Introduction
Hepatocellular carcinoma (HCC) is one of the most
life-threatening forms of solid tumor, with a mortality rate of
~650,000 individuals each year worldwide (1,2).
Factors contributing to HCC include hepatitis infection, alcohol
abuse and non-alcoholic fatty liver disease (3). In addition, with increasing
environmental pollution, its incidence is increasing rapidly in
China (4,5). It is difficult to detect HCC in its
early stage, resulting in a low 5-year-survival rate of patients,
of ~5%. The development, progression and chemoresistance of HCC
involves dysfunction of genes which are critical to cellular
proliferation, migration and metastasis. As one of systemic
treatment options for patients with HCC, chemotherapy is widely
used. However, long-term chemotherapy often fails to eliminate HCC
due to chemoresistance (5).
Previous studies on HCC chemoresistance have suggested that the
dysfunction of non-coding genes, particularly microRNAs (miRNAs),
is closely associated with HCC chemoresistance (6,7).
Non-coding miRNAs are a class of small
non-protein-coding RNAs (19–23 nucleotides) that negatively
regulate their downstream target mRNAs at a post-transcriptional
level by perfect or imperfect matching to the 5′ or 3′ untranslated
region, induce either degradation or translational inhibition of
mRNAs (8). miRNAs are critical for
the majority of cellular processes and have been predicted to
regulate the expression of >90% of human genes, including coding
and non-coding transcripts (9).
Previous reports have indicated the close association of miRNAs
with the evolution of chemoresistance, and miRNA expression
profiling can be correlated with the development of chemoresistance
(10–13), suggesting a novel mechanism of the
development of chemoresistance in an miRNA-mediated manner.
Among the miRNAs, miRNA-215 (miR-215) has been
identified as one of the miRNAs with the lowest levels of
expression in various types of tumor, including osteosarcoma, colon
cancer, renal carcinoma and ovarian cancer (14–17).
Karaayvaz et al (15)
reported that miR-215 is significantly reduced in colon tumor
tissues, compared with adjacent tissues, which results in the
overexpression of denticleless protein homolog, and the
downregulation of P53 and P21. This suggests that it is a
prognostic biomarker in stage II and III colon cancer. Mu et
al (18) observed that miR-215
is involved in transforming growth factor-β1-induced mesangial cell
phenotypic transition. White et al (16) demonstrated that over-expression of
miR-215 reduces cellular migration and invasion in a renal cell
carcinoma (RCC) cell line model, by targeting its direct and
indirect target genes. Notably, miR-215 has been recognized as a
novel inducer of chemoresistance (14). Although it directly targets
dihydrofolate reductase (DHFR) and thymidylate synthase (TS) mRNA,
reducing their levels of expression, the overexpression of miR-215
counter-intuitively reduces chemosensitivity to the DHFR inhibitor,
MTX, and the TS inhibitor, TDX (14).
In the present study, the expression profiling of
miR-215 in Adriamycin (ADM)-resistant HCC cells and tumor samples
was performed. The present study aimed to investigate the change in
the miR-215 expression level and the regulatory response to P53 and
P21.
Materials and methods
Patients
The present study involved the recruitment of 10
patients (3 males and 7 females; mean age, 48.1 years; age range,
41–64 years) at The Third People's Hospital of Chengdu (Southwest
Jiaotong University, Chengdu, China) with previously untreated and
histologically confirmed HCC. All patients were treated with
chemoradiotherapy and chemotherapy (ADM; Sigma-Aldrich, St. Louis,
Mo, USA; 800 mg/m2; orally; twice/day), which was
followed by surgical resection of the tumor. All patients provided
written informed consent and the study was approved by the local
ethics committee of Sichuan University (Chengdu, China). The
adjacent normal tissues were used as a control.
Cell cultures
The non-small cell lung cancer (NSCLC), HCC and RCC
cell lines were all purchased from American Type Culture Collection
(Manassas, VA, USA). The HepG2 (cat. no. HB-8065) and Hep3B (cat.
no. HB-8064) HCC cell lines were routinely maintained in minimum
essential medium (MEM; Gibco Life Technologies, Carlsbad, CA, USA).
The 786-O (cat. no. CRL1932) and ACHN (cat. no. CRL-1611) RCC cell
lines, and the A549 (cat. no. CRM-CCL-185) and H1299 (cat. no.
CRL-5803) NSCLC cell lines were all cultured in Dulbecco's modified
Eagle's medium (DMEM). All cultures were supplemented with 10%
fetal bovine serum (FBS; Gibco Life Technologies) and antibiotics
(50 U/ml penicillin and 50 µg/ml streptomycin; Gibco Life
Technologies) and incubated at 37°C in a humidified atmosphere
containing 5% CO2. The derived ADM-resistant sublines
were induced by gradual exposure of the cells to 0.1–2.5 mg/l ADM
in the culture medium. Starting at a concentration of 0.1 mg/l ADM,
the cells were maintained for 12 days, following which the ADM
concentration was increased by 0.15 mg/l/day for 12 days, until the
final ADM concentration reached 2.5 mg/l.
RNA isolation, reverse transcription (RT)
and quantification of mature miRNAs using RT-quantitative
polymerase chain reaction (RT-qPCR)
RNA was isolated from the cells and tissues using an
mirVanaTM miRNA Isolation kit (Ambion Life Technologies, Austin,
TX, USA). A TaqMan® MicroRNA RT kit (Applied Biosystems
Life Technologies, Foster City, CA, USA) was used for RT, using an
Applied Biosystems 9700 Thermocycler (Applied Biosystems Life
Technologies), according to the manufacturer's instructions. The
reaction contained 100 mM specific stem-loop primer (Sangon,
Shanghai, China), 1X reverse-transcriptional buffer (Thermo
Scientific, Waltham, MA, USA), 0.25 mM of each dNTP (Shenggong),
and 200 units reverse transcriptase (Thermo Scientific) in a total
volume of 25 µl. A TaqMan® PCR kit (Applied
Biosystems Life Technologies) was used for measuring the expression
levels of miR-215 on a Bio-Rad IQ5 Multi-Color Real-time PCR
Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The 20 µl PCR reaction mixture contained a Taqman probe for
miR-215 and 1X TaqMan® Universal PCR Master Mix (Applied
Biosystems Life Technologies), forward and reverse primers and the
RT product (Life Technologies, Grand Island, NY, USA). The reaction
program was as follows: 98°C for 5 min, followed by 40 cycles of
98°C for 10 sec and 60°C for 90 sec. The expression of small
nuclear U6 RNA was used as internal control for the miR-215
quantification assay (19). All
assays were performed in triplicate. All assays were performed in
triplicate. The data was analyzed by calculating the fold
difference individually for each housekeeping gene. Cycle threshold
(Ct) is defined as the number of PCR cycles at which the
fluorescence signal rises above the threshold value and is
inversely proportional to the amount of template present in the
reaction. Ct values of genes in tumor and adjacent tissue samples
(control) were compared, and the fold difference calculated by the
equation: Fold difference = 2ΔCt, where ΔCt =
CtTumor-CtControl.
SDS-PAGE and western blot analysis
Total cell extracts were mixed with loading buffer
(containing 0.05 M Tris-HCl, pH 8.0; 20% glycerol, 0.25% SDS and 5%
bromophenol blue), incubated for 10 min at 100°C and then loaded
onto a 10% acrylamide gel. Gels were run at 240 V constant voltage
until the tracing dye reached the bottom of the gel. The gels were
equilibrated for 30 min in blocking buffer (25 mM Tris-HCl, pH 7.6;
and 192 mM glycine) and then blotted onto polyvinylidene difluoride
membranes (Life Technologies). Images were obtained on X-ray films
(Life Technologies).
Cell apoptosis
The treated cells were harvested after 24 h by
trypsinization with 0.25% trypsin (Life Technologies) and analyzed
for the presence of apoptotic markers. The cell viability was
measured by cell-counting using 0.2% trypan blue staining
(Shenggong), in which stained cells were considered to be dead. The
viability results were interpreted The viability results were
interpreted as the ratio of viable cells following ADM treatment
and the final results were normalized with the untreated cells
(negative control). The presence of apoptotic markers was analyzed
using an ApoPercentage assay (Biocolor Ltd., Carrickfergus, UK), in
which cells undergoing apoptosis are stained on the inner plasma
membrane. The stained and unstained cells were then counted using
an XI71 microscope (Olympus, Tokyo, Japan).
Cell viability
Subsequent to interaction with 3-(4,
5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulphophenyl)-2H-tetrazolium
inner salt (MTS; Promega Corporation, Madison, WI, USA), dissolved
formazan production was measured at 490 nm for 1 h using Multiskan
spectrum microplate reader (Thermo Electron Corporation, Waltham,
MA, USA), according to the manufacturer's instructions.
Cell phase percentage assay
The percentages of cells in different phases of the
cell cycle were analyzed using flow cytometry (20). The trypsinized cells were washed
with phosphate-buffered saline (PBS) and fixed in 75% ethanol. The
cells (90% confluent) were then incubated with 100 µg/ml
RNase at 37°C for 30 min, and were then stained with propidium
iodide (Sigma-Aldrich) at a concentration of 50 µg/ml. The
cells were then analyzed on a FACScan flow cytometer (BD
Biosciences, Franklin Lakes, CA, USA).
Edu incorporation assay
EdU is a thymidine analogue, which is used to label
proliferating cells and can incorporate into replicated DNA when
cells are dividing (21). The
cells were assayed using a Cell-Light™ EdU DNA cell Proliferation
kit (RiboBio, Guangzhou, China) and Hoechst 33342 (Sigma-Aldrich)
according to the manufacturer's instructions. Each assay was
repeated three times.
Colony formation in soft agar
The cells were suspended in 1 ml 0.3% melted agar
(Shenggong) in medium containing 10% FBS and then plated in a
6-well plate with 0.6% agar in the same medium at 37°C. After 3
weeks, the colonies were stained with nitro blue tetrazolium
(Shenggong) and scanned 4 h after staining using an Epson
Perfection 3200 scanner (Seiko Epson Corporation, Suwa, Japan).
Statistical analysis
Data are presented as the mean ± standard deviation.
Differences were evaluated using Student's t-test and analysis was
performed using GraphPad Prism software, version 5 (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-215 is upregulated in ADM-resistant
HepG2 and Hep3B HCC cells and HCC tumor samples
The ectopic expression of miR-215 in osteosarcoma
and colon cancer cells is crucial in the development of
chemoresistance in different manners, including the suppression of
DHFR and TS, and the activation of P53 (14). In the present study, rather than
focusing on the ectopic expression of miR-215 alone, the mechanisms
of chemoresistance developed by endogenous miR-215 were
investigated in HepG2 and Hep3B HCC cells, A549 and H1299 NSCLC
cells, and ACHN and 786-O RCC cells. In each of these three pairs,
the first cell contains wild-type P53 and the second is P53-null,
to investigate the possible interactions between miR-215 and P53.
ADM-resistant sublines of HCC cells (HepG2/AR and Hep3B/AR), NSCLC
cells (A549/AR and H1299/AR) and RCC cells (ACHN/AR and 786-O/AR)
were formed by exposure to increasing concentrations of ADM, as
described above. The expression levels of miR-215 were detected in
these ADM-resistant cells and in paired untreated control cells.
The results demonstrated that, in HCC cells, the expression levels
of miR-215 in the ADM-resistant HCC cells were significantly
upregulated, however, this was not observed in the NSCLC or RCC
cells (Fig. 1A). Northern blotting
was performed to confirm the increased RNA level of miR-215
(Fig. 1B). Subsequently, the
expression of miR-215 was detected using RT-qPCR in 10 fresh HCC
tissue samples, which were obtained from ADM-treated HCC patients,
along with paired normal adjacent tissues. Compared with the
adjacent tissues, the expression levels of miR-215 were
significantly upregulated in the HCC samples (7/10; P<0.05,
Fig. 1C). U6 RNA was used as an
internal standard.
 | Figure 1miR-215 is upregulated in
ADM-resistant HCC cells and tumor samples. (A) RT-qPCR analysis of
the miR-215 and U6 levels in the A549 and H1299 NSCLC cell lines,
the ACHN and 786-O renal cell carcinoma cell lines and the HepG2
and Hep3B HCC cell lines, which were induced as AR sublines and
compared with the original non-resistant cells. (B) Northern blot
analysis of miR-215 and U6 RNA in the AR HepG2 and Hep3B, compared
with the original non-resistant cells. (C) RT-qPCR analysis of the
relative expression levels of miR-215 in HCC tissues, compared with
normal adjacent tissues. The relative expression values were
calculated using the equation: relative quantity =
2-ΔΔCt. *P<0.05 vs. adjacent tissue.
miR-215, microRNA-215; ADM, Adriamycin; AR, ADM resistant; HCC,
hepatocellular carcinoma; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; NSCLC,
non-small cell lung cancer; RCC, renal cell carcinoma. |
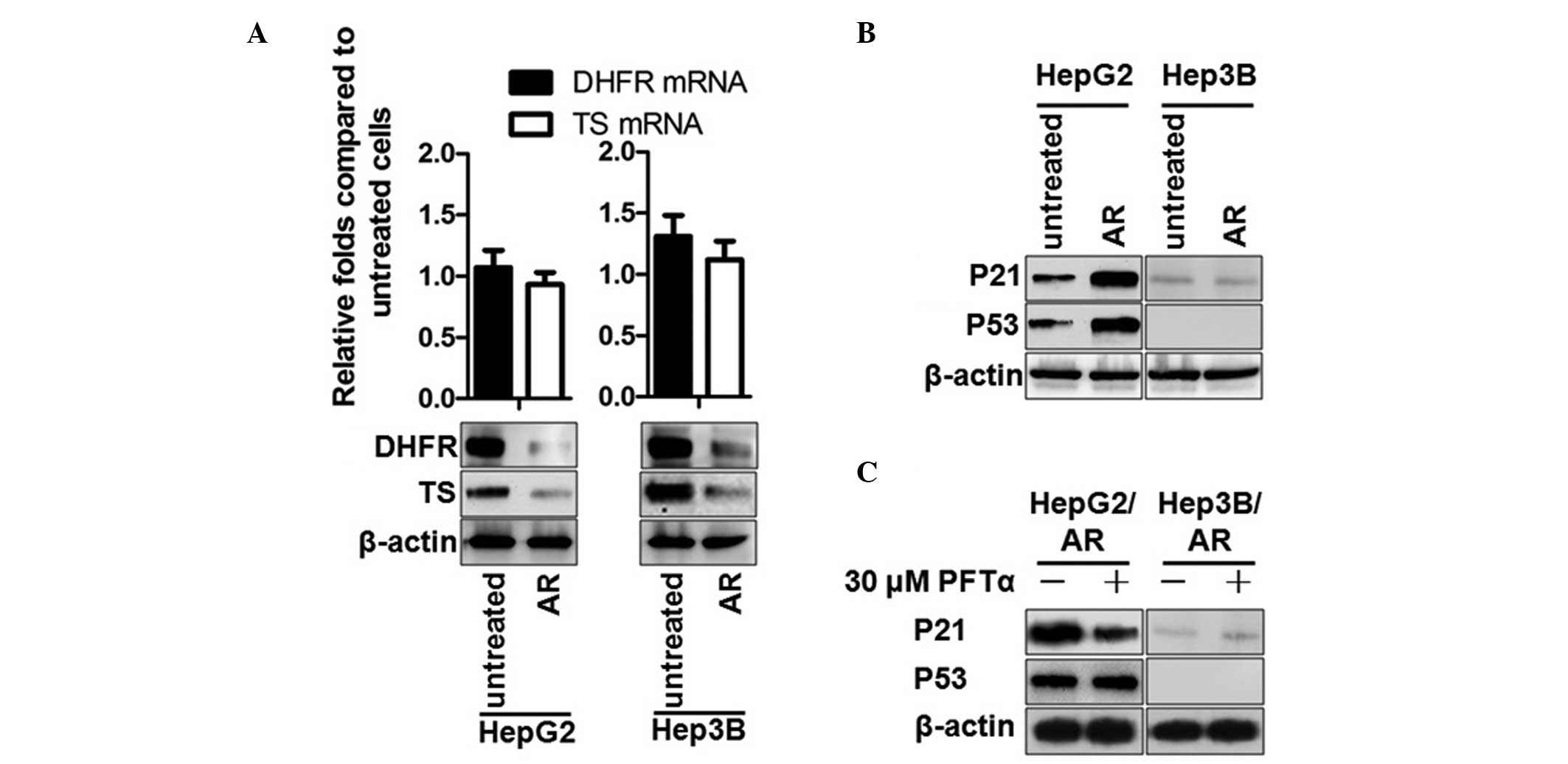
Upregulated miR-215 suppresses its direct
target genes, DHFR and TS, and indirectly activates the
transcriptional activity of P53 on P21
Due to the fact that DHFR and TS mRNAs are targeted
by miR-215, their mRNA and protein levels were measured in the
present study. Consistent with previous reports (6,7), the
protein levels of DHFR and TS reduced without mRNA levels being
altered, due to the imperfect matching (Fig. 2A). The P53 and P21 levels were then
further investigated in HepG2 and HepG2/AR cells. The results
demonstrated that the levels of P53 and P21 were upregulated in the
ADM-resistant cells (Fig. 2B).
Pifithrin-α treatment eliminated the increases in the protein level
of P2, indicating that the increase was caused by the
transcriptional activity of P53 (Fig.
2C).
Upregulated miR-215 develops
ADM-resistance in HCC and renders HCC cells insensitive to ADM
treatment
In order to detect the effect of the upregulation of
miR-215 in the HCC cells, the cells were treated with 2 mg/l ADM
for 24 h and the levels of apoptosis were analyzed. Compared with
sensitive cells, the HepG2/AR and Hep3B/AR cells exhibited a
reduced apoptotic ratio (Fig. 3A).
To determine whether the upregulation of miR-215 was sufficient and
necessary to develop ADM resistance, miR-215 mimics or antisense
oligonucleotides (ASOs) were introduced into the HCC and HCC/AR
cells. The results demonstrated that ectopic miR-215 mimics
rendered the HCC cells insensitive to ADM treatment (Fig. 3B and C, upper panel), indicating
that miR-215 is sufficient to induce ADM resistance. In addition,
the introduction of miR-215 ASOs eliminated ADM resistance in the
HCC/AR cells, further suggesting that miR-215 is necessary for ADM
resistance (Fig. 3B and C, lower
panel).
In HCC/AR cells, upregulated miR-215
inhibits cell proliferation and colony formation, in a
P53-dependent manner
The results of the present study demonstrated that,
in HepG2/AR cells, P53 and P21 were upregulated by miR-215,
although this was not observed in the Hep3b/AR cells (Fig. 2B). The present study also
investigated the effects of upregulated miR-215 on proliferation
and colony formation, which is tightly regulated by P53 and P21.
Cell phase percentage analysis indicated that the cell cycle of the
HepG2/AR cells was arrested at the G2 phase, compared
with HepG2 cells, whereas no detectable difference were observed
between the Hep3B/AR and Hep3B cells (Fig. 4A). Consistent with the cell phase
results, HepG2/AR exhibited significant differences in
proliferation and colony formation, compared with the HepG2 cells
(Fig. 4B-D). Taken together, in
HCC cells containing wild-type P53, miR-215 caused the inhibition
of proliferation and colony formation. This result indicated that
the upregulation of miR-215 in HCC with mutated P53 may increase
malignancy.
Discussion
The development of chemoresistance remains a major
challenge in the treatment of patients with HCC using
chemotherapeutic agents. Increasing evidence has indicated the
involvement of miRNAs in chemoresistance. Wu et al (22) reported that the overexpression of
miR-378 enhances cell survival and colony formation, in addition to
contributing to multi-drug resistance. Park et al (23) observed that, in chemoresistant
ovarian carcinoma, several miRNAs are over-expressed and are
closely associated with the development of chemoresistance. Robin
et al (24) also indicated
that the upregulation of miR-708 by EYA protein resulted in the
development of chemoresistance in sarcoma. MicroRNAs have also been
reported to work as anti-chemoresistance factors. Fujita et
al (25) reported that ectopic
expression of miR-34a results in cell cycle arrest and attenuates
chemoresistance to anticancer drugs. In addition, Huang et
al (26) identified miR-650 as
a novel prognostic marker in adenocarcinoma of the lung and
suggested its expression as a potential indicator of
chemosensitivity.
A previous study demonstrated that the
overexpression of miR-215 increases chemoresistance by targeting
two critical mRNAs (18), which
led the current study to investigate its possible role in the
development of HCC chemoresistance. The present study is the first,
to the best of our knowledge, to report that miR-215 is
significantly upregulated in ADM-resistant HCC tissues compared
with paired adjacent normal tissues, however, this was not observed
in the NSCLC or RCC cells. In ADM-resistant HepG2 and Hep3B
sublines, miR-215 was also significantly upregulated, compared with
the original cell lines. Endogenous miR-215 directly targeted DHFR
and TS mRNAs and reduced their protein levels without affecting
mRNA levels. Due to the fact that DHFR and TS proteins are the
targets of MTX and TDX, the reduction in protein levels caused by
miR-215 suggested that it had a positive effect on reducing
sensitivity to MTX and TDX. The expression of miR-215 mimics in
ADM-sensitive HCC cells increased the cell viability following ADM
treatment, indicating its key role in the mechanism of tumor
chemoresistance. By contrast, the expression of miR-215 AGOs in the
ADM-resistant HCC cells increased the sensitivity to ADM treatment,
suggesting the role of miR-215 in maintaining chemoresistance.
The present study also identified that upregulation
of miR-215 in ADM-resistant HCC cells exhibited differing effects,
in a P53-dependent manner. In the HepG2-AR cells, which expressed
wild-type P53, the indirect upregulation of P53 by miR-215
inhibited cell proliferation, colony formation and triggered cell
cycle arrest at the G2 phase, which was accompanied by
P53-dependent upregulation of P21. However, in the P53 null
Hep3B-AR cells, the proliferation, colony formation and cell phase
percentage exhibited no detectable differences, compared with the
Hep3B cells, which further confirmed the importance of P53 in
contributing these effects following upregulation of miR-215.
The present study demonstrated that a high
expression level of miR-215 was correlated with enhanced
chemoresistance in HCC cells and patients by targeting DHFR and TS
mRNAs. Furthermore, miR-215 affected the proliferation and colony
formation of the cells via regulating the expression of P21 by
indirectly targeting P53. Due to the small tissue sample sizes used
in the present study, further investigation of a larger patient
population is required in order to confirm the role of miR-215 in
the development of chemoresistance. In addition, the detection of
mutations of P53 in tumor samples is necessary to confirm the
association of miR-215, P53 and malignancy.
Acknowledgments
The authors would like to thank Dr Ziyi Zhao
(Sichuan University) for their assistance with English editing, Dr
Changjin Chen (Sichuan University) for his technical support and
Miss Jiao Lv (Sichuan University) for her technical assistance.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaw JJ and Shah SA: Rising incidence and
demographics of hepatocellular carcinoma in the USA: What does it
mean? Expert Rev Gastroenterol Hepatol. 5:365–370. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taylor-Robinson SD, Toledano MB, Arora S,
Keegan TJ, Hargreaves S, Beck A, Khan SA, Elliott P and Thomas HC:
Increase in mortality rates from intrahepatic cholangiocarcinoma in
England and Wales 1968–1998. Gut. 48:816–820. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kosaka N, Iguchi H, Hagiwara K, Yoshioka
Y, Takeshita F and Ochiya T: Neutral sphingomyelinase 2
(nSMase2)-dependent exosomal transfer of angiogenic microRNAs
regulate cancer cell metastasis. J Biol Chem. 288:10849–10859.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Allen KE and Weiss GJ: Resistance may not
be futile: microRNA biomarkers for chemoresistance and potential
therapeutics. Mol Cancer Ther. 9:3126–3136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma J, Dong C and Ji C: MicroRNA and drug
resistance. Cancer Gene Ther. 17:523–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng T, Wang J, Chen X and Liu L: Role of
microRNA in anticancer drug resistance. Int J Cancer. 126:2–10.
2010. View Article : Google Scholar
|
|
14
|
Song B, Wang Y, Titmus MA, Botchkina G,
Formentini A, Kornmann M and Ju J: Molecular mechanism of
chemoresistance by miR-215 in osteosarcoma and colon cancer cells.
Mol Cancer. 9:96–105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karaayvaz M, Pal T, Song B, Zhang C,
Georgakopoulos P, Mehmood S, Burke S, Shroyer K and Ju J:
Prognostic significance of miR-215 in colon cancer. Clin Colorectal
Cancer. 10:340–347. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
White NM, Khella HW, Grigull J, Adzovic S,
Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA,
et al: miRNA profiling in metastatic renal cell carcinoma reveals a
tumour-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ivan C, Hu W, Bottsford-Miller J, et al:
Comprehensive epigenetic analysis of Notch pathway in high-grade
serous ovarian cancer. Gynecol Oncol. 128:506–511. 2013. View Article : Google Scholar :
|
|
18
|
Mu J, Pang Q, Guo YH, Chen JG, Zeng W,
Huang YJ, Zhang J and Feng B: Functional implications of
microRNA-215 in TGF-β1-induced phenotypic transition of mesangial
cells by targeting CTNNBIP1. PLoS One. 8:e586222013. View Article : Google Scholar
|
|
19
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ou YC, Yang CR, Cheng CL, Raung SL, Hung
YY and Chen CJ: Indomethacin induces apoptosis in 786-O renal cell
carcinoma cells by activating mitogen-activated protein kinasesand
AKT. Eur J Pharmacol. 563:49–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chehrehasa F, Meedeniya AC, Dwyer P,
Abrahamsen G and Mackay-Sim A: EdU, a new thymidine analogue for
labelling proliferating cells in the nervous system. J Neurosci
Methods. 177:122–130. 2009. View Article : Google Scholar
|
|
22
|
Wu QP, Xie YZ, Deng Z, Li XM, Yang W, Jiao
CW, Fang L, Li SZ, Pan HH, Yee AJ, Lee DY, Li C, Zhang Z, Guo J and
Yang BB: Ergosterol peroxide isolated from Ganoderma lucidum
abolishes microRNA miR-378-mediated tumor cells onchemoresistance.
PLoS One. 7:e445792012. View Article : Google Scholar
|
|
23
|
Park YT, Jeong JY, Lee MJ, Kim KI, Kim TH,
Kwon YD, Lee C, Kim OJ and An HJ: MicroRNAs overexpressed in
ovarian ALDH1-positive cells are associated with chemoresistance. J
Ovarian Res. 6:182013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Robin TP, Smith A, McKinsey E, Reaves L,
Jedlicka P and Ford HL: EWS/FLI1 regulates EYA3 in Ewing sarcoma
via modulation of miRNA-708, resulting in increased cell survival
and chemoresistance. Mol Cancer Res. 10:1098–1108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fujita Y, Kojima K, Hamada N, Ohhashi R,
Akao Y, Nozawa Y, Deguchi T and Ito M: Effects of miR-34a on cell
growth and chemo-resistance in prostate cancer PC3 cells. Biochem
Biophys Res Commun. 377:114–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang JY, Cui SY, Chen YT, Song HZ, Huang
GC, Feng B, Sun M, De W, Wang R and Chen LB: MicroRNA-650 was a
prognostic factor in human lung adenocarcinoma and confers the
docetaxel chemoresistanceof lung adenocarcinoma cells via
regulating Bcl-2/Bax expression. PLoS One. 8:e726152013. View Article : Google Scholar
|