Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney malignancy. In addition it is the third most common type
of urological cancer after prostate and bladder cancer; however, it
has the highest mortality rate of the three at >40%. Among the
five subtypes of RCC, clear cell carcinoma (CCC) accounts for ~70%
of cases (1). Although the
majority of patients with early-stage RCC can be cured surgically,
~33% of patients present with synchronous metastatic disease for
which treat ment is usually not curative (2). The most common sites of metastatic
spread in RCC are the lung, bone, adrenal gland, liver and brain,
whereupon more than one organ system is often involved in the
metastatic process (3). In
addition, RCC is relatively resistant to radiotherapy and
chemotherapy, which result in the poor prognosis of patients with
RCC with metastatic or recurrent disease and a 5-year survival rate
of <20% (4). A number of
studies have identified putative oncogenes involved in the
carcinogenesis of RCC; however, the molecular mechanisms regulating
the aggressive properties of RCC remain poorly understood (5,6).
Hence, novel treatments are required to improve the prognosis of
patients with RCC.
MicroRNAs (miRNAs), which are a highly conserved
class of short non-coding endogenous RNAs comprising ~22
nucleotides, are endogenously expressed across mammals and other
species (7). Production and
function of miRNA requires a set of proteins collectively referred
to as the miRNA machinery (8).
Primary miRNA transcripts are first processed into precursor
microRNA (pre-miRNA). This step requires a 650-kDa microprocessor
complex that comprises of Drosha, RNase III endonuclease and DGCR8
(9–12). These pre-miRNAs are then actively
transported by Exportin-5 to the cytoplasm, where they are further
processed by the cytoplasmic RNase III enzyme Dicer (13–15).
Finally, Argonaute proteins are recruited with miRNAs into an
RNA-induced silencing complex for mRNA recognition (16). It has attracted attention for its
involvement in cell differentiation, development, apoptosis and
proliferation by targeting mRNAs for cleavage or translational
repression at the posttranscriptional level (17). The inappropriate expression of
miRNAs can lead to the aberrant expression of gene products that
may contribute to the acquisition of the hallmarks of cancer
(18). Upregulated miRNAs in
cancer may function as oncogenes by negatively regulating tumor
suppressors. By contrast, downregulated miRNAs may normally
function as tumor suppressor genes and inhibit cancer by regulating
oncogenes (19,20).
miR-204 has been reported to be frequently
downregulated in various types of cancer, including brain, kidney,
ovarian, hematological and colon cancer (20). However, the function of miR-204 has
not yet been investigated in RCC. The aim of this study was to
elucidate the effect of miR-204 on RCC and to investigate its
underlying mechanisms.
Materials and methods
Cell lines and cell culture
The 786-O and A498 human RCC cell lines were
purchased from the Shanghai Institute of Cell Biology, Chinese
Academy of Science (Shanghai, China). The cells were cultured in
RPMI-1640 (HyClone, Logan, UT, USA) medium supplemented with 10%
heat-inactivated fetal calf serum (Gibco-BRL, Grand island, NY,
USA), 100 U/ml penicillin and 100 mg/l streptomycin (Gibco-BRL)
under a humidified atmosphere of 5% CO2 at 37°C.
Transient transfection of miRNA mimics
and luciferase reporter plasmids
The miR-204 mimics, negative control (NC) and
luciferase reporter plasmid were designed and synthesized by
GenePharma (Shanghai, China). The insertion fragment was confirmed
by DNA sequencing. Cell transfection and cotransfection were
performed using Lipofectamine™ 2000 (Invitrogen Life Technologies,
Carlsbad, CA, USA) according to manufacturer's instructions.
Following transfection, cells were incubated at 37°C until
assessment.
Cell viability assay
Cell proliferation was measured using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT) method (Sigma-Aldrich). After 48 h transfection, the cells
were trypsinized (Gibco-BRL) and counted, respectively. Cells were
counted under a microscope (CKX41; Olympus, Tokyo, Japan). Cells
were plated in each well of 96-well plates at a density of 3,000
cells per well and incubated at 37°C. Cell proliferation was
documented every 24 h for 5 days according to the manufacturer's
instructions. Absorbance was measured at 490 nm using an automatic
multi-well spectrophotometer (Bio-Rad, Richmond, CA, USA). There
were six wells for every time point in each group. The growth
inhibition rate was calculated using the following equation: Growth
inhibition rate = (1-ODmiR-204/ODmiR-NC ×
100; where OD is the optical density. All the experiments were
performed in triplicate.
Migration and invasion assay
In vitro cell migration and invasion assays
were performed using 8 µm-pore polycarbonate membrane Boyden
chamber inserts in a Transwell apparatus (Costar, Cambridge, MA,
USA). The transfected cells (miR-204 mimics and negative control)
growing in the log phase were treated with trypsin/EDTA solution
(Gibco-BRL), washed once with no serum-containing medium,
centrifuged at 200 × g for 5 min and re-suspended as single-cell
solutions in no-serum containing medium. For the migration assays,
1×105 cells in 200 µl serum-free RPMI-1640 medium
were seeded on the upper chamber of transwell apparatus. For the
invasion assays, 1×105 cells were added to the upper
chamber of the transwell precoated with 30 µg Matrigel (BD
Biosciences, San Jose, CA, USA). In these assays, 600 µl
RPMI-1640 containing 20% fetal calf serum was added to the lower
chamber, serving as a chemoattractant. After 12–24 h at 37°C in a
5% CO2 incubator, the cells that had not migrated or
invaded through the pores were carefully removed with a cotton
swab. The filters were then fixed in 100% methanol for 2 min,
stained in 0.5% crystal violet (Beyotime Institute of
Biotechnology, Haimen, China) for 2 min, rinsed in
phosphate-buffered saline and then subjected to microscopic
inspection (CKX41; Olympus). Values for migration and invasion were
obtained by counting five fields per membrane and represent the
average of three independent experiments.
Western blot analysis
Primary antibodies used in this study including
rabbit anti-human monoclonal SOX4 (1:500; cat. no. BS8784) and
mouse anti-human monoclonal β-actin (1:1,000; cat. no. AP0060) were
purchased from Bioworld Technology (Louis Park, MN, USA). Total
protein of cells extracts were prepared in radioimmunoprecipitation
assay lysis buffer (Beyotime Institute of Biotechnology). Protein
concentration in the resulting lysate was performed using a
Bicinchoninic Acid Protein assay kit (Thermo Fisher Scientific,
Inc., Rockford, IL, USA) according to the manufacturer's
instructions. Briefly, equal quantities of protein were loaded onto
a 10% SDS-PAGE gel (Beyotime Institute of Biotechnology) and
electroblotted onto a polyvinylidene difluoride membrane
(Millipore, Billerica, MA, USA). The membranes were blocked in
phosphate-buffered saline containing 0.1% Tween-20 (Beyotime
Institute of Biotechnology) and 5% non-fat dry milk. The membranes
were incubated with primary antibody overnight at 4°C. Following
washing, the membranes were incubated with the corresponding
horseradish peroxidase-conjugated secondary antibody (Bioworld
Technology) in Tris-buffered saline with Tween-20. The bands were
then developed using an ECL solution (Pierce Biotechnology, Inc.,
Rockford, IL, USA) and images were captured using a FluorChem
imaging system (Alpha Innotech, San Leandro, CA, USA). The
intensity of each spot was read and analyzed with AlphaEaseFC
software. β-actin was used as a loading control.
Luciferase assay
TargetScan 5.2 (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/) were used to assess the
complementarity of miR-204 to the SOX4 3′-UTR. Luciferase reporter
assays were performed to evaluate whether SOX4 is a target of
miR-204. Cells were plated in a 12-well plate and transfected with
0.5 µg reporter plasmid, 40 nmol miR-204 mimics or their
negative control. Transfection was performed using Lipofectamine
2000. Each sample was also cotransfected with 0.05 µg
pRL-CMV plasmid expressing Renilla Luciferase (Promega
Corporation, Madison, WI, USA) as an internal control for
transfection efficiency. Relative luciferase activity was
calculated 48 h post-transfection by the Dual Luciferase Reporter
Assay kit (Promega Corporation). Firefly luciferase activity was
normalized to Renilla luciferase activity for each
transfected well. Each assay was replicated three times.
Statistical analysis
Data are presented as the mean ± standard deviation,
and compared using Student's t-test in Stata 10.0 (College Station,
Texas, USA). Double-tailed P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-204 suppresses cell proliferation in
RCC cell lines
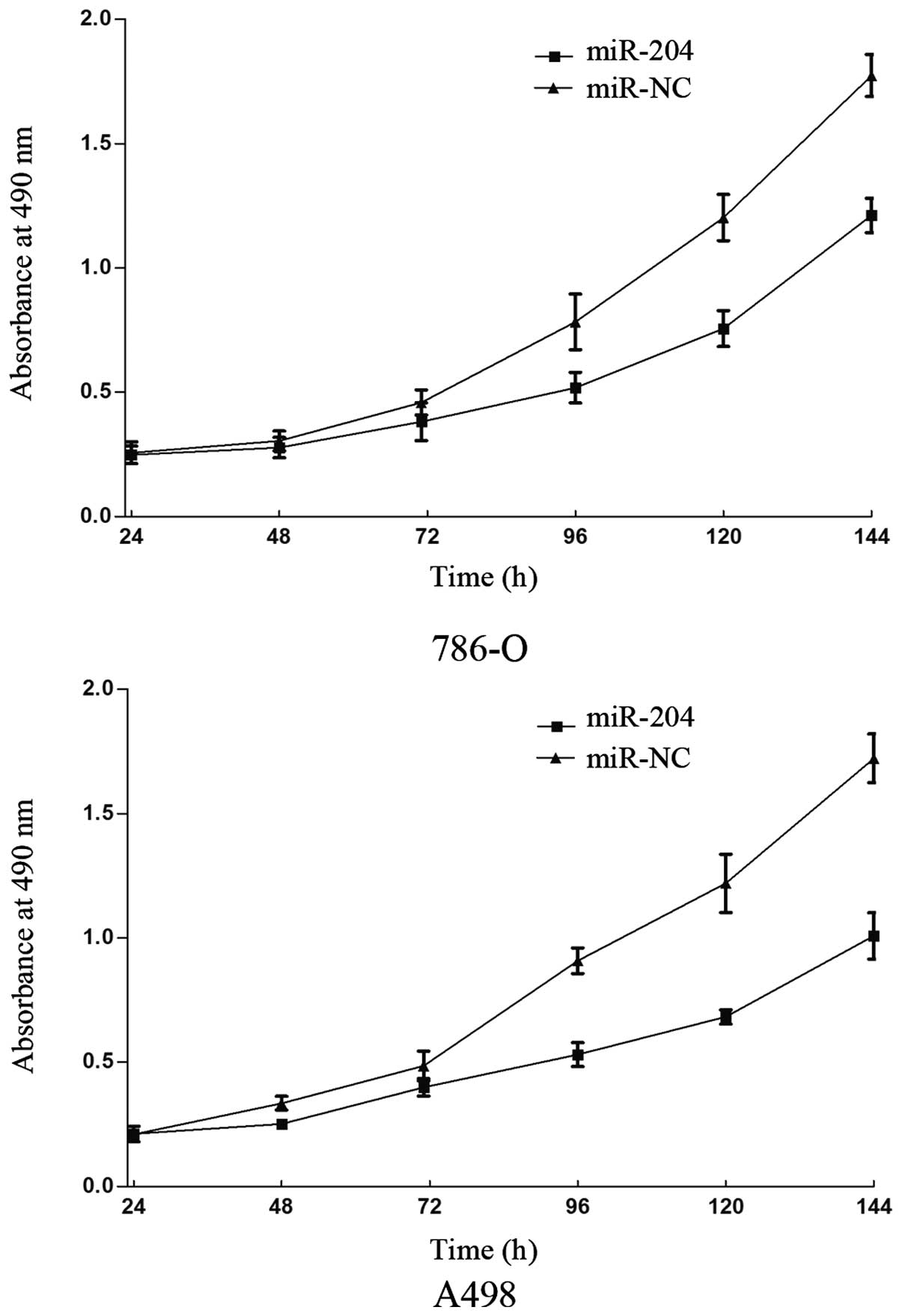
The effect of miR-204 on 786-O and A498 cell
proliferation was investigated using MTT assays. The data showed
significant cell growth inhibition in the miR-204 transfectant
compared with the control from 786-O and A498 cell lines
(P<0.05). As shown in Fig. 1,
MTT assays revealed that after 144 h of treatment, the suppression
rate of miR-204 reached 29.12±3.5% in 786-O cells and 36.68±4.5% in
A498 cells. These results indicated that miR-204 may be important
in 786-O and A498 cell lines.
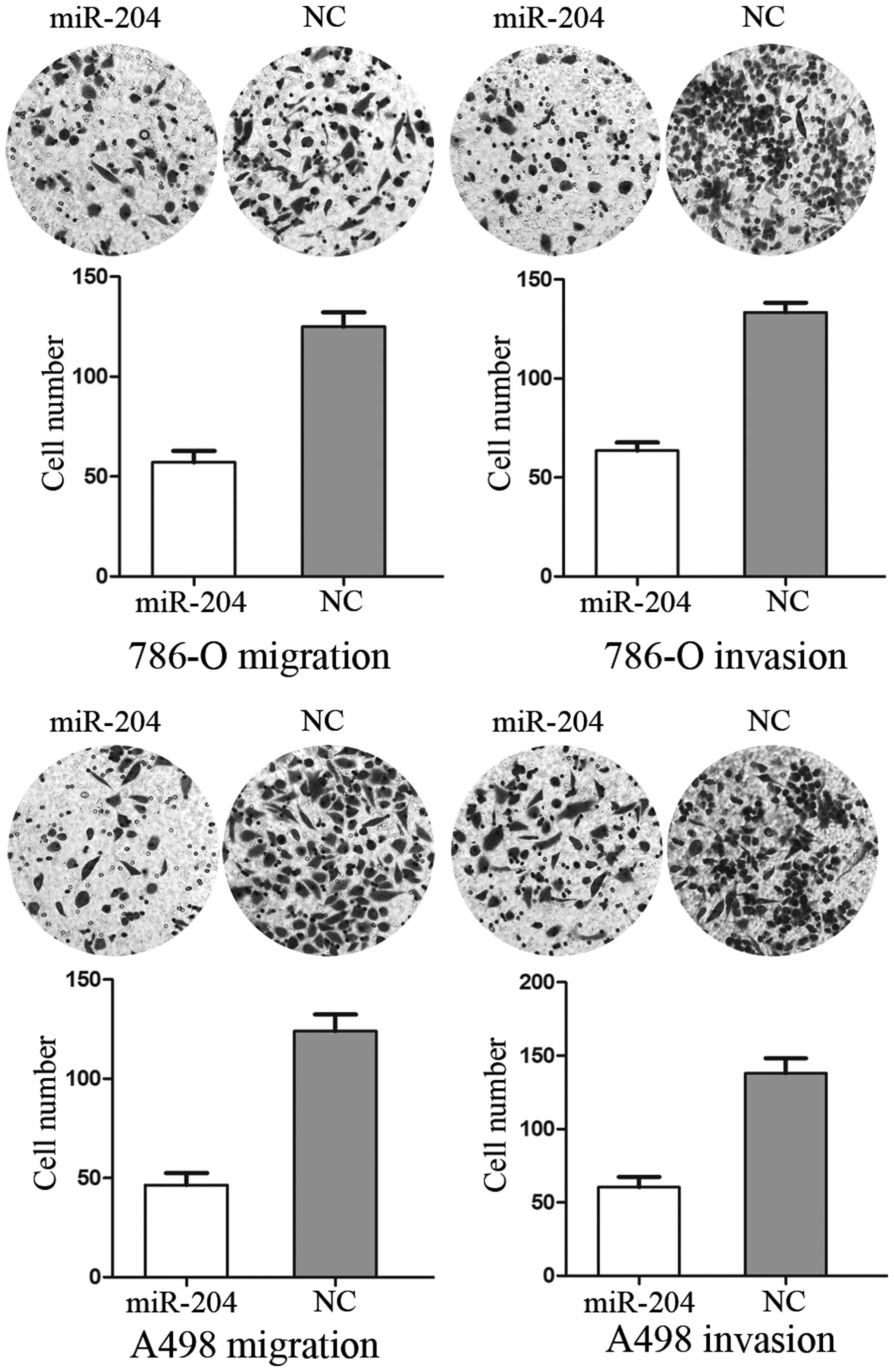
miR-204 inhibits cell migration and
invasion in RCC cell lines
The Transwell assay was performed to measure the
effect of miR-204 on tumor cell migration and invasion. As shown in
Fig. 2, cell migration and
invasion were significantly decreased in miR-204 group compared
with the control group (P<0.05). These results indicated that
miR-204 inhibits the cell migration and invasion in RCC cell
lines.
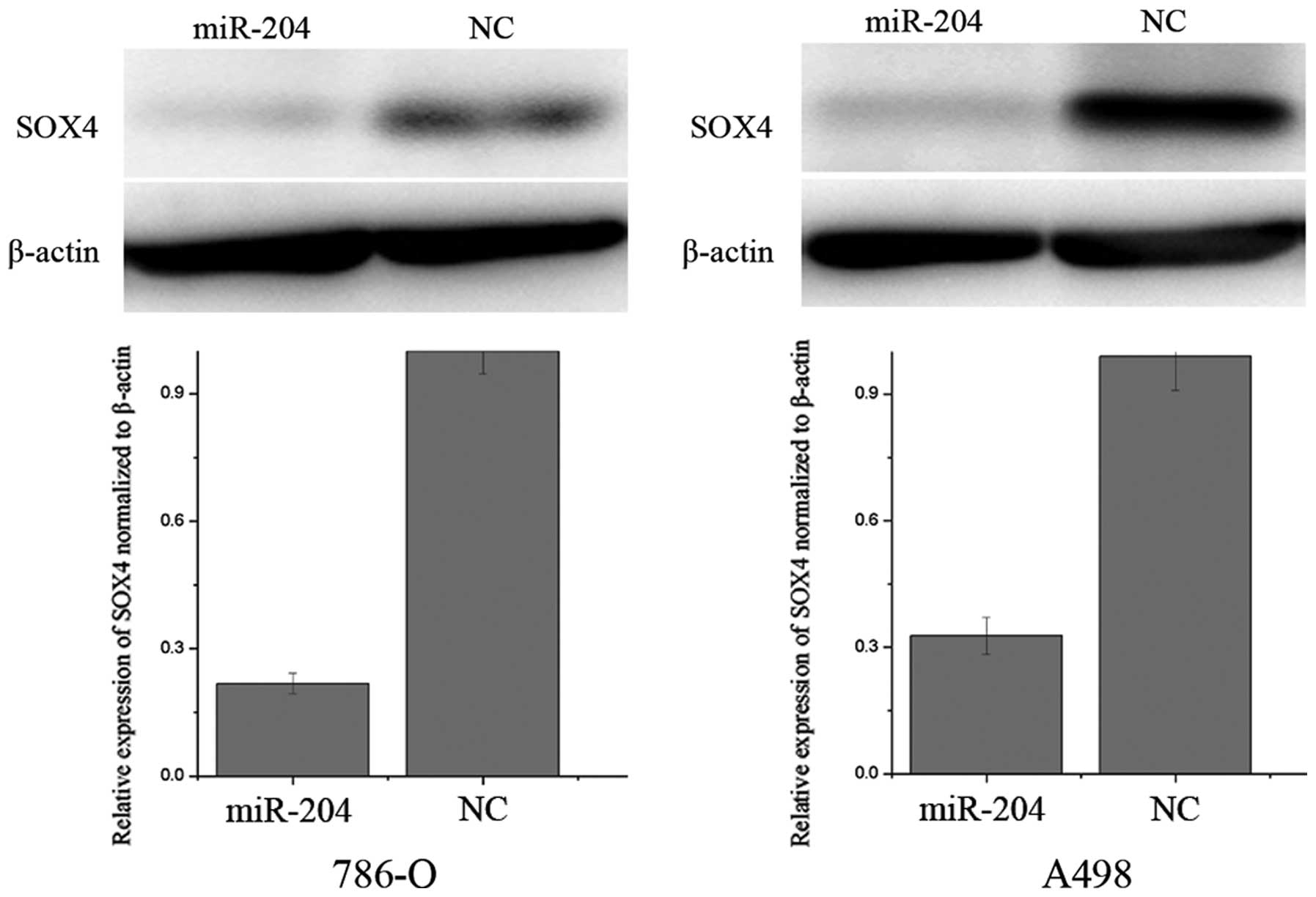
miR-204 suppresses the expression of SOX4
in RCC cell lines
Zhou et al (21) revealed that miR-204 may act as a
tumor suppressor in Helicobacter pylori-induced gastric
cancer by downregulation of SOX4. Western blot analysis was
performed to determine whether the SOX4 protein level decreased
following overexpression of miR-204. As shown in Fig. 3, SOX4 expression was significantly
decreased in 786-O and A498 cells after transfection of miR-204
(P<0.05). Thus, miR-204 reduces the protein level of SOX4 in
786-O and A498 RCC cell lines.
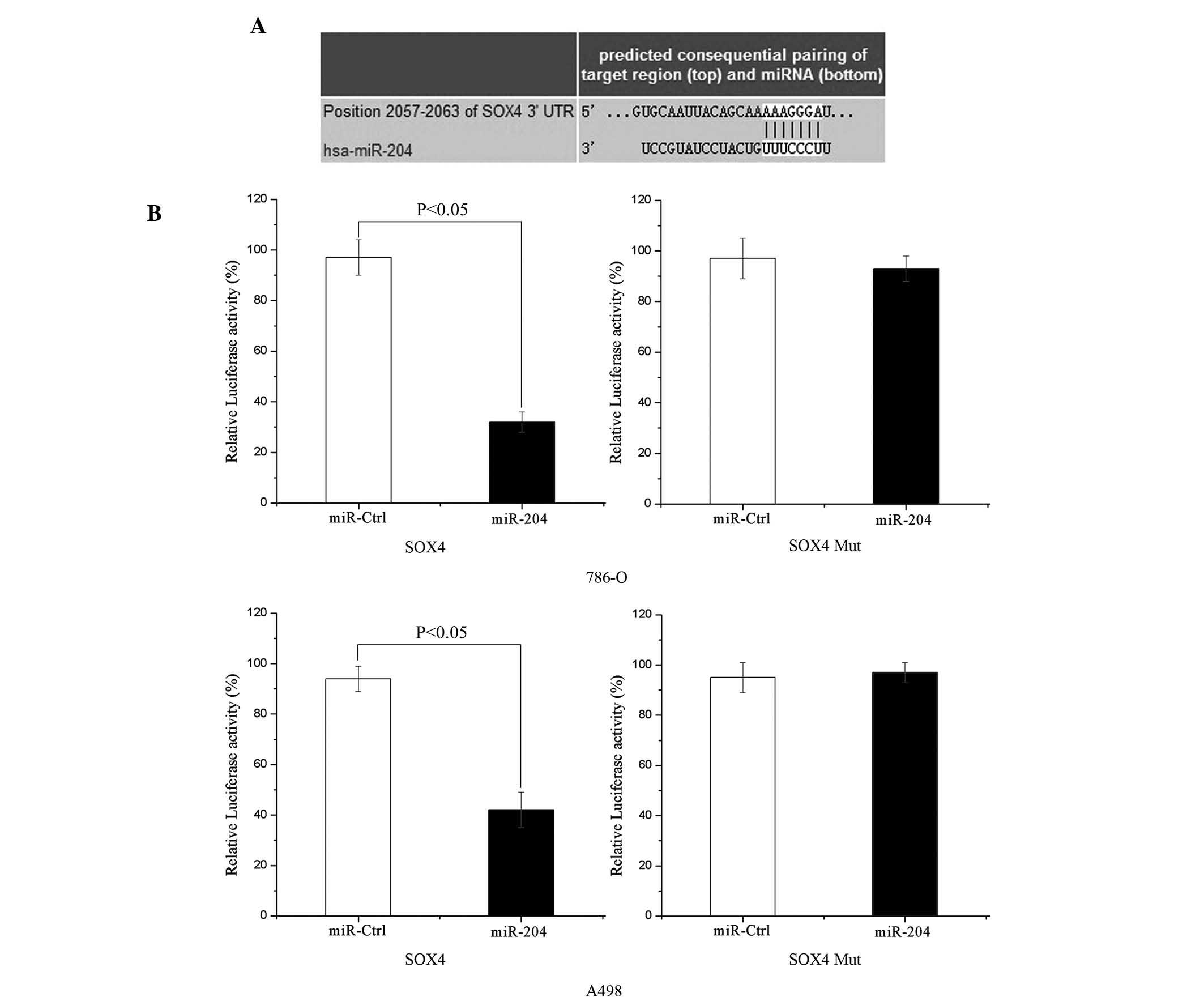
SOX4 is a direct target of miR-204
To determine whether miR-204 targets the SOX4
3′-untranslated region (UTR), TARGETSCAN 5.2 (http://www.targetscan.org/) and PICTAR (http://pictar.mdc-berlin.de/) were used to assess the
complementarity of miR-204 to the SOX4 3′-UTR. It was shown that
SOX4 mRNA contained an miR-204 seven-nucleotide seed match at
position 2057–2063 of the SOX4 3′-UTR (shown in Fig. 4A).
Luciferase reporter assays were performed to
evaluate whether the site could directly mediate expression
inhibition. As shown in Fig. 4B,
upregulation of miR-204 suppressed SOX4 3′UTR-luciferase activity
by 67% in 786-O cells and 55% in A498 cells (P<0.05). Thus, SOX4
may be a direct target of miR-204 in vitro.
Discussion
miRNAs have emerged as a novel mechanism of gene
regulation in recent years. To date, there are 1,527 human miRNAs
and 741 mouse miRNAs registered in the miRBase (http://www.mirbase.org/) (22–24).
However, thousands of miRNAs in various genomes and their targets
still require validation (23).
Investigation of the differentially expressed miRNAs in cancer
specimens has yielded important information on carcinogenesis
(25). Although RCC generally
carries a favorable prognosis, patients with metastatic RCC face a
poor prognosis and have limited therapeutic options. The median
survival rate in a recent cohort was only 1.5 years with <10% of
patients surviving 5 years after the initial diagnosis (26). Therefore, it is important to
determine the molecular pathways involved in RCC in order to
improve the diagnosis of and therapeutic options for the
disease.
miR-204, is located at the cancer-associated genomic
9q21.1-q22.3 locus and exhibits a high frequency of loss of
heterozygosity in certain types of tumor (27–29).
It is also located within the sixth intron of the host gene
transient receptor potential melastatin 3 cation channel and is
transcribed in the same direction as TRPM3 (30). The expression of miR-204 was
observed to be significantly decreased by 0.07–5%, in tumors in 5
of the 9 tissue types (brain, kidney, ovary, hematological cells,
and colon) compared with normal tissues (31). In addition, miR-204 expression was
observed to be downregulated in 60 tumor sample tissues compared
with 13 matched normal tissues (32). In addition, significant
downregulation of miR-204 was found in a subtype of acute myeloid
leukemia-bearing cytoplasmic mutated nucleophosmin and in 3 Burkitt
B-cell lymphoma cell lines (33).
In RCC, the miR-204 level was also found to be decreased as
compared with matched normal kidney tissue in paired and unpaired
analyses (34). These studies
strongly suggest that miR-204 functions as a tumor suppressor.
miR-204 appears to be an important regulator of cell
differentiation, apoptosis, stress response, inflammation, lens
development, retinal development, and in the maintenance of axonal
structure and function (35–38).
It has been shown to act as a tumor suppressor in a variety of
cancer types through different mechanisms (34,39,40).
It also reduced cell migration, invasion, and the formation of
metastatic tumors in a variety of squamous cell carcinomas but had
no effect on proliferation or viability (41). Identification of miR-204 target
genes is critical for understanding the role of miR-204 in
tumorigenesis, and is important for determining novel therapeutic
targets.
Several mRNA targets have been identified that are
important in normal cell development, including MEIS1, HOXA9,
MEIS2, RUNX2 and SIRT1 (41). In
breast cancer and ovarian cancer cells, miR-204 inhibits cell
invasion and metastasis by targeting the stemness-governing
transcription factor and the migration-promoting receptor (42). In endometrial cancer, miR-204 was
found to regulate cell migration and invasion by targeting the
FOXC1 gene (39). In the present
study, it was demonstrated that miR-204 trans fection resulted in
decreased cell proliferation, migration and invasion in RCC cell
lines by targeting SOX4. The results suggested that miR-204 may be
used for the development of novel molecular markers and therapeutic
approaches for RCC.
Sox4, a transcription factor of the sex-determining
gene on the Y chromosome, is characterized by a highly conserved
sequence in the high-mobility group (HMG) DNA-binding domain (DBD)
(43). SOX4 gene is located on
6p22.3 and encodes a protein of 474 amino acids with three major
domains: UA HMG box, a glycine-rich region, and a serine-rich
region (44). The HMG box acts as
DNA-binding region, whereas the SRR domain serves as
transactivation domain. The glycine-rich region, which is located
between the HMG box and SRR, is a part of the central domain, and
this region has a function in promoting apoptotic cell death
(45).
High levels of SOX4 expression have been reported in
hepatic cancer cells and a variety of human cancer types, such as
breast, brain, lung, pancreatic, salivary gland and ovarian cancer
(46). Sox4 is important in a
number of developmental processes, including embryonic cardiac,
thymocyte and nervous system development, through its
transcriptional activity (47).
Besides functioning as a transcription factor involved in the
regulation of developmental processes, SOX4 has been implicated in
cancer progression. In the case of bladder cancer, upregulated
expression of SOX4 was significantly correlated with increased
patient survival, and overexpression of SOX4 impaired cell
viability and promoted apoptosis in cancer cells (46). Similarly, in patients with
melanoma, reduced expression of SOX4 was significantly correlated
with poor prognosis and metastasis (48). By contrast, knockdown of SOX4
induced apoptosis in prostate and adenoid cystic cancer cells, and
suppressed tumor growth and local metastasis in hepatocellular
carcinoma (49,50). Results of the present study
indicated that miR-204 suppresses RCC cell proliferation, migration
and invasion via downregulation of SOX4, and thus decreasing SOX4
levels may represent a potential therapeutic strategy for RCC.
To the best of our knowledge, the present study is
the first to demonstrate that regulation of SOX4 by miR-204
inhibits RCC cell proliferation, migration and invasion. These
observations have therapeutic implications and may be exploited
further for the treatment of RCC. Future studies are required to
determine the potential of miR-204 in cancer treatment and
specifically, RCC.
References
|
1
|
Lian JH, Wang WH, Wang JQ, Zhang YH and Li
Y: MicroRNA-122 promotes proliferation, invasion and migration of
renal cell carcinoma cells through the PI3K/Akt signaling pathway.
Asian Pac J Cancer Prev. 14:5017–5021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Toma MI, Erdmann K, Diezel M, Meinhardt M,
Zastrow S, Fuessel S, Wirth MP and Baretton GB: Lack of ephrin
receptor A1 is a favorable independent prognostic factor in clear
cell renal cell carcinoma. PloS One. 9:e1022622014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang YQ and Chen J: Predictive role of
vascular endothelial growth factor polymorphisms in the survival of
renal cell carcinoma patients. Genet Mol Res. 13:5011–5017. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kardas I, Mrózek K, Babinska M, Krajka K,
Hadaczek P, Lubinski J, Roszkiewicz A, Kuziemska E and Limon J:
Cytogenetic and molecular findings in 75 clear cell renal cell
carcinomas. Oncol Rep. 13:949–956. 2005.PubMed/NCBI
|
|
6
|
Girolami F, Passerini I, Gargano D,
Frusconi S, Villari D, Nicita G and Torricelli F: Microsatellite
analysis of chromosome 3p region in sporadic renal cell carcinomas.
Pathol Oncol Res. 8:241–244. 2002. View Article : Google Scholar
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu D, Tao J, Xu B, Li P, Lu Q and Zhang W:
Downregulation of Dicer, a component of the microRNA machinery, in
bladder cancer. Mol Med Rep. 5:695–699. 2012.
|
|
9
|
Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee
JK, Sohn SY, Cho Y, Zhang BT and Kim VN: Molecular basis for the
recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell.
125:887–901. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee Y, Han J, Yeom KH, Jin H and Kim VN:
Drosha in primary microRNA processing. Cold Spring Harb Symp Quant
Biol. 71:51–57. 2006. View Article : Google Scholar
|
|
12
|
Yeom KH, Lee Y, Han J, Suh MR and Kim VN:
Characterization of DGCR8/Pasha, the essential cofactor for Drosha
in primary miRNA processing. Nucleic Acids Res. 34:4622–4629. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bohnsack MT, Czaplinski K and Gorlich D:
Exportin 5 is a RanGTP-dependent dsRNA-binding protein that
mediates nuclear export of pre-miRNAs. RNA. 10:185–191. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chendrimada TP, Gregory RI, Kumaraswamy E,
Norman J, Cooch N, Nishikura K and Shiekhattar R: TRBP recruits the
Dicer complex to Ago2 for microRNA processing and gene silencing.
Nature. 436:740–744. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lund E, Guttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar
|
|
16
|
Berdnik D, Fan AP, Potter CJ and Luo L:
MicroRNA processing pathway regulates olfactory neuron
morphogenesis. Curr Biol. 18:1754–1759. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fu W, Pang L, Chen Y, Yang L, Zhu J and
Wei Y: The microRNAs as prognostic biomarkers for survival in
esophageal cancer: A meta-analysis. Scientific World Journal.
2014:5239792014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan W, Sui C, Liu Q, Tang W, An H and Ma
J: Up-regulation of microRNA-145 associates with lymph node
metastasis in colorectal cancer. PloS One. 9:e1020172014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Li L, Su J and Zhang G: Decreased
miR-204 in H. pylori-associated gastric cancer promotes cancer cell
proliferation and invasion by targeting SOX4. PloS One.
9:e1014572014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar :
|
|
23
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar :
|
|
24
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar :
|
|
25
|
Zhou Y, Wu D, Tao J, Qu P, Zhou Z and Hou
J: MicroRNA-133 inhibits cell proliferation, migration and invasion
by targeting epidermal growth factor receptor and its downstream
effector proteins in bladder cancer. Scand J Urol. 47:423–432.
2013. View Article : Google Scholar
|
|
26
|
Xue YJ, Xiao RH, Long DZ, Zou XF, Wang XN,
Zhang GX, Yuan YH, Wu GQ, Yang J, Wu YT, et al: Overexpression of
FoxM1 is associated with tumor progression in patients with clear
cell renal cell carcinoma. J Transl Med. 10:2002012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bauer VL, Braselmann H, Henke M, Mattern
D, Walch A, Unger K, Baudis M, Lassmann S, Huber R, Wienberg J, et
al: Chromosomal changes characterize head and neck cancer with poor
prognosis. J Mol Med (Berl). 86:1353–1365. 2008. View Article : Google Scholar
|
|
28
|
Abou-Elhamd KE, Habib TN, Moussa AE and
Badawy BS: The role of genetic susceptibility in head and neck
squamous cell carcinoma. Eur Arch Otorhinolaryngol. 265:217–222.
2008. View Article : Google Scholar
|
|
29
|
Scully C, Field JK and Tanzawa H: Genetic
aberrations in oral or head and neck squamous cell carcinoma 2:
Chromosomal aberrations. Oral Oncol. 36:311–327. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lagos-Quintana M, Rauhut R, Meyer J,
Borkhardt A and Tuschl T: New microRNAs from mouse and human. RNA.
9:175–179. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang FE, Zhang C, Maminishkis A, Dong L,
Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S and Miller SS:
MicroRNA-204/211 alters epithelial physiology. FASEB J.
24:1552–1571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gong M, Ma J, Li M, Zhou M, Hock JM and Yu
X: MicroRNA-204 critically regulates carcinogenesis in malignant
peripheral nerve sheath tumors. Neuro Oncol. 14:1007–1017. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Garzon R, Garofalo M, Martelli MP,
Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG,
Schnittger S, Haferlach T, et al: Distinctive microRNA signature of
acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin.
Proc Natl Acad Sci USA. 105:3945–3950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J and
Czyzyk-Krzeska MF: VHL-regulated MiR-204 suppresses tumor growth
through inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and mesenchymal
progenitor cell differentiation. Stem cells. 28:357–364. 2010.
|
|
36
|
Courboulin A, Paulin R, Giguère NJ,
Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher
S, Côté J, et al: Role for miR-204 in human pulmonary arterial
hypertension. J Exp Med. 208:535–548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Conte I, Carrella S, Avellino R, Karali M,
Marco-Ferreres R, Bovolenta P and Banfi S: miR-204 is required for
lens and retinal development via Meis2 targeting. Proc Natl Acad
Sci USA. 107:15491–15496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Natera-Naranjo O, Aschrafi A, Gioio AE and
Kaplan BB: Identification and quantitative analyses of microRNAs
located in the distal axons of sympathetic neurons. RNA.
16:1516–1529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu
Y, Li J, Hasina R, Cheng C, Lingen MW, et al: Network modeling
identifies molecular functions targeted by miR-204 to suppress head
and neck tumor metastasis. PLoS Comput Biol. 6:e10007302010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chung TK, Lau TS, Cheung TH, Yim SF, Lo
KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, et al: Dysregulation
of microRNA-204 mediates migration and invasion of endometrial
cancer by regulating FOXC1. Int J Cancer. 130:1036–1045. 2012.
View Article : Google Scholar
|
|
41
|
Ryan J, Tivnan A, Fay J, Bryan K, Meehan
M, Creevey L, Lynch J, Bray IM, O'Meara A, Tracey L, et al:
MicroRNA-204 increases sensitivity of neuroblastoma cells to
cisplatin and is associated with a favourable clinical outcome. Br
J Cancer. 107:967–976. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H,
Li W, Hu B, Cheng SY and Li M: Loss of miR-204 expression enhances
glioma migration and stem cell-like phenotype. Cancer Res.
73:990–999. 2013. View Article : Google Scholar :
|
|
43
|
Penzo-Mendez AI: Critical roles for SoxC
transcription factors in development and cancer. Int J Biochem Cell
Biol. 42:425–428. 2010. View Article : Google Scholar :
|
|
44
|
Gunes S, Yegin Z, Sullu Y, Buyukalpelli R
and Bagci H: SOX4 expression levels in urothelial bladder
carcinoma. Pathol Res Pract. 207:423–427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hur W, Rhim H, Jung CK, Kim JD, Bae SH,
Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, et al: SOX4
overexpression regulates the p53-mediated apoptosis in
hepatocellular carcinoma: Clinical implication and functional
analysis in vitro. Carcinogenesis. 31:1298–1307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Aaboe M, Birkenkamp-Demtroder K, Wiuf C,
Sørensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjøt L and
Ørntoft T: SOX4 expression in bladder carcinoma: Clinical aspects
and in vitro functional characterization. Cancer Res. 66:3434–3442.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jang SM, Kang EJ, Kim JW, Kim CH, An JH
and Choi KH: Transcription factor Sox4 is required for
PUMA-mediated apoptosis induced by histone deacetylase inhibitor,
TSA. Biochem Biophys Res Commun. 438:445–451. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jafarnejad SM, Wani AA, Martinka M and Li
G: Prognostic significance of Sox4 expression in human cutaneous
melanoma and its role in cell migration and invasion. Am J Pathol.
177:2741–2752. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC,
Wang JL, Horng JT, Hsiao M and Tsou AP: Identification of SOX4
target genes using phylogenetic footprinting-based prediction from
expression microarrays suggests that overexpression of SOX4
potentiates metastasis in hepatocellular carcinoma. Oncogene.
27:5578–5589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pramoonjago P, Baras AS and Moskaluk CA:
Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3
cells. Oncogene. 25:5626–5639. 2006. View Article : Google Scholar : PubMed/NCBI
|