Introduction
Salivary adenoid cystic carcinoma (SACC) is one of
the most common types of malignancy, which is known to be
associated with persistent slow growth, perineural invasion, high
rates of recurrence and the formation of distant metastases
(1). Despite the use of aggressive
surgery, the 5-year survival rate of SACC is ~75%, whereas the
long-term survival rate is only 39.6% (2). The underlying molecular mechanisms of
SACC carcinogenesis remain to be fully elucidated.
Changes in DNA methylation is commonly observed in
human cancer (3,4). The hypomethylation of oncogenes can
result in their aberrant activation, and hypermethylation of tumour
suppressor genes can result in their silencing (5). DNA methylation inhibitors can be
divided into two categories: Nucleoside inhibitors and
non-nucleoside inhibitors. Nucleoside inhibitors, including
5-aza-2′-deoxycytidine, are associated with substantial toxic
effects and have short half-lives in aqueous solutions (6). Epigallocatechin-3-gallate (EGCG) is a
non-nucleoside inhibitor (7),
which mediates the inhibition of DNA methylation by binding to the
catalytic pocket of human DNA methyltransferase (DNMT) and inhibit
the hypermethylation of newly synthesised DNA, resulting in the
reversal of hypermethylation and the re-expression of silenced
genes.
Several oncogene and tumour suppressor gene
candidates have been suggested to be involved in SACC, including
supra-basin (8), aquaporin 1
(9), phosphatase and tensin
homolog deleted on chromosome 10 (10), cyclin-dependent kinase inhibitors
(11) and RAS-associated domain
family protein 1A (12). The
reversion-inducing cysteine-rich protein with Kazal motifs (RECK)
gene is anchored to the cell surface by
glycosylphosphatidylinositol, which can suppress tumour invasion
and metastasis via degradation of the extracellular matrix
(13). RECK is expressed in
various types of normal human tissue, and is downregulated in
several types of human cancer, including pancreatic cancer
(14), colorectal cancer (15), breast cancer (16) and hepatocellular carcinoma
(17). Our previous study
demonstrated that the expression of RECK was significantly lower in
SACC, compared with normal tissue (18); however, the underlying mechanism
remains to be fully elucidated.
The present study aimed to determine the methylation
status of the RECK promoter in the human SACC83 SACC cell line, and
aimed to investigate the effects of EGCG on the expression of RECK
and the invasiveness of the SACC83 cells.
Materials and methods
Cell lines and cell culture
The SACC83 SACC cell line [kindly provided by
Professor Shenglin Li (Department of Oral and Maxillofacial
Surgery, School of Stomatology, Beijing University, Beijing,
China)] was cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS; GE Healthcare Life
Sciences, Logan, UT, USA), 50,000 units penicillin and 50 mg
streptomycin (both from Sigma-Aldrich, St. Louis, MO, USA) at 37°C
in a humidified incubator containing 5% CO2.
EGCG treatment
To determine the dose- and time-dependent changes
associated with EGCG treatment, the SACC83 cells were treated in
triplicate with EGCG (Wako Pure Chemical Industries, Ltd., Osaka,
Japan), as described previously (19). Briefly, the cells were seeded at
low density (5×105/100 mm dish) 24 h prior to treatment
with EGCG. To determine the dose-dependent changes, the cells were
treated with 0, 5, 15 or 45 µm EGCG for 6 days. EGCG was
added, in new culture medium, to the cells on days 1, 3 and 5. For
the assessment of time-dependent changes, the cells were treated
with 45 µm EGCG for 0, 36, 72 or 144 h.
Bisulphite modification and
methylation-specific polymerase chain reaction (PCR; MSP)
Genomic DNA was isolated and modified using a
CpGenome™ Direct Prep Bisulfite Modification kit (EMD Millipore,
Billerica, MA, USA), according to the manufacturers' instructions.
The DNA concentration was determined using spectrophotometry. For
MSP, the following primer sets (Takara Bio Inc., Dalian, China)
were used for methylated DNA: M_RECK, forward
5′-ATAAAGAGTTTTGGTACGGGGTAC-3′ and reverse
5′-AAAACCGCGAAATACTCGAA-3′); and the primer sets for unmethylated
DNA were as follows: U_RECK, forward
5′-TAAAGAGTTTTGGTATGGGGTATGT-3′ and reverse
5′-CTCCAAACCACAAAATACTCAAA-3′. The MSP reactions were performed in
a mixture of 12.5 µl 2X Taq PCR Master mix (Tiangen
Biotech, Co., Ltd., Beijing, China) reverse primers (1 µl),
DNA (<1 µg) and distilled water in 25-µl volumes
in a S1000 (Bio-Rad Laboratories, Inc., Hercules, CA. USA) under
the following conditions: 94°C for 3 min; followed by 30 cycles at
94°C for 30 sec, 55°C for 30 sec and 72°C for 60 sec; and finally 5
min at 72°C. The PCR product lengths for the methylated and
unmethylated RECK were 195 and 199 bp, respectively. Universal
Methylated DNA (EMD Millipore) and normal human blood DNA were used
as positive controls for methylated and unmethylated statuses,
respectively. The blood was donated by the authors of the present
study, and ethical approval was obtained from the ethics committee
of Weifang People's Hospital (Weifang, China). A water blank was
used as a negative control. The positive and negative controls were
used appropriately in each round of MSP. All assays were performed
in triplicate. The PCR products were separated on 2% agarose gels
(Zeta Biotechnology, Co., Ltd., Guangzhou, China) and visualised
using ethidium bromide (EtBr; Haoran Biological Technology Co.,
Ltd., Shanghai, China) staining.
RNA extraction and reverse transcription
(RT)-PCR
Total RNA was extracted using a Total RNA Extraction
kit (Takara Biotechnology Co., Ltd., Dalian, China), according to
the manufacturer's instructions. The quantity and quality of the
RNA samples were measured using a spectrophotometer and
electrophoresis. RECK cDNA was synthesised from 1 µg of
total RNA using a PrimeScript™ RT Reagent kit with gDNA Eraser
(Takara Biotechnology Co., Ltd.). The following primers (Takara Bio
Inc.) were used for PCR: RECK, forward 5′-CCTCAGTGAGCACAGTTCAGA-3′
and reverse 5′-GCAGCACACACACTGCTGTA-3′. Reactions were performed in
20 µl volumes containing 2X Mater Mix (10µl), forward
and reverse primers (1 µl), cDNA (2 µl) and distilled
water under the following conditions: 95°C for 10 min; followed by
35 cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec;
and finally 7 min at 72°C. GAPDH was used as an internal control to
estimate the efficiency of cDNA synthesis using the primer: GAPDH,
forward 5′-GCAGCACACACACTGCTGTA-3′ and reverse
5′-TGTGGTCATGAGTCCTTCCA-3′. The predicted size for the PCR products
of RECK and GAPDH were 477 and 512 bp, respectively. The PCR
products were separated on 2% agarose gel, stained with EtBr and
visualised under ultraviolet light and images were captured using a
Bio-Rad VersaDoc 3000 Imaging system (Bio-Rad Laboratories, Inc.).
Image J software (version 1.48u; National Institutes of Health,
Bethesda, MD, USA) was used for grey value analysis. The relative
mRNA expression levels of RECK were normalised against GAPDH.
Western blot analysis
Following treatment with EGCG, the SACC83 cells were
washed twice with cold phosphate-buffered saline (PBS) and treated
with extraction buffer [50 M Tris-HCl (pH 7.4), 150 M NaCl, 2 M
EDTA and 1% NP-40]. The cell extractions were subsequently
centrifuged at 12,000 × g for 15 min at 4°C, and the supernatants
were collected. The protein concentration was quantified using a
Bicinchoninic Acid Protein Measurement kit (Shenneng Bocai Biology,
Co., Ltd., Shanghai, China). Equal quantities (40 µg) of
cellular proteins were subjected to 8% SDS-PAGE, and transferred
onto polyvinylidene fluoride membranes (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA). The membranes were blocked with 5% (w/v)
non-fat milk in PBS containing 0.1% Tween-20 (Shenneng Bocai
Biology, Co., Ltd., Shanghai, China) and then incubated with
primary antibodies (mouse anti-human RECK polyclonal, cat. no.
ab88249; anti-GAPDH, cat. no. sc-365062) at 1:1,000 and room
temperature for 1 h. Subsequently, the membranes were incubated
with an appropriate horseradish peroxidase-conjugated goat
anti-mouse immunoglobulin G secondary antibody (cat. no. sc-2005)
at room temperature for 1 h. The immuno-detected proteins were
visualised using enhanced chemiluminescence. Image J software was
used for grey value analysis. The anti-RECK antibody was purchased
from Abcam (Cambridge, MA, USA) and the anti-GAPDH and secondary
antibodies were obtained from Santa Cruz Biotechnology, Inc.
In vitro invasion assay
To assess the invasive ability of the cells treated
with EGCG, Matrigel invasion assays were performed using 8-mm pore
filter inserts in 24-well plates (Sigma-Aldrich) coated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The cells were
incubated with EGCG (0, 5, 15 or 45 µm) for 72 h, and were
subsequently collected, washed three times with PBS and
re-suspended in serum-free DMEM. A total of 1×105
cells/200 µl medium were plated in the upper chamber of the
Transwell unit and allowed to invade for 24 h at 37°C. The lower
chamber of the Transwell unit was filled with 500 µl medium
supplemented with 10% FBS. At the end of the incubation period, the
non-invaded cells on the upper surface of the membrane were
carefully removed using a cotton swab. The invaded cells on the
bottom surface of the membrane were fixed in 4% formaldehyde for 20
min and stained with 0.1% crystal violet for 5 min. Subsequently,
the invaded cells on the lower surface of the membrane were
visualised in five randomly-selected fields under a microscope
(Olympus BH2; Olympus, Tokyo, Japan; magnification, ×200). All
assays were performed in triplicate and the mean number of invaded
cells was used for analysis.
Statistical analysis
Statistical analysis was performed using SPSS 13.0
for Windows (SPSS, Inc., Chicago, IL, USA). Data are expressed as
the mean ± standard deviation. Differences between the groups were
assessed using one-way analysis of variance with Dunnett's post-hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Methylation status of the RECK gene in
SACC83 cells
The RECK gene in SACC83 cells exhibited marked
expression of methylated and weak expression of unmethylated
promoter, determined using MSP (Fig.
1A). Normal human blood DNA was used as a positive control for
unmethylation, and Universal Methylated DNA was used as a positive
control for methylation. H2O was used as a negative
control.
 | Figure 1Detection of the methylation status of
the RECK gene in SACC83 cells. (A) Methylation status of the RECK
gene in SACC83 cells, NBD positive control for unmethylatiom, UMN
positive control for methylation and H2O negative
control groups. (B) Changes in the methylation status of the RECK
gene in SACC83 cells following treatment with 0, 5, 15 or 45
µm EGCG for 144 h, or treatment with 45 µm EGCG for
0, 36, 72 or 144 h. RECK, reversion-inducing cysteine-rich protein
with Kazal motifs; EGCG, epigallocatechin-3-gallate; SACC, salivary
adenoid cystic carcinoma; M, methylation-specific band; U,
unmethylation-specific band; NBD, normal blood DNA; UMD, Universal
Methylated DNA; Bp, base pairs. |
EGCG reverses the hypermethylation status
of RECK in the SACC83 cell line
The present study aimed to examine the time-and
dose-dependent effects of EGCG in SACC83 cells. Following treatment
of the cells with 0, 5, 15 or 45 µm EGCG for 144 h,
methylation-specific bands of the RECK gene were weak in
appearance, whereas unmethylation-specific bands of the RECK gene
appeared markedly enhanced. Following treatment of the cells with
45 µm EGCG for 0, 36, 72 or 144 h, the
unmethylation-specific bands of RECK were more marked, whereas the
methylation-specific bands of RECK were almost undetectable
(Fig. 1B).
Treatment with EGCG enhances the mRNA and
protein expression levels of RECK in SACC83 cells
To determine the effects of EGCG on the mRNA
expression of RECK, RT-qPCR was performed to detect the mRNA
expression levels of RECK in the SACC83 cells following treatment
with a range of concentrations of EGCG for different durations
(Fig. 2). The relative mRNA
expression levels of RECK increased in a dose- and time-dependent
manner (Fig. 2A and B).
As shown in Fig. 3,
the protein expression levels of RECK were relatively low in the
untreated SACC83 cells. Following treatment of the cells with
different doses of EGCG for 6 days, and with 45 µm EGCG for
different periods of time, the protein expression levels of RECK
increased in the SACC83 cells (Fig. 3A
and B).
 | Figure 3Western blotting was used to detect
the protein expression levels of RECK in SACC83 cells following
treatment with (A) 0, 5, 15 or 45 µm EGCG, or treatment with
45 µm EGCG for (B) 0, 36, 72 or 144 h. R/G indicates the
density ratio of the RECK protein (106 kDa), vs. GAPDH (36 kDa).
Data are presented as the mean ± standard deviation.
*P<0.05, compared with the control. RECK,
reversion-inducing cysteine-rich protein with Kazal motifs; EGCG,
epigallocatechin-3-gallate; SACC, salivary adenoid cystic
carcinoma. |
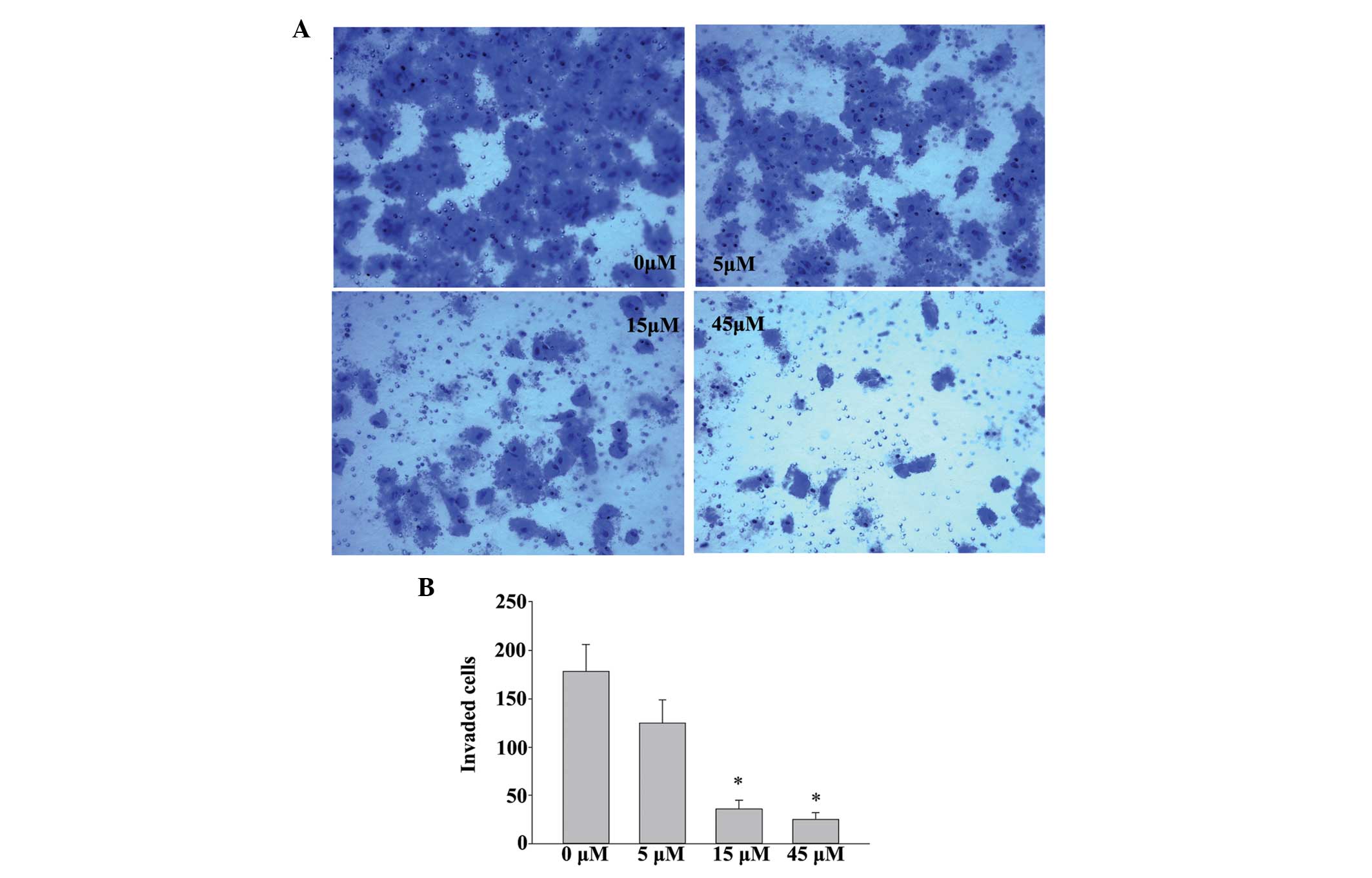
EGCG reduces the invasiveness of human
SACC83 cells
The present study also investigated the effects of
EGCG on cell invasion. As shown in Fig. 4, treatment with 5, 15 or 45
µm EGCG markedly suppressed the invasive ability of the
SACC83 cells in a dose-dependent manner. These results suggested
that restoration of the expression of RECK by EGCG is important for
inhibiting the invasiveness of SACC83 cells.
Discussion
RECK is an important matrix metalloproteinase
inhibitor, which is involved in the regulation of various
physiological and pathological processes. Oh et al (20) previously reported that mice lacking
the expression of RECK succumb to mortality in utero due to
developmental defects in blood vessels, the neural tube and
mesenchymal tissues. In addition, RECK has been identified as a
target of myogenic regulatory factors and is involved in the
control of myogenesis (21).
Several studies have demonstrated that RECK mRNA and
protein are highly expressed in human tissue and untransformed
cells (22); however, the
expression of RECK is lost or undetected in the majority of tumour
cells. Several hypotheses have been suggested regarding the
mechanism underlying the low expression levels of RECK in tumour
tissues. DNA methylation, which is a critical epigenetic
alteration, is associated with the silencing of tumour suppressor
genes in several types of cancer. Cho et al (22) demonstrated that downregulation of
the mRNA and protein expression levels of RECK in colon tumour
tissue are significantly correlated with methylation of the RECK
promoter. Furthermore, Chang et al (23) reported that downregulation of the
metastasis suppressor RECK is due to promoter methylation in
non-small cell lung cancer. In the present study, the methylation
status of the RECK promoter was determined using MSP, and the
SACC83 cells were found to exhibit weak expression levels of
unmethylated promoter and marked expression levels of methylated
promoter. These results suggested that a decrease or deficiency in
the expression of RECK in SACC83 cells may be caused by the
methylation of CpG islands in the RECK promoter region.
EGCG is a major polyphenol in green tea, and a key
active ingredient (19). Previous
studies (24,25) have demonstrated that EGCG inhibits
DNMT and inhibits the hypermethylation of newly synthesised DNA,
resulting in the reversal of hypermethylation and the re-expression
of silenced genes, with fewer side effects and toxicity. The
present study demonstrated that almost no RECK protein was detected
in untreated SACC83 cells. However, following treatment of the
cells with EGCG, the mRNA and protein expression levels of RECK
increased in a dose- and time-dependent manner in the SACC83 cells.
Furthermore, following treatment of SACC83 cells with EGCG, their
invasive capability was significantly reduced.
In conclusion, the results of the present study
demonstrated that EGCG inhibited cancer cell invasion through
reversal of the hypermethylation status of RECK. This may offer a
potential therapeutic strategy for the chemotherapeutic treatment
of SACC. Further investigations are required to fully elucidate the
underlying molecular mechanisms by which EGCG inhibits tumour
invasion.
Acknowledgments
This study was supported by grants from the Science
and Technology Development Planning Project of Jining City (no.
2014jnwk03).
References
|
1
|
Gondivkar SM, Gadbail AR, Chole R and
Parikh RV: Adenoid cystic carcinoma: A rare clinical entity and
literature review. Oral Oncol. 47:231–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fordice J, Kershaw C, El-Naggar A and
Goepfert H: Adenoid cystic carcinoma of the head and neck:
Predictors of morbidity and mortality. Arch Otolaryngol Head Neck
Surg. 125:149–152. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ehrlich M: DNA hypomethylation in cancer
cells. Epigenomics. 1:239–259. 2009. View Article : Google Scholar
|
|
4
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Latham KE: X chromosome imprinting and
inactivation in the early mammalian embryo. Trends Genet.
12:134–138. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lyko F and Brown R: DNA methyltransferase
inhibitors and the development of epigenetic cancer therapies. J
Natl Cancer Inst. 97:1498–1506. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H,
Welsh W and Yang CS: Tea polyphenol (−)-epigallocatechin-3-gallate
inhibits DNA methyltransferase and reactivates methylation-silenced
genes in cancer cell lines. Cancer Res. 63:7563–7570.
2003.PubMed/NCBI
|
|
8
|
Shao C, Tan M, Bishop JA, Liu J, Bai W,
Gaykalova DA, Ogawa T, Vikani AR, Agrawal Y, Li RJ, et al:
Suprabasin is hypomethylated and associated with metastasis in
salivary adenoid cystic carcinoma. PLoS One. 7:e4855822012.
View Article : Google Scholar
|
|
9
|
Shao C, Sun W, Tan M, Glazer CA, Bhan S,
Zhong X, Fakhry C, Sharma R, Westra WH, Hoque MO, et al:
Integrated, genome-wide screening for hypomethylated oncogenes in
salivary gland adenoid cystic carcinoma. Clin Cancer Res.
17:4320–4330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan X, Chen B, Xu J, Zhang H, Deng F and
Xiang X: Methylation status of the PTEN gene in adenoid cystic
carcinoma cells. Mol Med Rep. 3:775–779. 2010.
|
|
11
|
Daa T, Kashima K, Kondo Y, Yada N, Suzuki
M and Yokoyama S: Aberrant methylation in promoter regions of
cyclin-dependent kinase inhibitor genes in adenoid cystic carcinoma
of the salivary gland. APMIS. 116:21–26. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Williams MD, Chakravarti N, Kies MS,
Maruya S, Myers JN, Haviland JC, Weber RS, Lotan R and El-Naggar
AK: Implications of methylation patterns of cancer genes in
salivary gland tumors. Clin Cancer Res. 12:7353–7358. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clark JC, Thomas DM, Choong PF and Dass
CR: RECK - a newly discovered inhibitor of metastasis with
prognostic significance in multiple forms of cancer. Cancer
Metastasis Rev. 26:675–683. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Masui T, Doi R, Koshiba T, Fujimoto K,
Tsuji S, Nakajima S, Koizumi M, Toyoda E, Tulachan S, Ito D, et al:
RECK expression in pancreatic cancer: Its correlation with lower
invasiveness and better prognosis. Clin Cancer Res. 9:1779–1784.
2003.PubMed/NCBI
|
|
15
|
Takemoto N, Tada M, Hida Y, Asano T, Cheng
S, Kuramae T, Hamada J, Miyamoto M, Kondo S and Moriuchi T: Low
expression of reversion-inducing cysteine-rich protein with Kazal
motifs (RECK) indicates a shorter survival after resection in
patients with adenocarcinoma of the lung. Lung Cancer. 58:376–383.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Figueira RC, Gomes LR, Neto JS, Silva FC,
Silva ID and Sogayar MC: Correlation between MMPs and their
inhibitors in breast cancer tumor tissue specimens and in cell
lines with different metastatic potential. BMC Cancer. 9:202009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang C, Ling Y, Xu Y, Gao L, Li R, Zhu J,
Fan L and Wei L: The silencing of RECK gene is associated with
promoter hypermethylation and poor survival in hepatocellular
carcinoma. Int J Biol Sci. 8:451–458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou X, Huang S, Jiang L, Zhang S, Li W,
Chen Z and Zhang D: Expression of RECK and MMP-2 in salivary
adenoid cystic carcinoma: Correlation with tumor progression and
patient prognosis. Oncol Lett. 7:1549–1555. 2014.PubMed/NCBI
|
|
19
|
Kato K, Long NK, Makita H, Toida M,
Yamashita T, Hatakeyama D, Hara A, Mori H and Shibata T: Effects of
green tea polyphenol on methylation status of RECK gene and cancer
cell invasion in oral squamous cell carcinoma cells. Br J Cancer.
99:647–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oh J, Takahashi R, Kondo S, Mizoguchi A,
Adachi E, Sasahara RM, Nishimura S, Imamura Y, Kitayama H,
Alexander DB, et al: The membrane-anchored MMP inhibitor RECK is a
key regulator of extracellular matrix integrity and angiogenesis.
Cell. 107:789–800. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Echizenya M, Kondo S, Takahashi R, Oh J,
Kawashima S, Kitayama H, Takahashi C and Noda M: The
membrane-anchored MMP-regulator RECK is a target of myogenic
regulatory factors. Oncogene. 24:5850–5857. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cho CY, Wang JH, Chang HC, Chang CK and
Hung WC: Epigenetic inactivation of the metastasis suppressor RECK
enhances invasion of human colon cancer cells. J Cell Physiol.
213:65–69. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang HC, Cho CY and Hung WC:
Downregulation of RECK by promoter methylation correlates with
lymph node metastasis in non-small cell lung cancer. Cancer Sci.
98:169–173. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hellebrekers DM, Griffioen AW and van
Engeland M: Dual targeting of epigenetic therapy in cancer. Biochim
Biophys Acta. 1775:76–91. 2007.
|
|
25
|
Shaw RJ, Hall GL, Lowe D, Bowers NL,
Liloglou T, Field JK, Woolgar JA and Risk JM: CpG island
methylation phenotype (CIMP) in oral cancer: Associated with a
marked inflammatory response and less aggressive tumour biology.
Oral Oncol. 43:878–886. 2007. View Article : Google Scholar : PubMed/NCBI
|