Introduction
Green tea is one of the most popular beverages
consumed worldwide, and its tumor-suppressive effects have been
reported in various types of cancer, including human head and neck
squamous cell carcinoma (1).
Epigallocatechin-3-gallate (EGCG), the most abundant polyphenol in
green tea, is considered to have the predominant anticancer and
cancer-preventive effects (2).
EGCG inhibits carcinogen activity, tumorigenesis,
proliferation and angiogenesis, and induces cell death. These
effects are associated with modulation of reactive oxygen species
(ROS) production and nuclear factor-κB, mitogen-activated protein
kinase (MAPK), epidermal growth factor receptor and insulin-like
growth factor-I signaling pathways, affecting diverse processes,
such as proliferation, differentiation, apoptosis, angiogenesis,
metastasis and migration (3). In
addition, EGCG could also induce epigenetic modification by
inhibition of DNA methyltransferase activity and regulation of
histone acetylation, leading to an upregulation of apoptosis
(4).
Alterations in DNA methylation, including
hypomethylation of oncogenes and hypermethylation of tumor
suppressor genes, particularly the hypermethylation of promoter CpG
islands are critical in cancer progression (5). Oral squamous cell carcinoma (OSCC)
represents the eighth most common type of malignancy in males and
the 13th most common type in females worldwide, accounting for ~38%
of all head and neck tumors (6).
Previous studies have demonstrated that EGCG has
cancer-preventative activity in OSCC development (7–9).
Mechanistically, this may occur via epigenetic modulation. To date,
several studies have investigated DNA methylation and EGCG
(10,11). These have predominantly focused on
the ability of EGCG to inhibit DNA methyltransferase (DNMT), which
leads to the reactivation of genes silenced by promoter or enhancer
methylation, including p16INK4a, RARβ, MGMT, hMlH1,
WIF-1 and hTERT (12,13). Although these studies were not
specific to the development of OSCC, they have indicated that EGCG
is capable of affecting genes involved in carcinogenesis, via an
epigenetic pathway.
To better understand the mechanisms underlying the
effect of EGCG on DNA methylation and its chemopreventative action
in OSCC, DNA methylation and mRNA expression profiling in CAL-27
cells treated with EGCG was analyzed.
Materials and methods
Cell culture and drug treatment
The CAL-27 human OSCC cell line,, was purchased from
the American Type Culture Collection (Manassas, VA, USA). Cells
were cultured in Dulbecco's modified Eagle's medium (Hyclone, GE
Healthcare, Little Chalfont, UK) supplemented with 10%
heat-inactivated fetal bovine serum (Hyclone, GE Healthcare),
penicillin (100 IU ml−1) and streptomycin (100 µg
ml−1), and maintained at 37°C in a 5% CO2
atmosphere. EGCG (14) was
obtained from Sigma Aldrich (Carlsbad, CA, USA) (E4143 EGCG, ≥95%)
and freshly prepared each time prior to use. For microarray
analysis, cells were left untreated or were treated with 100
µM EGCG for 24 h, then genomic DNA and RNA were
isolated.
Cell proliferation assay
The effect of EGCG on CAL-27 cell growth was
assessed by an MTT cell proliferation and cytotoxicity detection
kit (KeyGen Biotech, Nanjing, China). Cells (7×104) were
seeded onto 96-well plates and grown to 80% confluence prior to
treatment with the indicated concentrations of EGCG (0–200
µM). After 24 h, cell viability was determined by measuring
absorbance at 490 nm using a microplate reader (Bio-Rad, Richmond,
CA, USA). All assays were performed in triplicate.
Genome-wide methylation assay and
statistical analysis
The Infinium II Methylation assay (Illumina, San
Diego, CA, USA) was used to detect 27,578 CpG sites genome-wide,
spanning 14,495 genes. Illumina chip technology quantifies
methylation levels at specific loci within the genome. Following
bisulfite treatment, the unmethylated cytosine is converted into
uracil and methylated cytosine remains unchanged. Following
hybridization with the methylation-specific probe or the
non-methylation probe, single-base extension with labelled
dideoxynucleotides is performed. Finally, the fluorescence
intensities of the methylated and unmethylated signals are measured
at each targeted cytosine position to estimate DNA methylation
level. Genomic DNA was extracted using the Genomic DNA Mini
Preparation kit with Spin Columns (Beyotime Biotech., Jiangsu,
China). Bisulfite conversion of genomic DNA from EGCG-treated (n=3)
and control (n=3) samples, was performed using the Zymo EZ DNA
methylation kit (Illumina). Samples were run using the automated
process on the Infinium methylation BeadChips, in accordance with
the manufacturer's instructions. Data were analyzed using
BeadStudio Gene Expression Module v3.4 (Illumina).
All differential methylation analysis algorithms
compared a group of samples with EGCG treatment to a control group.
This comparison was made using the following error models: Illumina
Custom Model. P-values were calculated using the following
formula:
Where z is the two-sided tail probability of the standard normal
distribution; Sref and Scond are standard
deviations of probe signals; and Nref and
Ncond denote the number of samples in the reference
(treatment without EGCG) and condition (treatment with EGCG)
groups, respectively. β=methylated signal/(unmethylated
signal+methylated signal+100). To identify possible cellular
functions of these genes, the Gene Ontology analysis tool AmiGO was
used ()
().
Gene expression and statistical
analysis
Total RNA was extracted using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA). The
concentration of RNA was determined by NanoDrop 8000 (Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). For amplification and
labeling of the RNA with the Illumina TotalPrep RNA Amplification
kit, 200 ng RNA from each sample was used. The Illumina Sentrix
arrays were processed according to the manufacturer's instructions.
Slides were scanned immediately using Illumina BeadStation iScan.
Data were analyzed using GenomeStudio Gene Expression Module v1.0
(Illumina).
All differential expression algorithms compared a
group of samples with EGCG treatment with a control group. The
comparison was performed using unpaired Welch's t-test for unequal
variance. Genes of which expression was significantly different
between the compared groups were selected based on a fold change
>2 in gene expression as determined by a t-test, and a P<0.05
and corrected for multiple testing using the Benjamin-Hochberg
method.
Results
Effect of EGCG on CAL-27 cell
proliferation
The effect of various doses of EGCG on CAL-27 cell
proliferation was investigated after 24 h exposure. As shown in
Fig. 1, EGCG treatment
significantly inhibited cell growth in a concentration-dependent
manner. Inhibition of proliferation was clearly observed at a
concentration of 100 µM EGCG, and this concentration was
therefore utilized in methylation profiling experiments.
Genome-wide differential DNA methylation
profiling in EGCG-treated cells
To identify loci that were differentially methylated
in CAL-27 cells in response to treatment with EGCG, the Infinium II
Methylation assay was used, which interrogated 27,578 loci,
covering >14,000 genes. Comparison of methylation profiles
between EGCG-treated and control samples identified 677 genes
(P<0.05) and 84 genes (P<0.01) that were differentially
methylated. Of the 84 genes altered in response to EGCG treatment,
57 were hypermethylated and 24 were hypomethylated (Fig. 2A and B). To identify possible
cellular functions of these genes, gene ontology (GO) analysis was
performed. Biological functions of hyper-methylated genes include
transport, cell cycle, transduction, oxidative processes and
apoptosis, whereas hypomethylated gene loci were enriched for genes
involved in apoptosis, transduction, oxidative processes and cell
adhesion (Fig. 2A and B).
Differentially methylated genes are
enhanced for specific signaling pathways
To identify signaling pathways associated with EGCG
treatment, pathway analyses were performed. Of the 677 genes
(P<0.05) differentially methylated following EGCG treatment, 229
genes were associated with signaling pathways. The top 10 enriched
pathways were: Metabolic pathways, cell cycle/mitotic, axon
guidance, MAPK signaling, cytokine-cytokine receptor interaction,
signaling by nerve growth factor, protein processing in endoplasmic
reticulum, signaling by G-protein-coupled receptor, Wnt signaling
pathway and cell cycle (Fig. 3A).
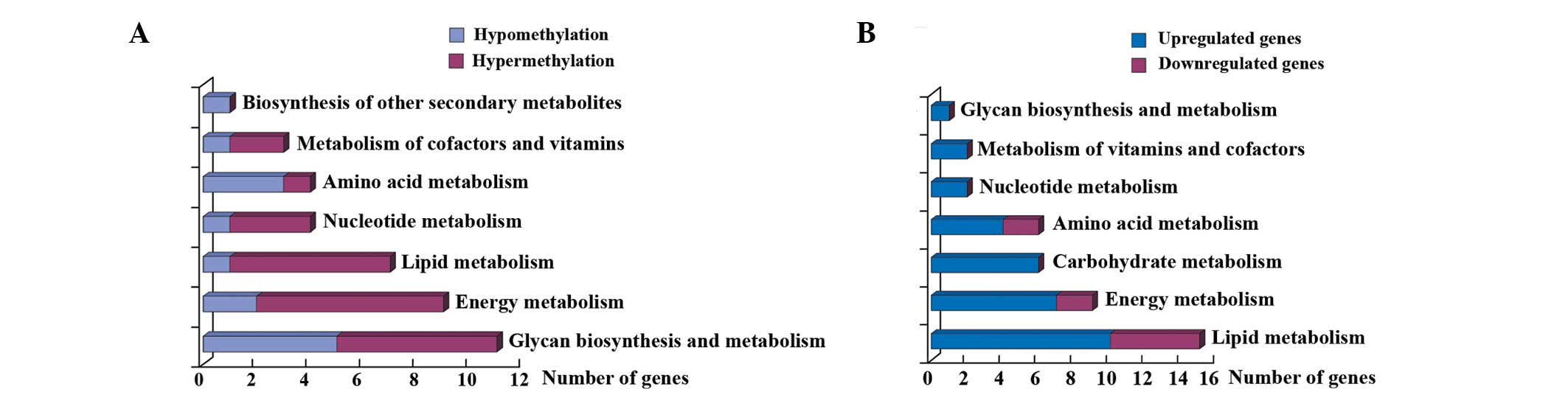
Analysis of metabolic pathway genes affected by EGCG treatment
identified an enrichment of genes involved in glycan biosynthesis
and metabolism, energy metabolism and lipid metabolism (Fig. 4A). The other 84 genes (P<0.01)
were also analyzed but no enriched pathways were identified.
Differential gene RNA expression
Whole-genome gene expression analysis was performed
on RNA samples isolated from CAL-27 cells with and without EGCG
treatment. One hundred and eighty-four transcripts showed a fold
change difference >2 in gene expression after EGCG treatment in
CAL-27 cells from 47,317 detected probes in the array. One hundred
and fifteen transcripts were upregulated and 69 down-regulated
among the 184 transcripts that were significantly changed after
EGCG treatment. Up- and downregulated genes were involved in the
following biological functions: Apoptosis, transport,
transcription, cell proliferation, transferase activity, cell
adhesion, oxidative processes and the cell cycle (Fig. 2C and D).
Pathway analyses indicated that the main pathways
associated with the differentially expressed genes were: Metabolism
pathway, MAPK, hypoxia signaling pathway, apoptosis, transforming
growth factor β signaling, interferon signaling, Wnt, p53
signaling, Notch signaling, cytoskeletal signaling and cell cycle
(Fig. 3B).
Analysis of metabolic pathway genes affected by EGCG
treatment identified an enrichment of genes involved in lipid,
energy and carbohydrate metabolism (Fig. 4B).
Discussion
A number of studies using cell culture and animal
models have suggested that EGCG is a prospective candidate for use
in the chemoprevention of cancer (16–22).
Anticancer effects of EGCG include: Inhibition of carcinogen
activity and tumorigenesis; inhibition of tumor proliferation and
angiogenesis; inhibition of tumor migration and invasion; and
induction of cell death (23).
These effects have also been found in OSCC, including in the CAL-27
cell line (24–26). However, to the best of our
knowledge, the mechanisms underlying EGCG action have not been
clarified. DNA methylation is the most extensively investigated
epigenetic modification. Hypermethylation on the DNA molecule
limits the binding of transcription factors to promoters, resulting
in the recruitment of additional proteins and gene silencing
(27). This methylation is
mediated by DNMT. EGCG is known to be an inhibitor of DNMT by
direct inhibitory interaction with the catalytic site of DNMT
(28). Several studies have found
that EGCG reverses the methylation-mediated downregulation of the
tumor suppressors and then reduces cell growth and colony formation
(29). However, few studies have
investigated the effect of EGCG on global methylation. The
demethylation effect of EGCG on the matrix metal-loproteinase
inhibitor, RECK, is the only report of EGCG modulating DNA
methylation in oral carcinoma cells (30).
In the present study, EGCG treatment significantly
inhibited the proliferation of CAL-27 cells after 24 h, in a
concentration-dependent manner. These results are consistent with
our previous findings that, at 100 µM EGCG, the survival
rate of CAL-27 cells rapidly declined; however, the survival rate
of human gingival fibroblasts was not affected, even after 100
µM EGCG treatment for 72 h. Hence, 100 µM EGCG was
used to treat CAL-27 cells, which does not harm normal oral cells
but inhibits the growth of oral cancer cells.
This study represents the first genome-wide
methylation analysis of genes affected by EGCG. EGCG treatment of
CAL-27 cells leads to hypermethylation and hypomethylation of gene
loci, while EGCG treatment was associated with higher levels of
hypermethylation compared with hypomethylation (57 vs. 27 genes).
Whole genome expression analysis indicated that the expression of
184 transcripts was altered. Although the majority of genes with
altered methylation showed no significant changes in expression,
the function and pathway analyses indicated the anti-tumor effects
of EGCG. Analysis of genes with changed methylation status revealed
their functions are involved in the regulation of the cell cycle,
transport, oxidative processes, apoptosis, transcription,
transferase activity and cell adhesion. These functions are
consistent with the analysis of genes with changed expression
levels and strongly indicate the anticancer properties of EGCG.
Integrated analysis of DNA methylation and mRNA
expression showed that four pathways were significantly changed by
EGCG treatment: Metabolism, MAPK, Wnt and cell cycle pathways. A
large portion of genes that were altered upon treatment with EGCG
were involved in metabolism pathways. The ability of EGCG to affect
metabolism may explain why it is considered to be beneficial in the
prevention and/or treatment of cardiovascular and metabolic
diseases, such as obesity and diabetes mellitus (31). Given that metabolism pathways are
essential for cancer cells, they may be an important target from
EGCG, contributing to its inhibitory effect on tumorigenesis.
MAPKs are composed of extracellular signal-regulated
kinase, p38 MAPK, and c-Jun N-terminal kinase, and the deregulation
of MAPK cascades contributes to cancer. MAPK signaling has
previously been identified as a target for cancer prevention by
EGCG (32), the present data show
that it may be regulated via an epigenetic mechanism.
Aberrant regulation of the Wnt signaling pathway
exhibits an important role in cancer biology. Kim et al
(33) found that EGCG inhibits Wnt
signaling and the Wnt target gene c-MYC in breast cancer cells by
inducing the HBP1 transcriptional repressor and inhibiting aspects
of invasive breast cancer. According to the data of the present
study, EGCG changed the methylation status of CSNK1E, CSNK2A1,
LRP6, MYC, NFATC4, SMSD4, TCF7 and TCF7L1, and altered gene
expression of ANGPTL4, DAB2, NDRG1 and CXXC5, which was associated
with the Wnt signaling pathway.
The cell cycle is a series of events that takes
place in a cell leading to its division and duplication. Regulation
of the cell cycle involves processes crucial to the survival of a
cell, including the detection and repair of genetic damage as well
as the prevention of uncontrolled cell division. EGCG downregulated
the cell cycle regulatory proteins CCND1 and PPP2R2B, and
upregulated CDKN1C. This may have occurred via epigenetic
mechanisms. Other pathways, such as apoptosis and p53 signaling,
are classical antitumor pathways, their dysregulation following
EGCG treatment strongly indicated the antitumorigenic activity of
EGCG.
In conclusion, to the best of our knowledge, the
present study reported for the first time the genome-wide analysis
of promoter methylation and expression profiling in the OSCC cell
line treated with EGCG. Additionally, the changes in several
important signaling pathways may reveal the antitumor mechanism of
EGCG.
Acknowledgments
This study was sponsored by the National Science
Foundation of China (grant no. NFSC-81371949, 30970726), and
Shanghai (grant nos. 114119a3700 and 13411951201).
References
|
1
|
Yang CS, Wang X, Lu G and Picinich SC:
Cancer prevention by tea: animal studies, molecular mechanisms and
human relevance. Nat Rev Cancer. 9:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shimizu M, Shirakami Y and Moriwaki H:
Targeting receptor tyrosine kinases for chemoprevention by green
tea catechin, EGCG. Int J Mol Sci. 9:1034–1049. 2008. View Article : Google Scholar
|
|
3
|
Khan N, Afaq F, Saleem M, Ahmad N and
Mukhtar H: Targeting multiple signaling pathways by green tea
polyphenol (-)-epigal-locatechin-3-gallate. Cancer Res.
66:2500–2505. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nandakumar V, Vaid M and Katiyar SK:
(-)-Epigallocatechin-3-gallate reactivates silenced tumor
suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA
methylation and increasing histones acetylation in human skin
cancer cells. Carcinogenesis. 32:537–544. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lund AH and van Lohuizen M: Epigenetics
and cancer. Genes Dev. 18:2315–2335. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hwang YS, Park KK and Chung WY:
Epigallocatechin-3 gallate inhibits cancer invasion by repressing
functional invadopodia formation in oral squamous cell carcinoma.
Eur J Pharmacol. 715:286–295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen PN, Chu SC, Kuo WH, Chou MY, Lin JK
and Hsieh YS: Epigallocatechin-3 gallate inhibits invasion,
epithelial-mesen-chymal transition and tumor growth in oral cancer
cells. J Agric Food Chem. 59:3836–3844. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ho YC, Yang SF, Peng CY, Chou MY and Chang
YC: Epigallocatechin-3-gallate inhibits the invasion of human oral
cancer cells and decreases the productions of matrix
metallo-proteinases and urokinase-plasminogen activator. J Oral
Pathol Med. 36:588–593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Wang X, Han L, Zhou Y and Sun S:
Green tea polyphenol EGCG reverse cisplatin resistance of A549/DDP
cell line through candidate genes demethylation. Biomed
Pharmacother. 69:285–290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin H, Chen JX, Wang H, Lu G, Liu A, Li G,
Tu S, Lin Y and Yang CS: NNK-induced DNA methyltransferase 1 in
lung tumorigenesis in A/J mice and inhibitory effects of
(-)-epigallo-catechin-3-gallate. Nutr Cancer. 67:167–176. 2015.
View Article : Google Scholar :
|
|
12
|
Lee WJ, Shim JY and Zhu BT: Mechanisms for
the inhibition of DNA methyltransferases by tea catechins and
bioflavonoids. Mol Pharmacol. 68:1018–1030. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao Z, Xu Z, Hung MS, Lin YC, Wang T, Gong
M, Zhi X, Jablon DM and You L: Promoter demethylation of WIF-1 by
epigallocatechin-3-gallate in lung cancer cells. Anticancer Res.
29:2025–2030. 2009.PubMed/NCBI
|
|
14
|
Lai SM, Gu JY, Huang BH, Chang CM and Lee
WL: Preparative separation and purification of epigallocatechin
gallate from green tea extracts using a silica adsorbent containing
β-cyclodextrin. J Chr omatogr B Analyt Technol Biomed Life Sci.
887–888:112–121. 2012. View Article : Google Scholar
|
|
15
|
An IS, An S, Park S, Lee SN and Bae S:
Involvement of microRNAs in epigallocatechin gallate-mediated UVB
protection in human dermal fibroblasts. Oncol Rep. 29:253–259.
2013.
|
|
16
|
Berletch JB, Liu C, Love WK, Andrews LG,
Katiyar SK and Tollefsbol TO: Epigenetic and genetic mechanisms
contribute to telomerase inhibition by EGCG. J Cell Biochem.
103:509–519. 2008. View Article : Google Scholar :
|
|
17
|
Xu Q, Yang CH, Liu Q, Jin XF, Xu XT, Tong
JL, Xiao SD and Ran ZH: Chemopreventive effect of
epigallocatechin-3-gallate (EGCG) and folic acid on the
N-methyl-N'-nitro-N-nitrosoguanidine (MNNG)-induced
gastrointestinal cancer in rat model. J Dig Dis. 12:181–187. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li GX, Chen YK, Hou Z, Xiao H, Jin H, Lu
G, Lee MJ, Liu B, Guan F, Yang Z, Yu A, et al: Pro-oxidative
activities and dose-response relationship of
(-)-epigallocatechin-3-gallate in the inhibition of lung cancer
cell growth: a comparative study in vivo and in vitro.
Carcinogenesis. 31:902–910. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen X, Zhang Y, Feng Y, Zhang L, Li J,
Xie YA and Luo X: Epigallocatechin-3-gallate inhibits cell growth,
induces apoptosis and causes S phase arrest in hepatocellular
carcinoma by suppressing the AKT pathway. Int J Oncol. 44:791–796.
2014.PubMed/NCBI
|
|
20
|
Lim YC and Cha YY:
Epigallocatechin-3-gallate induces growth inhibition and apoptosis
of human anaplastic thyroid carcinoma cells through suppression of
EGFR/ERK pathway and cyclin B1/CDK1 complex. J Surg Oncol.
104:776–780. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maruyama T, Murata S, Nakayama K, Sano N,
Ogawa K, Nowatari T, Tamura T, Nozaki R, Fukunaga K and Ohkohchi N:
(-)-Epigallocatechin-3-gallate suppresses liver metastasis of human
colorectal cancer. Oncol Rep. 31:625–633. 2014.
|
|
22
|
Hsu YC and Liou YM: The anti-cancer
effects of (-)-epiga-locathine-3-gallate on the signaling pathways
associated with membrane receptors in MCF-7 cells. J Cell Physiol.
226:2721–2730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Singh BN, Shankar S and Srivastava RK:
Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms,
perspectives and clinical applications. Biochem Pharmacol.
82:1807–1821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu X, Zhang DY, Zhang W, Zhao X, Yuan C
and Ye F: The effect of green tea extract and EGCG on the signaling
network in squamous cell carcinoma. Nutr Cancer. 63:466–475. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin HY, Hou SC, Chen SC, Kao MC, Yu CC,
Funayama S, Ho CT and Way TD: (-)-Epigallocatechin gallate induces
Fas/CD95-mediated apoptosis through inhibiting constitutive and
IL-6-induced JAK/STAT3 signaling in head and neck squamous cell
carcinoma cells. J Agric Food Chem. 60:2480–2489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang CM, Chang PY, Tu MG, Lu CC, Kuo SC,
Amagaya S, Lee CY, Jao HY, Chen MY and Yang JS: Epigallocatechin
gallate sensitizes CAL-27 human oral squamous cell carcinoma cells
to the anti-metastatic effects of gefitinib (Iressa) via
synergistic suppression of epidermal growth factor receptor and
matrix metalloproteinase-2. Oncol Rep. 28:1799–1807.
2012.PubMed/NCBI
|
|
27
|
Ling Y, Zhang C, Xu Y, Zhu J, Zhu C, Lu M,
Liu Y and Zhou T: Promoter methylation-associated silencing of
p27kip1 gene with metastasis in esophageal squamous cell carcinoma.
Mol Med Rep. 9:1075–1079. 2014.PubMed/NCBI
|
|
28
|
Lee WJ, Shim JY and Zhu BT: Mechanisms for
the inhibition of DNA methyltransferases by tea catechins and
bioflavonoids. Mol Pharmacol. 68:1018–1030. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H,
Welsh W and Yang CS: Tea polyphenol (-)-epigallocatechin-3-gallate
inhibits DNA methyltransferase and reactivates methylation-silenced
genes in cancer cell lines. Cancer Res. 63:7563–7570.
2003.PubMed/NCBI
|
|
30
|
Kato K, Long NK, Makita H, Toida M,
Yamashita T, Hatakeyama D, Hara A, Mori H and Shibata T: Effects of
green tea polyphenol on methylation status of RECK gene and cancer
cell invasion in oral squamous cell carcinoma cells. Br J Cancer.
99:647–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thielecke F and Boschmann M: The potential
role of green tea catechins in the prevention of the metabolic
syndrome-a review. Phytochemistry. 70:11–24. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim HS, Kim MH, Jeong M, Hwang YS, Lim SH,
Shin BA, Ahn BW and Jung YD: EGCG blocks tumor promoter-induced
MMP-9 bexpression via suppression of MAPK and AP-1 activation in
human gastric AGS cells. Anticancer Res. 24:747–753.
2004.PubMed/NCBI
|
|
33
|
Kim J, Zhang X, Rieger-Christ KM,
Summerhayes IC, Wazer DE, Paulson KE and Yee AS: Suppression of Wnt
signaling by the green tea compound (-)-epigallocatechin 3-gallate
(EGCG) in invasive breast cancer cells. Requirement of the
transcriptional repressor HBP1. J Biol Chem. 281:10865–10875. 2006.
View Article : Google Scholar : PubMed/NCBI
|