Introduction
Major depressive disorder (MDD) is a prevalent,
debilitating mood disorder with a lifetime prevalence of 16% that
contributes to increased rates of disability and suicide (1). The pathoetiology of MDD is complex
and likely involves a combination of environmental and genetic
factors. After several years of research, investigators have
discovered several genetic polymorphisms associated with MDD; in
particular, a comprehensive 2007 meta-analysis by López-León et
al (2) provided statistically
significant evidence for six MDD susceptibility genetic
polymorphisms: APOE, DRD4, GNB3, MTHFR, SLC6A3 and SLC6A4.
One such polymorphism, namely that of apolipoprotein
E (APOE), was initially discovered by Ramachandran et al
(3) in 1996. APOE has a key role
in transporting lipoproteins, fat-soluble vitamins and cholesterol
through binding to low-density lipoprotein (LDL) and APOE receptors
(4). The APOE gene is polymorphic
and possesses three alleles: ε2, ε3 and ε4 (5). This polymorphism leads to six unique
APOE protein isoforms (6,7). The APOE phenotyping method, an
isoelectric focusing (IEF) technique that is based on
simultaneously determining the charge differences (pI) between
distinct APOE polypeptides, is a complex procedure requiring
considerable expertise (8). By
contrast, genotyping methods that detect sequence differences in
the APOE alleles [single nucleotide polymorphisms (SNPs)] are
simpler and more accurate than IEF; as a result, increasing numbers
of PCR-based genotyping techniques have been applied to determine
APOE genotypes (9–15).
Several common PCR-based genotyping techniques are
currently in use, including amplification refractory mutation
system PCR (ARMS-PCR), PCR restriction-fragment length polymorphism
(PCR-RFLP), single-stranded conformational polymorphism (SSCP) and
real-time PCR. However, not all PCR-based genotyping techniques are
equally efficacious; for example, ARMS-PCR and PCR-RFLP have been
found to be more accurate than SSCP (16,17).
Since several different techniques have been employed to assess the
mutation status of APOE with little evidence of their comparative
accuracy, the present study comparatively evaluated two modern APOE
genotyping methods: ARMS-PCR versus optimized PCR-RFLP (a modified
PCR-RFLP method using the restriction enzymes AflIII and
HaeII), in blood samples taken from 708 MDD patients in
order to find a more accurate and cost-effective APOE genotyping
method for MDD screening in large populations.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Chongqing Medical
University (Chongqing, China). Written informed consent was
obtained from all individuals prior to inclusion in this study.
Subject recruitment, blood sampling and
genomic DNA extraction
In order to identify MDD candidates for recruitment,
a structured clinical interview assessing the relevant Diagnostic
and Statistical Manual of Mental Disorders (DSM-IV, 4th edition)
(18) criteria was performed to
diagnose candidates with a single depressive episode (19), and the 17-item version of the
observer-rated Hamilton Depression Rating Scale (HDRS) was applied
to define the severity of their depression (20). Only depressed candidates with HDRS
scores >17 were recruited for the present study, while those
with one or more confounding factors, including physical or mental
disorders, were excluded.
A total of 708 peripheral blood samples (271 males,
437 females; age range, 15–79 years) were obtained from 708 MDD
patients at the First Affiliated Hospital of Chongqing Medical
University (Chongqing, China). Venous blood samples were collected
in 10-ml Vacutainer tubes (BD Bioscience, Franklin Lakes, NJ, USA)
containing the chelating agent EDTA and then separated into
200-µl blood samples that were stored individually at −80°C.
Genomic DNA was extracted from 0.2 ml of each blood sample using
the QIAamp® DNA Blood Mini kit (Qiagen, Hilden,
Germany). To remove RNA from the eluted DNA, 2 µl RNase (10
mg/ml) was added to the samples followed by incubation at 37°C for
15 min.
APOE genotyping by ARMS-PCR
APOE genotyping by ARMS-PCR was performed with
specific Cys primers (Cys112 and Cys158) as well as Arg primers
(Arg112 and Arg158) (Table I). PCR
was performed in a 20-µl reaction volume including 100 ng
genomic DNA, 0.4 µl Cys primers (10 µM) or Arg
primers (10 µM), 0.8 µl ARMS-reverse primer (common
primer; 10 µM), 1.6 µl dimethylsulfoxide, 10
µl Green mix (GoTaq® Green Master Mix; M7122;
Promega Corporation, Madison, WI, USA) and 4.8 µl
nuclease-free water. PCR amplification was initiated by
denaturation at 95°C for 5 min, followed by amplification
comprising 35 cycles of 95°C for 30 sec, 63°C for 30 sec and 72°C
for 30 sec, and final extension at 72°C for 15 min. Amplified
nucleotides were resolved by 2% agarose gel electrophoresis with a
2,000 + 1.5 Kbp ladder (Biomed, Beijing, China; DM0103) as a marker
and stained with Gold View (1:20,000). Negative controls were used
throughout the experiment as appropriate. The theoretical results
determined using Primer Premier 5.0 software (Premier Biosoft, Palo
Alto, CA, USA) are shown in Table
II. For this genotyping method, the detection of each sample
was repeated in triplicate.
 | Table IPrimers for amplification refractory
mutation system polymerase chain reaction. |
Table I
Primers for amplification refractory
mutation system polymerase chain reaction.
| Primer name | Primer sequence | Product length
(bp) |
|---|
| Arg112 (forward) |
5′-CGCGGACATGGAGGACGTTC-3′ | 588 |
| Arg158 (forward) |
5′-ATGCCGATGACCTGCAGACGC-3′ | 451 |
| Common primer
(reverse) |
5′-GTTCAGTGATTGTCGCTGGGCA-3′ | |
| Cys112 (forward) |
5′-CGCGGACATGGAGGACGTTT-3′ | 588 |
| Cys158 (forward) |
5′-ATGCCGATGACCTGCAGACGT-3′ | 451 |
 | Table IITheoretical results of amplification
refractory mutation system polymerase chain reaction. |
Table II
Theoretical results of amplification
refractory mutation system polymerase chain reaction.
| Genotype | Cys112 (bp) | Cys158 (bp) | Arg112 (bp) | Arg158 (bp) |
|---|
| ε2ε2 | 588 | 451 | 0 | 0 |
| ε4ε4 | 0 | 0 | 588 | 451 |
| ε3ε3 | 588 | 0 | 0 | 451 |
| ε2ε3 | 588 | 451 | 0 | 451 |
| ε3ε4 | 588 | 0 | 588 | 451 |
| ε2ε4 | 588 | 451 | 588 | 451 |
APOE genotyping by optimized
PCR-RFLP
Optimized PCR-RFLP was performed in a 25-µl
reaction mixture containing 100 ng purified genomic DNA, 0.2
µM apoE-forward and apoE-reverse primers
(5′-ACAGAATTCGCCCCGGCCTGGTACACTGCCA-3′ and
5′-TCCAAGGAGCTGCAGGCGGCGCA-3′, respectively; product length, 227
bp) 12.5 µl Green mix, and nuclease-free water. PCR
amplification was initiated by denaturation at 95°C for 5 min,
followed by 36 cycles of 95°C for 30 sec, 69°C for 30 sec and 72°C
for 30 sec, and a final extension at 72°C for 15 min. The
AflIII digestion mixture contained 10 µl PCR products
and five units of AflIII (R0541L; New England Biolabs,
Ipswich, WI, USA) in the buffer supplied by the manufacturer (NEB 3
buffer). Similarly, the HaeII digestion mixture contained 10
µl PCR products mixed with 10 U HaeII (R0107L; New
England Biolabs) in the buffer supplied by the manufacturer (NEB 4
buffer). The two reactions were allowed to proceed for at least
three hours at 37°C. The resulting fragments were separated on a 4%
agarose gel with a 50-bp marker (Biomed; DM0903), and the bands
were visualized by Gold View [3,6-Bis(dimethylamino) acridine zinc
chloride hydrochloride] staining (1:20,000). The gel images were
captured by a ChemiDoc XRS gel imaging system (1000 Alfred Nobel
Driver, 94547; Bio-Rad Laboratories Inc., Hercules, CA, USA).
Negative controls were used as appropriate. The theoretical results
which obtained by the Primer Premier 5.0 software are shown in
Table III. Each of the 708
samples was analyzed three times using this genotyping method.
 | Table IIITheoretical results of optimized
polymerase chain reaction restriction-fragment length
polymorphism. |
Table III
Theoretical results of optimized
polymerase chain reaction restriction-fragment length
polymorphism.
| Genotype | AflIII
digestion fragment (bp) | HaeII
digestion fragment (bp) |
|---|
| ε2/ε2 | 177, 50 | 227 |
| ε4/ε4 | 227 | 195, 32 |
| ε3/ε3 | 177, 50 | 195, 32 |
| ε2/ε3 | 177, 50 | 227, 195, 32 |
| ε3/ε4 | 227, 177, 50 | 195, 32 |
| ε2/ε4 | 227, 177, 50 | 227, 195, 32 |
APOE genotyping by Sanger sequencing
APOE genotyping results from ARMS-PCR and optimized
PCR-RFLP were confirmed against the gold standard for gene
sequencing, Sanger sequencing (Invitrogen Life Technologies,
Carlsbad, CA, USA). PCR was performed in a 25-µl reaction
volume including 100 ng purified genomic DNA, 0.2 µM
APOE-forward primer, 5′-TAAGCTTGGCACGGCTGTCCAAGGA-3′, and
APOE-reverse primer, 5′-ACAGAATTCGCCCCGGCCTGGTACAC-3′ (9), 12.5 µl Green Mix
(GoTaq® Green Master Mix; M7122; Promega Corporation)
and nuclease-free water. PCR amplification was initiated by
denaturation at 95°C for 5 min, followed by 36 cycles of 95°C for
30 sec, 65°C for 30 sec, and 72°C for 30 sec, and final extension
at 72°C for 15 min. Negative controls were used as appropriate. The
detection of each sample was repeated in triplicate.
Statistical analysis
Data were analyzed using SPSS 19.0 (International
Business Machines, Armonk, NY, USA). Pearson's χ2 test
was applied to assess the diversity of the two genotyping methods.
Comparing ARMS-PCR results with DNA sequencing results, the Pearson
χ2 value was 0.96, while comparing optimized PCR-RFLP
results to DNA sequencing results, the Pearson χ2 value
was 0.001. This indicated that ARMS-PCR results and PCR-RFLP
results were not wholly consistent.
Results
APOE genotyping by ARMS-PCR
The results of the ARMS-PCR analysis showing the six
unique APOE genotypes are displayed in Fig. 1. Amplification of the ε3/ε3
genotype, which contains Cys at codon 112 and Arg at codon 158,
generated a 588-bp product when the Cys primers were used and a
451-bp product when the Arg primers were used. Amplification of the
ε4/ε4 genotype, which carries Arg at codons 112 and 158, resulted
in 588- and 451-bp products when the Arg primers were used, while
no products were obtained when using the Cys primers. From the
heterozygote ε2/ε3 genotype, which contains Cys at codon 112 as
well as Cys and Arg at codon 158, 588- and 451-bp products were
generated by using the Cys primers and a 451-bp product was
obtained with the Arg primers. From the ε3/ε4 genotype, which
contains Cys and Arg at codon 112 and Arg at codon 158, a 588-bp
product was obtained with Cys primers, while 588- and 451-bp
products were obtained with Arg primers. Amplification of the ε2/ε4
genotype, which possesses Cys and Arg at codons 112 and 158,
generated 588- and 451-bp products when Cys or Arg primers were
used. Our experimental results are consistent with the results
reported by Kim et al (21).
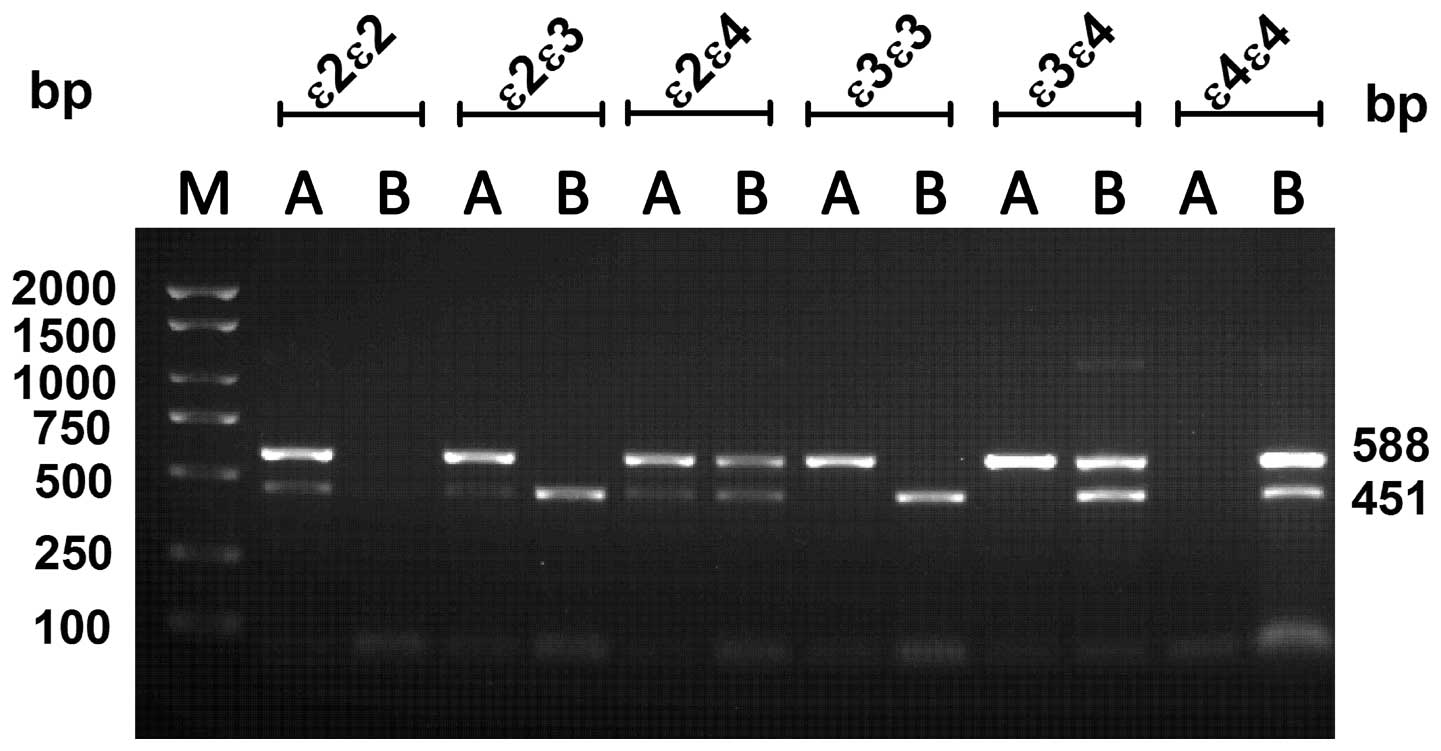 | Figure 1Gel showing ARMS-PCR products.
ARMS-PCR was performed with Cys primers (lane A) containing Cys112
(588 bp) and Cys158 (451 bp) primers or Arg primers (lane B)
containing Arg112 (588 bp) and Arg158 (451 bp) primers. Every
apolipoprotein E genotype was amplified with allele-specific
primers, including ε2/ε2 (Cys112 and Cys158), ε3/ε3 (Cys112 and
Arg158), ε4/ε4 (Arg112 and Arg158), ε2/ε3 (Cys112, Cys158, and
Arg158), ε2/ε4 (Cys112, Cys158, Arg112 and Arg158), and ε3/ε4
(Cys112, Arg112 and Arg158). Lane M: 2,000 + 1.5 Kbp ladder. All
products were separated on a 2% agarose gel. ARMS-PCR,
amplification refractory mutation system polymerase chain
reaction. |
APOE genotyping by optimized
PCR-RFLP
Optimized PCR-RFLP is a modified method using two
restriction enzymes, AflIII and HaeII, by which ε2,
ε3 and ε4 alleles can be identified in a simple and unambiguous
manner. As shown in Fig. 2,
undigested 227-bp PCR fragments were separated from the 195- and
177-bp restriction products, with each genotype presenting a unique
pattern following digestion with the two enzymes.
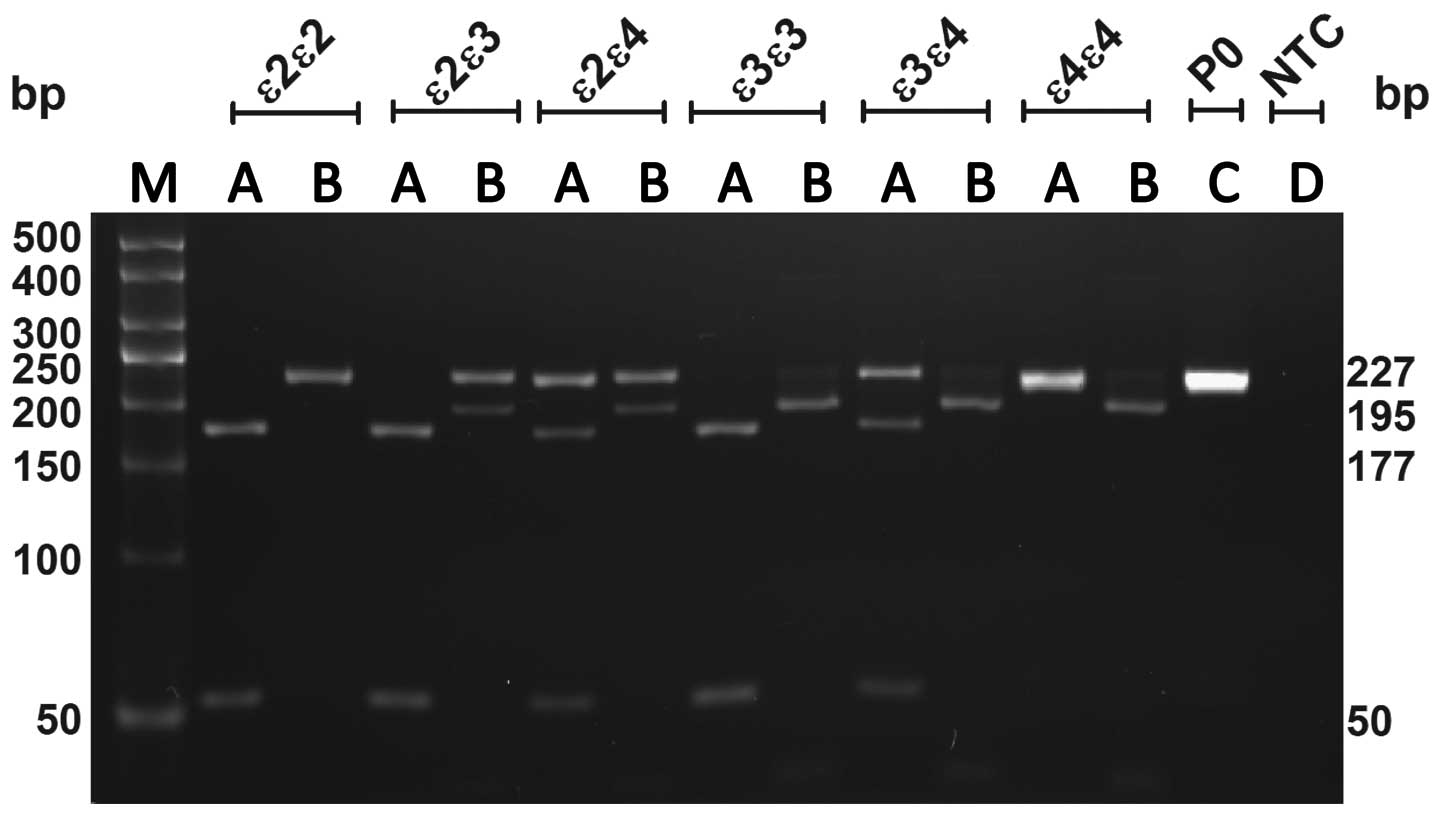 | Figure 2Gel showing optimized PCR
restriction-fragment length polymorphism products. Lanes: A, PCR
fragments digested by AflIII; B, PCR fragments digested by
HaeII [namely, ε2/ε2: AflIII (177 and 50 bp) and
HaeII (227 bp); ε3/ε3: AflIII (177 and 50 bp) and
HaeII (195 and 32 bp); ε4/ε4 AflIII (227 bp) and
HaeII (195 and 32 bp); ε2/ε3 (177 and 50 bp) and
HaeII (227, 195 and 32 bp); ε2/ε4 AflIII (227, 177
and 50 bp) and HaeII (227, 195 and 32 bp); ε3/ε4
AflIII (227, 177 and 50 bp) and HaeII (195 and 32
bp)]; C, undigested PCR fragment (227 bp); D, template control; M,
50-bp ladder. All products were separated on a 4% agarose gel. PCR,
polymerase chain reaction. |
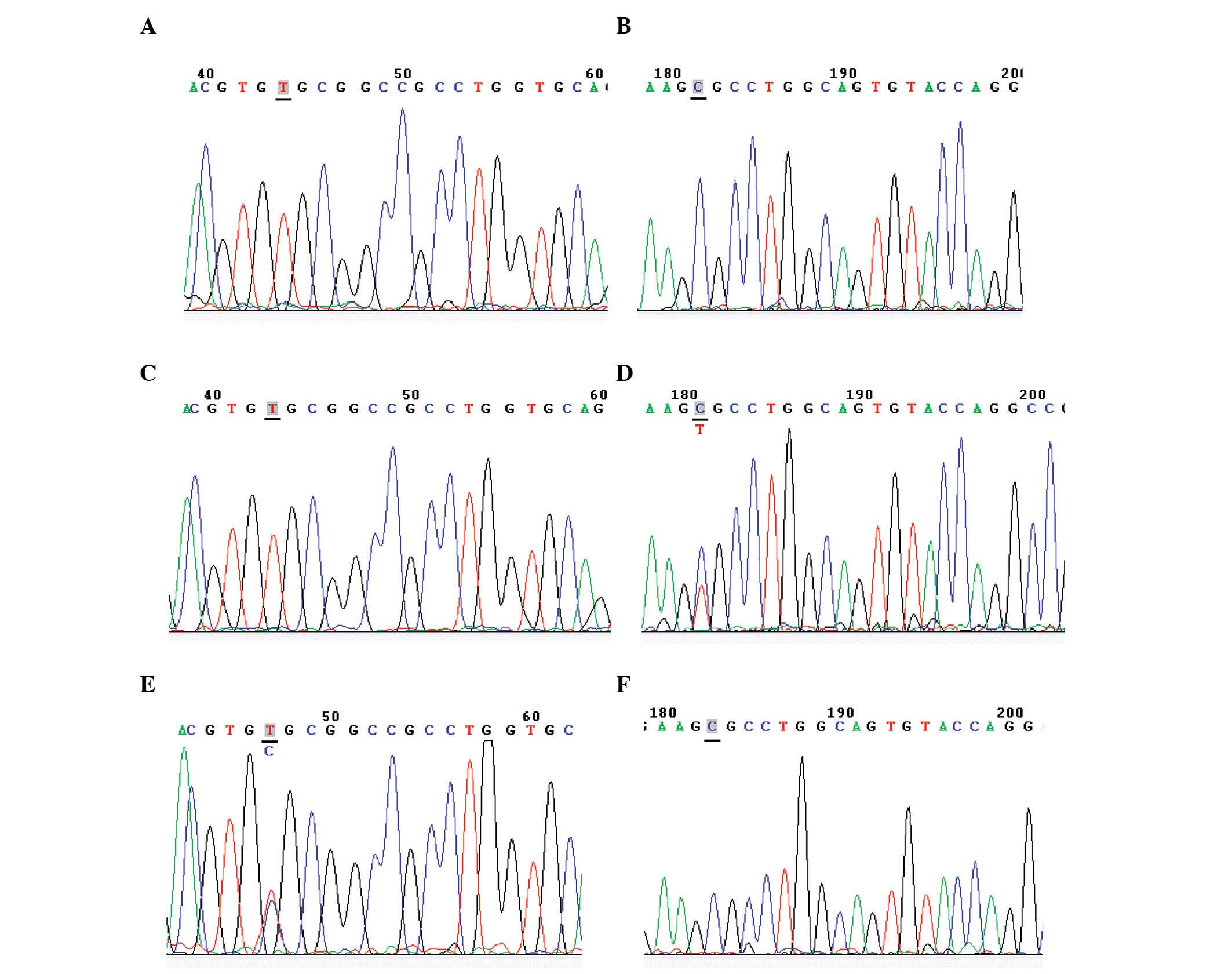
APOE genotyping by Sanger sequencing
As Sanger sequencing is considered the gold standard
for genotyping, all results obtained from ARMS-PCR and optimized
PCR-RFLP were compared against those from Sanger sequencing. Three
representative APOE genotypes determined by Sanger sequencing are
shown in Fig. 3. The APOE
frequencies obtained by ARMS-PCR, optimized RFLP-PCR and Sanger
sequencing are shown in Table
IV.
 | Table IVApolipoprotein E genotype frequencies
[n (%)] in the major depressive disorder study population
(n=708). |
Table IV
Apolipoprotein E genotype frequencies
[n (%)] in the major depressive disorder study population
(n=708).
| Method | ε3/ε3 | ε3/ε4 | ε2/ε3 | ε2/ε2 | ε2/ε4 | ε4/ε4 |
|---|
| ARMS-PCRa | 465 (65.7) | 113 (16) | 105 (14.8) | 5 (0.7) | 16 (2.3) | 4 (0.6) |
| Optimized
PCR-RFLPb | 402 (56.8) | 114 (16.1) | 146 (20.6) | 6 (0.8) | 37 (5.2) | 3 (0.4) |
| Sanger
sequencing | 460 (65) | 111 (15.7) | 115 (16.2) | 4 (0.6) | 13 (1.8) | 5 (0.7) |
APOE genotyping by ARMS-PCR is more
accurate than optimized PCR-RFLP
Pearson's χ2 test was applied to assess
the diversity of the two genotyping methods, the results of which
were not fully consistent (Table
IV). After comparing the accuracy of the two genotyping methods
against Sanger sequencing, ARMS-PCR (94%) was found to be more
accurate than optimized PCR-RFLP (82%) in detecting APOE genotypes
in these MDD patients (Table
V).
 | Table VAccuracy of ARMS-PCR versus optimized
PCR-RFLP based on Sanger sequencing (n=708). |
Table V
Accuracy of ARMS-PCR versus optimized
PCR-RFLP based on Sanger sequencing (n=708).
| Method | True cases (n) | False casesa (n) | Accuracy (%) |
|---|
| ARMS-PCR | 664 | 44 | 94 |
| Optimized
PCR-RFLP | 581 | 127 | 82 |
Discussion
In order to find a more accurate and lower-cost APOE
genotyping method for MDD screening in large populations, the
present study comparatively evaluated two genotyping methods,
ARMS-PCR and optimized PCR-RFLP, in blood samples from 708 MDD
patients. Although the two APOE genotyping methods were able to
detect all six APOE genotypes, Pearson's χ2 test
revealed that the APOE genotyping results of ARMS-PCR and optimized
PCR-RFLP were not fully consistent. Comparison of the two methods
with Sanger sequencing demonstrated that ARMS-PCR was significantly
more accurate than optimized PCR-RFLP.
After years of research and development, several
genotyping techniques for APOE have been introduced. The earliest
methods for detecting APOE isoforms were based on protein
isoelectric focusing electrophoresis (IEF) (22). Since IEF requires considerable
expertise and expensive instrumentation, it was not particularly
practical for small laboratories or population-based screening
programs. Thereafter, molecular genetic techniques (23) based on PCR amplification and
HhaI restriction enzyme digestion were introduced (9,15,24).
However, these HhaI-based assays were difficult to interpret
as HhaI digestion yielded several small fragments; in
addition, incomplete digestion by HhaI produced ambiguous
results. Through utilizing two distinct restriction enzymes
(AflIII and HaeII), the quality of the results was
significantly improved but the cost of running the assay was also
increased.
From these early methods, more advanced PCR-based
APOE genotyping techniques have been recently developed, including
allele-specific PCR (e.g. ARMS-PCR), single-stranded conformational
polymorphism (SSCP) and real-time PCR. SSCP requires higher
separation systems, and real-time PCR requires expensive reagents
and instruments, and therefore, the cost of these methods is
prohibitively high for small laboratories or population-based
screening programs. Compared with these advanced genotyping
methods, ARMS-PCR does not rely on restriction enzyme digestion,
other treatment steps, or expensive reagents and instrumentation
(25). In a BRAFV600E genotyping
study, ARMS-PCR was found to be more sensitive and cost-effective
than real-time PCR for BRAF mutational screening (26). Furthermore, ARMS-PCR was found to
be more sensitive than automated dideoxy sequencing in detecting
low BRAFV600E allele burdens in formalin-fixed and
paraffin-embedded tumor specimens (27). The present study demonstrated that
ARMS-PCR (94%) was significantly more accurate than optimized
PCR-RFLP (82%) in detecting APOE genotypes in a population of 708
MDD patients. Furthermore, ARMS-PCR has distinct advantages over
PCR-RFLP in terms of its cost of reagents and instrumentation, time
consumption and simplicity of experimental processing, but faces a
singular disadvantage to other genotyping methods in being unable
to detect novel genetic mutations (25). Overall, this combination of factors
make ARMS-PCR a superior APOE genotyping method for MDD screening
in large populations.
It should be pointed out that the present study had
several limitations: First, only blood samples from MDD patients
were used; therefore the accuracy of APOE genotyping in healthy
individuals or neuropsyciatric patients with clinical presentations
similar to MDD, including bipolar disorder and schizophrenia, was
not evaluated in the present study. Second, the entire population
of 708 MDD patients in the present study was of Han Chinese
ethnicity residing in the Chongqing metropolitan area; thus, future
studies should use a more ethnically heterogeneous population
sampled from multiple clinical sites in order to improve the
validity of the conclusions. Third, the present study only compared
ARMS-PCR to PCR-RFLP for genotyping of APOE polymorphisms in MDD
patients; therefore, future studies should include other modern
genotyping methods, including SSCP and real-time PCR, and assess
other genetic polymorphisms associated with MDD, including DRD4,
GNB3, MTHFR, SLC6A3 and SLC6A4.
In conclusion, the present study showed that
ARMS-PCR was significantly more accurate than optimized PCR-RFLP in
APOE genotyping of MDD patients. ARMS-PCR should prove useful in
quickly verifying ambiguous results obtained by other APOE
genotyping methods and can be cost-effectively performed in the
setting of a small laboratory or a population-based screening
program.
Acknowledgments
The authors would like to thank the scientific
editors at Impactys for editing and proofreading this manuscript.
The authors also express their gratitude to Dr Ke Cheng,
(Department of Neurology, First Affiliated Hospital of Chongqing
Medical University). This work was supported by the National Basic
Research Program of China (973 Program; grant no. 2009CB918300) and
the National Natural Science Foundation of China (grant no.
31271189).
References
|
1
|
Bakish D: New standard of depression
treatment: Remission and full recovery. J Clin Psychiatry. 62(Suppl
26): 5–9. 2001.
|
|
2
|
López-León S, Janssens AC,
González-Zuloeta Ladd AM, Del-Favero J, Claes SJ, Oostra BA and van
Duijn CM: Meta-analyses of genetic studies on major depressive
disorder. Mol Psychiatry. 13:772–785. 2008. View Article : Google Scholar
|
|
3
|
Ramachandran G, Marder K, Tang M,
Schofield PW, Chun MR, Devanand DP, Stern Y and Mayeux R: A
preliminary study of apolipoprotein E genotype and psychiatric
manifestations of Alzheimer's disease. Neurology. 47:256–259. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Willnow TE: The low-density lipoprotein
receptor gene family: Multiple roles in lipid metabolism. J Mol Med
(Berl). 77:306–315. 1999. View Article : Google Scholar
|
|
5
|
Das HK, McPherson J, Bruns GA,
Karathanasis SK and Breslow JL: Isolation, characterization, and
mapping to chromosome 19 of the human apolipoprotein E gene. J Biol
Chem. 260:6240–6247. 1985.PubMed/NCBI
|
|
6
|
Emi M, Wu LL, Robertson MA, Myers RL,
Hegele RA, Williams RR, White R and Lalouel JM: Genotyping and
sequence analysis of apolipoprotein E isoforms. Genomics.
3:373–379. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Utermann G, Langenbeck U, Beisiegel U and
Weber W: Genetics of the apolipoprotein E system in man. Am J Hum
Genet. 32:339–347. 1980.PubMed/NCBI
|
|
8
|
Cartier R and Sassolas A: Apolipoprotein E
phenotyping by isoelectric focusing in immobilized pH gradients and
silver staining. Electrophoresis. 13:252–257. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hixson JE and Vernier DT: Restriction
isotyping of human apolipoprotein E by gene amplification and
cleavage with HhaI. J Lipid Res. 31:545–548. 1990.PubMed/NCBI
|
|
10
|
Srinivasan JR, Kachman MT, Killeen AA,
Akel N, Siemieniak D and Lubman DM: Genotyping of apolipoprotein E
by matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry. Rapid Commun Mass Spectrom. 12:1045–1050. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ben-Avi L, Durst R, Shpitzen S,
Leitersdorf E and Meiner V: Apolipoprotein E genotyping: Accurate,
simple, high throughput method using ABI Prism SNaPshot Multiplex
System. J Alzheimers Dis. 6:497–501. 2004.PubMed/NCBI
|
|
12
|
Aozaki R, Kawaguchi R, Ogasa U, Hikiji K,
Kubo N and Sakurabayashi I: Rapid identification of the common apo
E isoform genotype using polymerase chain reaction-single strand
conformation polymorphism (PCR-SSCP). Mol Cell Probes. 8:51–54.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nauck M, Hoffmann MM, Wieland H and März
W: Evaluation of the Apo E genotyping kit on the LightCycler. Clin
Chem. 46:722–724. 2000.PubMed/NCBI
|
|
14
|
Donohoe GG, Salomäki A, Lehtimäki T,
Pulkki K and Kairisto V: Rapid identification of apolipoprotein E
genotypes by multiplex amplification refractory mutation system PCR
and capillary gel electrophoresis. Clin Chem. 45:143–146.
1999.PubMed/NCBI
|
|
15
|
Chapman J, Estupiñan J, Asherov A and
Goldfarb LG: A simple and efficient method for apolipoprotein E
genotype determination. Neurology. 46:1484–1485. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen J, Zhang X, Gao P and Jiang Q:
Comparison between two PCR-based bacterial identification methods
through artificial neural network data analysis. J Clin Lab Anal.
22:14–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Health Quality O; Health Quality Ontario:
Epidermal growth factor receptor mutation (EGFR) testing for
prediction of response to EGFR-targeting tyrosine kinase inhibitor
(TKI) drugs in patients with advanced non-small-cell lung cancer:
An Evidence-Based Analysis. Ont Health Technol Assess Ser. 10:1–48.
2010.
|
|
18
|
American Psychiatric Association:
Diagnostic and statistical manual of mental disorders. 4th.
Washington DC: 1994
|
|
19
|
Persson ML, Runeson BS and Wasserman D:
Diagnoses, psychosocial stressors and adaptive functioning in
attempted suicide. Ann Clin Psychiatry. 11:119–128. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fava GA, Kellner R, Munari F and Pavan L:
The Hamilton Depression Rating Scale in normals and depressives.
Acta Psychiatr Scand. 66:26–32. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim SW, Heo JH, Kim CH, Yoo DC, Won DH,
Lee SG, Cho KJ, Song JH, Park SJ, Yang YG and Choi DW: Rapid and
direct detection of apolipoprotein E genotypes using whole blood
from humans. J Toxicol Environ Health A. 73:1502–1510. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Utermann G, Hees M and Steinmetz A:
Polymorphism of apolipoprotein E and occurrence of
dysbetalipoproteinaemia in man. Nature. 269:604–607. 1977.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paik YK, Chang DJ, Reardon CA, Davies GE,
Mahley RW and Taylor JM: Nucleotide sequence and structure of the
human apolipoprotein E gene. Proc Natl Acad Sci USA. 82:3445–3449.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kontula K, Aalto-Setälä K, Kuusi T,
Hämäläinen L and Syvänen AC: Apolipoprotein E polymorphism
determined by restriction enzyme analysis of DNA amplified by
polymerase chain reaction: Convenient alternative to phenotyping by
isoelectric focusing. Clin Chem. 36:2087–2092. 1990.PubMed/NCBI
|
|
25
|
Yang YG, Kim JY, Park SJ, Kim SW, Jeon OH
and Kim DS: Apolipoprotein E genotyping by multiplex tetra-primer
amplification refractory mutation system PCR in single reaction
tube. J Biotechnol. 131:106–110. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Machnicki MM, Glodkowska-Mrowka E,
Lewandowski T, Ploski R, Wlodarski P and Stoklosa T: ARMS-PCR for
detection of BRAF V600E hotspot mutation in comparison with
Real-Time PCR-based techniques. Acta Biochim Pol. 60:57–64.
2013.PubMed/NCBI
|
|
27
|
Huang T, Zhuge J and Zhang WW: Sensitive
detection of BRAF V600E mutation by Amplification Refractory
Mutation System (ARMS)-PCR. Biomark Res. 16:32013. View Article : Google Scholar
|