to synthesize the complementary DNA (cDNA) using 1 µg
of each RNA sample. Using the synthesized cDNA as a template, qPCR
amplification was performed using the GeneAmp 9700 PCR System
(Applied Biosystems Life Technologies, Foster City, CA, USA) and
the following primers from Jingmei Biotech Co., Ltd. (Shenzhen,
China), which were designed to detect the KIF4A gene: Forward
5′-TCAAGCAGAAACTGACCCTC-3′ and reverse 5′-CGTTCAACAGTGCCCAAG-3′.
Based on standard amplification procedures, the PCR cycling
conditions involved 25 cycles of 94°C for 45 sec, 56°C for 45 sec
and 72°C for 45 sec. β-actin was used as an internal control, and 5
µl each PCR product was analyzed using 1% agarose gel
(Jingmei Biotech Co., Ltd.) electrophoresis.
Western blot analysis
The tissues and HepG2 cells were lysed with 1X lysis
buffer, sonicated on ice with the Ultrasonic Instrument (1 sec/ml;
5 times at setting 5; Shanghai Hao Chong Instrument Co., Ltd.,
Shanghai, China) and centrifuged at 13,000 × g for 5 min at 4°C.
The concentrations of protein in the sample supernatants were
quantified using the Coomassie Brilliant Blue G-250 (Beinuo Biotech
Co., Ltd., Shanghai, China) method. For each western blot, 30
µg protein from each sample was mixed with an equal volume
of 5X loading buffer (0.5 mol/l Tris·HCl, 2.5 ml; DTT, 0.39 g; SDS,
0.5 g; bromophenol blue, 0.025 g; glycerol, 2.5 ml), boiled for 5
min in a 100°C water bath, and separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis on a 12% gel. Following
electrophoresis, the proteins were transferred onto a
nitrocellulose membrane (Sigma-Aldrich). This membrane was blocked
for 2 h at room temperature in PBS Tween-20 (PBST; O'BioLab)
containing 5% nonfat milk, incubated with 1:600 polyclonal KIF4A
antibody for 2 h at room temperature, washed three times with PBST,
incubated with 1:5,000 HRP-conjugated goat anti-rabbit secondary
antibody for 1 h, and then washed four times with PBST. An enhanced
chemiluminescence system (Biomart, Beijing, China) was used to
examine the chromogenic signal from the membrane.
Statistical analysis
The SPSS 13.0 software package (SPSS, Inc., Chicago,
IL, USA) was used to process and analyze the experimental data. The
data are presented as the mean ± standard deviation, and
differences between the groups were compared using Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
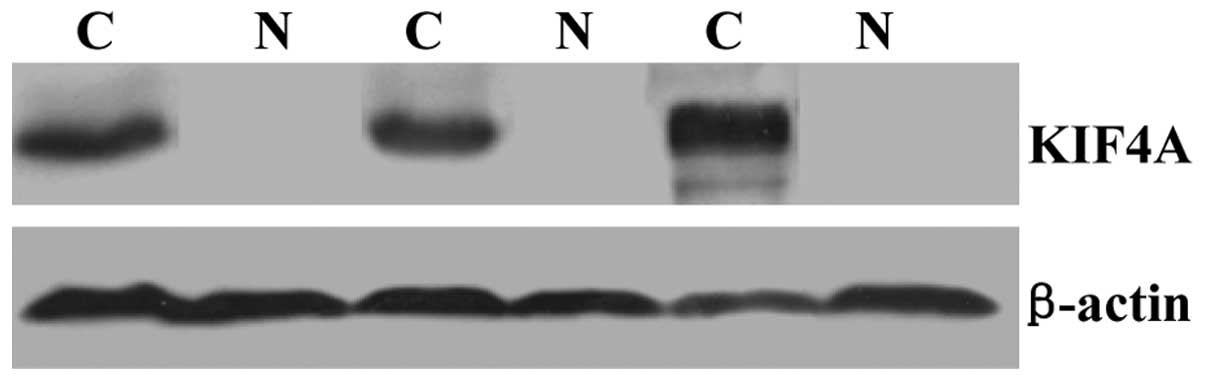
Elevated expression levels of KIF4A in
cancer tissues of patients with HCC
In our previous study, DNA microarrays were used to
screen for differentially expressed genes in the paracancerous and
cancerous tissues of patients with HCC, who presented with
histories of chronic HBV infection (7). This screen revealed increased
expression levels of KIF4A in the cancerous tissues, compared with
the paracancerous tissues. To further confirm these results, the
present study analyzed paracancerous and cancerous tissue samples
from three patients with HCC using RT-qPCR and western blotting, to
determine the differences in the mRNA and protein expression levels
of KIF4A, respectively. The results of the RT-qPCR and western blot
analysis revealed almost no expression of KIF4A in the
paracancerous tissues, however significantly higher expression
levels of KIF4A were observed in cancerous tissues (Figs. 1 and 2).
Activation of the KIF4A gene promoter by
HBV
To investigate the effect of HBV on the expression
of KIF4A, the HepG2 cells were co-transfected with a plasmid
encoding the pHBV1.3 HBV infectious clone and pKIF4A-Luc, in which
the luciferase gene is driven by the KIF4A gene promoter. In
addition, HepG2 cells were also transfected with an empty vector,
pBlue-ks, and were included as a control group. The activities of
luciferase in each group were subsequently measured to determine
the activation of the KIF4A gene promoter by pHBV1.3. The results
demonstrated that transfection with the pBlue-ks empty vector
produced 157.5±12.4 RLU/µg protein of luciferase activity,
whereas transfection with the pHBV1.3 vector produced 694.6±36.5
RLU/µg protein of luciferase activity (Fig. 3). Statistical analysis confirmed
the statistical significance of this difference (P<0.001),
indicating that HBV activated the KIF4A gene promoter in the HepG2
cells.
Different concentrations of pHBV1.3 or pBlue-ks (0,
0.2, 0.4, 0.6 or 0.8 µg/ml) were co-transfected with the
pKIF4A-Luc construct to determine the concentration dependence of
the expression of KIF4A on pHBV1.3 in the HepG2 cells. The
luciferase activity experiments demonstrated that luciferase
activity increased as the concentration of pHBV1.3 increased. In
particular, at pHBV1.3 concentrations of 0, 0.2, 0.4, 0.6 and 0.8
µg/ml, luciferase activities of 126.8±13.4, 219.8±16.7,
387.6±21.5, 586.5±228.9 and 657.6±35.5 RLU/µg protein were
observed, respectively. By contrast, transfection with different
concentrations of empty vector produced little change in luciferase
activity, as pBlue-ks concentrations of 0.2, 0.4, 0.6 and 0.8
µg/ml resulted in luciferase activities of 123.6±13.8,
131.8±14.6, 129.7±13.5, 135.3±13.4 and 127.1±12.7 RLU/µg
protein, respectively. Statistical analysis demonstrated that
transfection with pHBV1.3 produced significantly higher levels of
luciferase activity, compared with transfection with the empty
vector (P<0.001), and that activation of the KIF4A gene promoter
by HBV was dose-dependent (Fig.
4).
Upregulation of the mRNA expression of
KIF4A by HBV
To investigate the effect of HBV on the mRNA
expression of KIF4A, the HepG2 cells were transfected with
different concentrations of pHBV1.3, an infectious clone of HBV.
RT-qPCR was used to detect changes in the mRNA expression levels of
KIF4A 48 h after transfection. The results indicated that HBV
upregulated the mRNA expression of KIF4A, and that the mRNA
expression of KIF4A increased as the concentration of pHBV1.3
increased (Fig. 5). These findings
were consistent with those of the luciferase activity analysis,
described above.
Upregulation of the protein expression of
KIF4A by HBV
To examine the effects of HBV on the protein
expression of KIF4A, different concentrations of pHBV1.3 were used
to transfect the HepG2 cells. Western blot anaylsis was used to
measure the protein expression of KIF4A 48 h after transfection.
The results revealed that HBV increased the protein expression of
KIF4A in the HepG2 cells in a dose-dependent manner (Fig. 6).
Discussion
HBV infection is recognized as one of the major
causes of primary HCC. Globally, 60–80% of primary HCC cases are
caused by HBV, and ~0.4–0.6% of patients with chronic HBV
infections are diagnosed with HCC each year (4,5,14).
To examine the pathogenic and carcinogenic mechanisms of HBV, our
prelimanry study used cDNA microarrays to screen for genes, which
were expressed at different levels in the cancerous and
paracancerous tissues of patients with HCC. The results of this
examination indicated that KIF4A was expressed at significantly
higher levels in cancerous tissues than in paracancerous
tissues.
As DNA microarrays can produce false positive
results, the present study obtained cancerous and paracancerous
tissue samples from three patients with HCC and histories of
chronic HBV infection to confirm the KIF4A-associated results.
RT-qPCR and western blotting were used to examine the expression of
KIF4A, and the results indicated that KIF4A was expressed at
significantly higher levels in the cancerous tissues, comapared
with the paracancerous tissue. These findings were consistent with
the microarray data.
Following transfection into the HepG2 cells,
pHBV1.3, an infectious clone of HBV, can induce the packaging of
mature viral particles (15,16).
To further examine the molecular mechanisms underlying the
upregulation of KIF4A by HBV, the present study constructed
pKIF4A-Luc, which contained a luciferase reporter gene driven by
the KIF4A gene promoter. The HepG2 cells were co-transfected with
pKIF4A-Luc and pHBV1.3, and luciferase reporter assays were
performed to determine the role of HBV in regulating the KIF4A gene
promoter. Notable, these assays used luciferin as the substrate,
which is oxidized in a luciferase-catalyzed reaction to produce
bioluminescence that can be quantified using a luminometer
(17). The regulatory effects were
evaluated based on the levels of luciferase activity following the
addition of different quantities of pHBV1.3. The results
demonstrated that pHBV1.3 activated the KIF4A gene promoter in a
dose-dependent manner. The RT-qPCR and western blotting results
revealed that HBV induced the expression of KIF4A in a
dose-dependent manner at the mRNA and protein levels, respectively.
These findings were consistent with the luciferase activity
results.
KIF4A is a multifunctional protein. Previous studies
have suggested that KIF4A is involved in responses to DNA damage
and DNA repair pathways (12). In
particular, KIF4A can interact with the BRCA2 breast cancer
susceptibility gene to regulate the BRCA2/Rad51 pathway, which is
involved in the response to DNA damage (18). Thus, KIF4A may be important in the
incidence and development of breast cancer. KIF4A is overexpressed
in cervical and lung cancer, downregulated in gastric cancer, and
treatment of non-small cell lung carcinoma cells with specific
siRNA to knockdown the expression of KIF4A results in suppression
of cancer cell growth (11,19,20).
In the present study, elevated expression levels of
KIF4A were observed in tumor tissues from primary tumors of
patients with HCC, who presented with histories of chronic HBV
infection. A possible mechanism for this observation is that, upon
HBV infection, the gene expression of KIF4A is upregulated in liver
tissue, which contributes to liver carcinogenesis. However, the way
in which HBV-dependent regulation of KIF4A is associated with liver
tumor occurrence requires further investigation.
In conclusion, the present study examined the
regulation of KIF4A by HBV at the molecular level and presented
preliminary evidence for at least one of the potential carcinogenic
mechanisms of HBV.
Acknowledgments
This study was supported by research grants from the
Major State Basic Research Development Program (973 Program; grant.
no. 2012CB518900), the National Clinical Key Subject (grant. no.
2010305), the National Science Foundation of China (grant. no.
81101485 to Dr Cheng-Liang Zhu and no. 31270206 to Dr Kai-Lang Wu),
the Open Research Program of the State Key Laboratory of Virology
of China (grant. nos. 2011009, 2012007 and 2013004) and the China
Postdoctoral Foundation (grant. no. 201104485).
References
|
1
|
Gomaa AI, Khan SA, Toledano MB, Waked I
and Taylor-Robinson SD: Hepatocellular carcinoma: Epidemiology,
risk factors and pathogenesis. World J Gastroenterol. 14:4300–4308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lim SG, Mohammed R, Yuen MF and Kao JH:
Prevention of hepatocellular carcinoma in hepatitis B virus
infection. J Gastroenterol Hepatol. 24:1352–1357. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim BK, Han KH and Ahn SH: Prevention of
hepatocellular carcinoma in patients with chronic hepatitis B virus
infection. Oncology. 81(Suppl 1): 41–49. 2011. View Article : Google Scholar
|
|
4
|
Nguyen VT, Law MG and Dore GJ: Hepatitis
B-related hepatocellular carcinoma: epidemiological characteristics
and disease burden. J Viral Hepat. 16:453–463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su CH, Lin Y and Cai L: Genetic factors,
viral infection, other factors and liver cancer: an update on
current progress. Asian Pac J Cancer Prev. 14:4953–4960. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cougot D, Neuveut C and Buendia MA: HBV
induced carcinogenesis. J Clin Virol. 34(Suppl 1): 75–78. 2005.
View Article : Google Scholar
|
|
7
|
Zhang R, Cao Y, Bai L, et al: The collagen
triple helix repeat containing 1 facilitates hepatitis B
virus-associated hepatocellular carcinoma progression by regulating
multiple cellular factors and signal cascades. Mol Carcinog. Sep
27–2014.Epub a head of print. View
Article : Google Scholar
|
|
8
|
Martinez NW, Xue X, Berro RG, Kreitzer G
and Resh MD: Kinesin KIF4 regulates intracellular trafficking and
stability of the human immunodeficiency virus type 1 Gag
polyprotein. J Virol. 82:9937–9950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu G and Chen PL: Structural requirements
of chromokinesin Kif4A for its proper function in mitosis. Biochem
Biophys Res Commun. 372:454–458. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ha MJ, Yoon J, Moon E, Lee YM, Kim HJ and
Kim W: Assignment of the kinesin family member 4 genes (KIF4A and
KIF4B) to human chromosome bands Xq13.1 and 5q33.1 by in situ
hybridization. Cytogenet Cell Genet. 88:41–42. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Taniwaki M, Takano A, Ishikawa N, et al:
Activation of KIF4A as a prognostic biomarker and therapeutic
target for lung cancer. Clin Cancer Res. 13:6624–6631. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu G, Zhou L, Khidr L, et al: A novel role
of the chromokinesin Kif4A in DNA damage response. Cell Cycle.
7:2013–2020. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nunes Bastos R, Gandhi SR, Baron RD,
Gruneberg U, Nigg EA and Barr FA: Aurora B suppresses microtubule
dynamics and limits central spindle size by locally activating
KIF4A. J Cell Biol. 202:605–621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan YJ: Hepatitis B virus infection and
the risk of hepatocellular carcinoma. World J Gastroenterol.
17:4853–4857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu C, Zhang R, Liu L, et al: Hepatitis B
virus enhances interleukin-27 expression both in vivo and in vitro.
Clin Immunol. 131:92–97. 2009. View Article : Google Scholar
|
|
16
|
Li Y, Xie J, Xu X, et al: Inducible
interleukin 32 (IL-32) exerts extensive antiviral function via
selective stimulation of interferon lambda1 (IFN-lambda1). J Biol
Chem. 288:20927–20941. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu L, Wei L, Peng G, et al: NS3 protein of
hepatitis C virus regulates cyclooxygenase-2 expression through
multiple signaling pathways. Virology. 371:61–70. 2008. View Article : Google Scholar
|
|
18
|
Lee YM and Kim W: Association of human
kinesin superfamily protein member 4 with BRCA2-associated factor
35. Biochem J. 374:497–503. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao J, Sai N, Wang C, et al:
Overexpression of chromokinesin KIF4 inhibits proliferation of
human gastric carcinoma cells both in vitro and in vivo. Tumour
Biol. 32:53–61. 2011. View Article : Google Scholar
|
|
20
|
Narayan G, Bourdon V, Chaganti S, et al:
Gene dosage alterations revealed by cDNA microarray analysis in
cervical cancer: identification of candidate amplified and
overexpressed genes. Genes Chromosomes Cancer. 46:373–384. 2007.
View Article : Google Scholar : PubMed/NCBI
|