Introduction
Renal cell carcinoma (RCC) is a kidney cancer that
originates in the lining of the proximal convoluted tubule and is
the most common type of kidney carcinoma (1,2). The
incidence of RCC varies substantially with respect to ethnicity and
gender (1). Although numerous risk
factors, including smoking, diabetes, hypertension and family
history of malignancy have been identified to be associated with
RCC (3–5), only a small proportion of the
individuals exposed to these environmental factors develop RCC in
their lifetime suggesting that genetic factors may be important in
RCC oncogenesis.
MicroRNAs (miRNAs), an abundant class of
~22-nucleotide-long non-protein coding RNAs, are initially
transcribed from genomic DNA to long primary transcripts
(pri-miRNAs) and are then cleaved by nuclear Drosha into 60–70
nucleotides hairpin-shaped precursor RNAs (pre-miRNAs) (6,7). To
date, miRNAs are considered to be able to modulate the expression
of up to a third of all protein-coding genes by binding to the 3′
untranslated region (3′UTR) of target gene messenger RNA (mRNA),
causing translational repression and/or mRNA degradation (8). miRNAs have been reported to be
involved in the regulation of various biological process, including
the cell cycle, differentiation, apoptosis and metastasis (8). Previous studies have demonstrated
that single nucleotide polymorphisms (SNP) or mutations in miRNA
sequences may affect cancer susceptibility by altering miRNA
expression, maturation or miRNA-mRNA interaction (9,10).
Aberrant miRNA expression has been consistently reported to be
associated with oncogenesis (11,12)
and the deregulated expression of miRNAs and their targets may
result from functional polymorphisms in the miRNA sequence
(13–17).
A functional variant (rs2910164) in the miR-146a
precursor has been demonstrated to be able to alter the processing
of miR-146a and is associated with various types of cancer,
including breast or ovarian cancer, papillary thyroid cancer,
hepatocellular cancer, esophageal squamous cell cancer, gastric
cancer and prostate cancer (11,15–17).
Jazdewski et al (11)
reported that the C allele of rs2910164 may interfere with the
processing of microRNA, leading to a reduction of mature miR-146a
and less inhibition of its target genes, including tumor necrosis
receptor-associated factor 6 (TRAF6) and interleukin-1
receptor-associated kinase 1 (IRAK1).
A previous study demonstrated that the expression of
miR-146a is significantly elevated in RCC tissue (18). In the present study, potential
target genes of miR-146a were computationally searched for by
interrogating online miRNA target predicting tool (www.mirdb.org), and inducible nitric oxide synthase
(iNOS) was identified as a candidate target gene. iNOS has been
validated as a target gene of miR-146a in the mouse RCC cell line
RENCA and miR-146a is hypothesized to be involved in the
carcinogenesis of RCC by exerting an inhibitory effect on iNOS
(19). Nitric oxide synthases
(NOS) are a family of enzymes catalyzing the production of nitric
oxide (NO) from L-arginine. The inducible isoform, iNOS, is
involved in the regulation of cell differentiation, proliferation
and immune response (20–22). Several lines of investigation have
demonstrated that the expression of iNOS is significantly decreased
in RCC compared with normal kidney tissue (23–26).
To the best of our knowledge, there are currently no
studies the association between pre-miR-146a polymorphisms and the
susceptibility or prognosis of RCC. Based on our knowledge
regarding the new polymorphism and biological function of miR-146a,
it was hypothesized that the pre-miR-27a polymorphism was
associated with RCC susceptibility or prognosis. In order to assess
this hypothesis, this particular SNP (rs895819) was genotyped and
the association with the risk and prognosis of RCC was assessed in
a Chinese population.
Materials and methods
Patients
In total, 421 patients with histologically confirmed
primary RCC and 432 cancer-free controls were recruited at the
General Hospital of Jinan Military Region (Jinan, China). All
participants were ethnic Han Chinese individuals and were grouped
into two subgroups based on histological diagnosis. The Fuhrman
scale was used to assess tumor nuclear grade (27). The controls were recruited from
individuals in the outpatient departments at the same hospital. The
cancer-free controls were frequency matched by gender and age to
the cases without an individual history of cancer and family
unrelated to the cases. Demographic data and information on known
and suspected RCC risk factors were collected through interviewer
administered questionnaires. The present study was approved by
investigational review committees at Qingdao University (Qingdao,
China) and written informed consent was obtained from each
participant.
Analysis of hsa-miR-146a and iNOS
expression
In total, 82 surgically removed renal tissue samples
of different genotypes were collected. Total RNA was isolated using
a TRIzol one-step RNA isolation kit (Invitrogen Life Technologies,
Carlsbad, CA, USA). miR-146a, iNOS and U6 (as an internal
control)-specific cDNA were synthesized from total RNA using
gene-specific primers according to the manufacturer's instructions
of the TaqMan microRNA assay (Applied Biosystems, Foster City, CA,
USA). Relative quantification of target miRNA expression was
measured by the ΔΔ cycle threshold (ΔΔCt) method. Each sample was
examined in triplicate and the raw data are presented as the
relative quantity of target miRNA, normalized to U6.
Genotyping of the miR-146a rs2910164
SNP
Peripheral blood (5 ml) was drawn from each
participant and genomic DNA for genotyping was isolated using a DNA
extraction kit [Shunhua Bioengineer (Shanghai) Co. Ltd., Shanghai,
China]. Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific,
Inc., Wilmington, DE, USA) and 0.6% agarose electrophoresis were
used to assess the concentration and purity of the DNA. The
genotyping protocol was performed as described previously (11). Briefly, DNA samples were amplified
by polymerase chain reaction (PCR; Eppendorf
Mastercycler® pro; Eppendorf, Hauppauge, NY, USA) using
the following primer set: 5′-ATTTTACAGGGCTGGGACAG-3′ and
5′-TCTTCCAAGCTCTTCAGCAG-3′. The PCR products were purified using a
USB ExoSAP-IT purification kit (Affymetrix, London, UK) and then
was sent to the core facility for sequencing in both directions
with the ABI sequencing system (Applied Biosystems).
Cell survival and proliferation
assay
The human RCC cell line A498 was used in the present
study and was obtained from ATCC® (Manassas, VA, USA).
Untreated or pre-treated cells were cultured in serum-free
RPMI-1640 medium (Gibco-BRL, Eggenstein, Germany) for 3 days. To
examine cell viability, cells in RPMI-1640 medium were supplemented
with 10% FBS (Gibco-BRL) in 6-well plates for 48 h and the cell
number was counted using the trypan blue exclusion method.
Knockdown of iNOS in RCC cells
A498 cells were cultured until the confluence
reached 70–80% and miR-146a mimics (miR10000449-1-5) and inhibitors
(miR30000449-1-10 and iNOS specific siRNA (Q000004843-1-A) were
purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
Lipofectamine 2000 (Invitrogen Life Technologies) was used to
transfect the small molecules into the A498 cells and the silencing
effect was evaluated using quantitative (q)PCR and western blot
analysis.
Transwell invasion and migration
assay
A Transwell insert invasion assay was performed in
24-well fitted inserts with membranes (8 mm pore size; CoStar,
Cambridge, MA, USA). Cell invasion was examined using a
polycarbonate membrane cell culture insert (Corning Inc., Corning,
NY, USA) coated with growth factor reduced Matrigel (BD
Biosciences, Bedford, MA, USA). The treated or untreated cells were
placed on top of the wells at a density of 2.5×105
cells/well. Invaded cells on the lower surface of the membrane were
stained and counted.
Western blot analysis
The target proteins were separated with
SDS-polyacrylamide gel electrophoresis and then the proteins were
transferred onto a polyvinylidene fluoride membrane followed by
being blocked with Tris-buffered saline and Tween 20 (10 mM Tris-Cl
pH 8.0, 150 mM NaCl and 0.05% Tween 20) containing 5% non-fat dry
milk powder under room temperature for 1 h. The membrane was then
incubated with the rabbit anti-iNOS polyclonal antibody (cat. no.
ab3523; Abcam, Cambridge, MA, USA; 1:2,000) at 4°C overnight,
followed by incubation with horseradish peroxidase-conjugated goat
anti-rabbit secondary antibody (cat. no. ab6721; Abcam; 1:10,000)
at room temperature for 2 h. A chemical fluorescence signal was
detected using an ECL kit (GE Healthcare, Little Chalfont,
Buckinghamshire, UK) according to the manufacturer's instructions.
The target bands were densitometrically analyzed and normalized to
β-actin.
Statistical analysis
Fisher's exact χ2 test and Student's
t-test were used to compare the frequency distribution of age,
gender and smoking status between the cases and controls.
Hardy-Weinberg equilibrium of the genotype
frequencies of the controls was assessed by a χ2
goodness-of-fit test. The association between the SNP rs895819
polymorphism and RCC risk were estimated by computing odds ratios
(ORs) and their 95% confidence intervals (CIs) from unconditional
logistic regression analysis with the adjustment for possible
confounders.
The Kaplan-Meier method was used to estimate overall
survival (OS), defined as the time between study registration and
patient mortality. The OS periods were compared with log-rank test.
Univariate and multivariate Cox proportional hazard regression
analyses were performed to estimate the effect of miR-146a
polymorphisms on survival, the expression levels of miR-146 and
Notch1 in the presence of other known prognostic factors, including
age, gender and smoking status. Analyses were performed using SPSS
19.0 software (SPSS, Inc, Chicago, IL, USA) and P<0.05 was
considered to indicate a statistically significant difference.
Results
A total of 421 RCC patients and 432 healthy controls
were recruited in the present study and the demographic
characteristics and tumor grading information are presented in
Table I. No difference was
identified regarding age (P=0.444), gender (P=0.521) and drinking
status (P=0.338). RCC patients exhibited a higher body mass index
(BMI; P=0.077). Significantly more current smokers, ex-smokers and
diabetic patients were found among the RCC patients. Approximately
142 (35%), 112 (27%), 63 (14%) and 104 (24%) patients were in stage
I, II, III and IV, respectively (Table
I).
 | Table IDistribution of selected variables
between renal cell carcinoma cases and control subjects. |
Table I
Distribution of selected variables
between renal cell carcinoma cases and control subjects.
| Variable | Cases (n=421) | Control (n=432) | P-value |
|---|
| Gender (%) | | | 0.521 |
| Male | 264 (63) | 280 (65) | |
| Female | 157 (37) | 152 (35) | |
| Age (years) | 57.1±9.9 | 57.6±9.2 | 0.444 |
| Body mass index
(kg/m2) | 25.2±3.2 | 24.8±3.4 | 0.077 |
| Drinking status
(%) | | | 0.338 |
| Yes | 118 (28) | 134 (31) | |
| No | 303 (72) | 298 (69) | |
| Smoking status
(%) | | | 0.010 |
| Yes | 307 (73) | 280 (65) | |
| No | 114 (27) | 152 (35) | |
| Hypertension
(%) | | | 0.089 |
| Yes | 109 (26) | 108 (25) | |
| No | 312 (74) | 324 (75) | |
| Diabetes(%) | | | 0.038 |
| Yes | 68 (16) | 48 (11) | |
| No | 353 (84) | 384 (89) | |
| Stage (%) | | | |
| I | 142 (35) | | |
| II | 112 (27) | | |
| III | 63 (14) | | |
| IV | 104 (24) | | |
Genotype distributions of the pre-miR-146a rs2910164
polymorphism in patients and controls are presented in Table II. Following adjusting for
possible confounding factors (age, gender, BMI, smoking status,
drinking status, hypertension and diabetes), logistic regression
analysis revealed that the pre-miR-146a rs2910164 polymorphism was
not associated with RCC risk (adjusted OR=1.28, 95% CI=0.94–1.73).
However, it was found that the association was significant in
senior subjects (adjusted OR=1.59, 95% CI=1.04–2.43; Table III). Additionally, the
association between the pre-miR-146a rs2910164 polymorphism and the
clinical characteristics of RCC was also examined, however, no
association was noted (data not shown). The present study further
evaluated the association between the SNP and the survival rate of
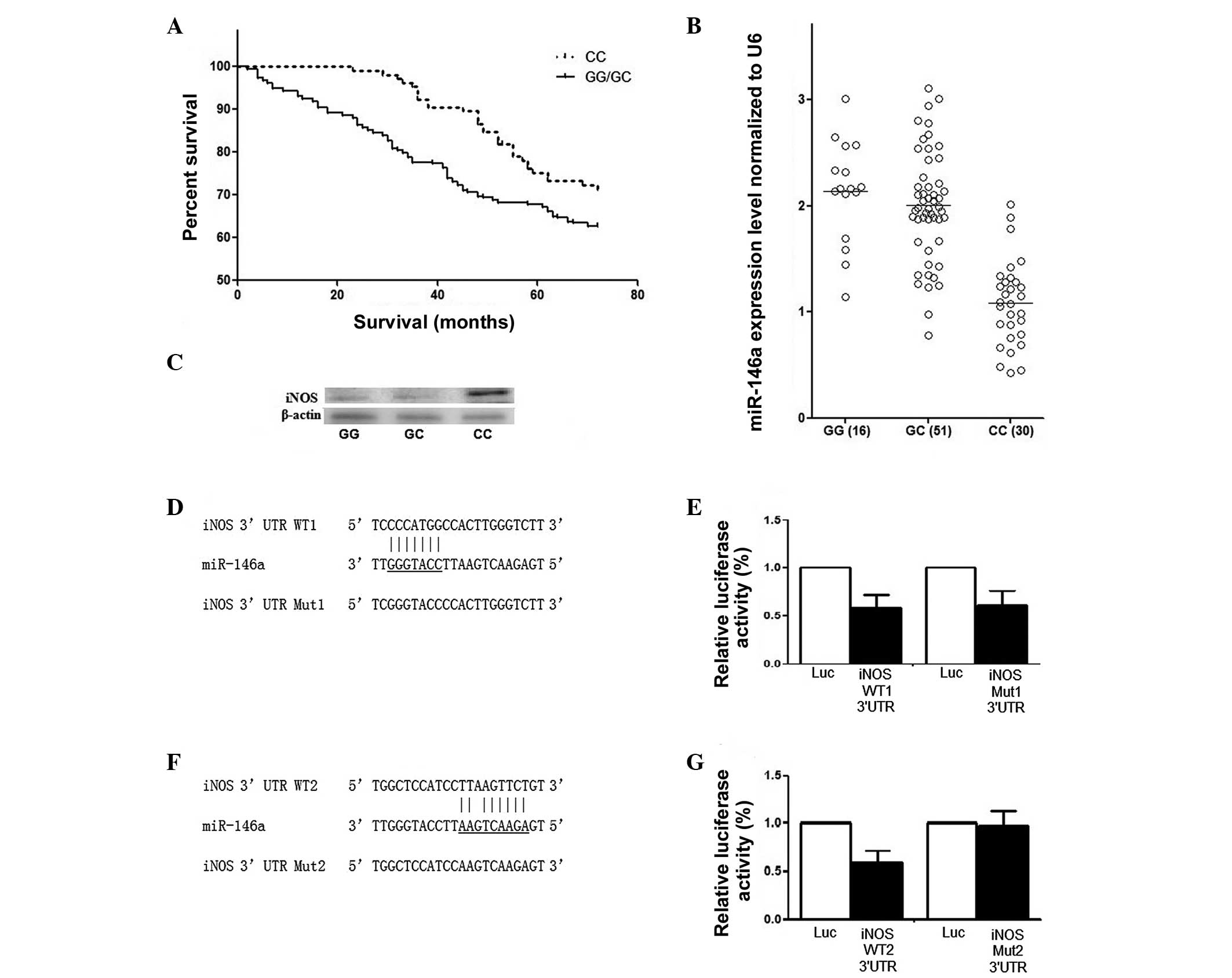
RCC patients. As shown in Fig. 1A,
participants carrying the rs2910164 GG/GC genotype had a
significantly decreased survival rate compared with the CC
genotype, following adjusting for age, gender and smoking status
(P=0.002; Fig. 1A).
 | Table IIGenotype and allele frequencies of
the rs895819 polymorphism among cases and controls and the
association with RCC risk. |
Table II
Genotype and allele frequencies of
the rs895819 polymorphism among cases and controls and the
association with RCC risk.
| rs2910164
polymorphism | Control
(n=432) | RCC (n=421) | OR (95% CI) | P-value |
|---|
| CC | 129 (30%) | 105 (25%) | 1.00 | |
| GC | 234 (54%) | 236 (56%) | 1.24
(0.90–1.69) | 0.181 |
| GG | 69 (16%) | 80 (19%) | 1.42
(0.94–2.15) | 0.092 |
| GC/CC | 303 (7%) | 316 (75%) | 1.28
(0.94–1.73) | 0.316 |
 | Table IIIStratification analyses between the
genotypes of the rs895819 polymorphism and renal cell carcinoma
risk. |
Table III
Stratification analyses between the
genotypes of the rs895819 polymorphism and renal cell carcinoma
risk.
| Variable | Genotype
(case/control)
| GC/GG vs. CC
|
|---|
| CC (n/%) | GC/GG (n/%) | OR (95% CI) | P-value |
|---|
| Total
(case/control) | 105/129 | 316/303 | 0.85
(0.64–1.15) | 0.316 |
| Age (years) | | | | |
| ≤57 | 056/060 | 129/138 | 0.99
(0.64–1.55) | 0.994 |
| >57 | 049/069 | 187/165 | 0.62
(1.04–2.43) | 0.029 |
| Gender | | | | |
| Male | 059/083 | 205/197 | 0.68
(0.46–1.01) | 0.053 |
| Female | 046/046 | 111/106 | 0.84
(0.52–1.36) | 0.495 |
| Smoking | | | | |
| No | 029/053 | 085/099 | 0.73
(0.37–1.09) | 0.100 |
| Yes | 075/076 | 231/204 | 0.87
(0.60–1.26) | 0.466 |
| Drinking | | | | |
| No | 076/088 | 227/210 | 0.79
(0.55–1.14) | 0.221 |
| Yes | 029/041 | 089/093 | 0.73
(0.42–1.29) | 0.287 |
| Hypertension | | | | |
| Yes | 027/038 | 082/070 | 0.60
(0.34–1.09) | 0.095 |
| No | 078/091 | 234/233 | 0.85
(0.59–1.21) | 0.378 |
| Diabetes | | | | |
| Yes | 017/013 | 050/036 | 0.94
(0.40–2.18) | 0.888 |
| No | 088/096 | 265/288 | 0.99
(0.71–1.39) | 0.982 |
A total of 82 surgically removed RCC renal tissue
samples with different rs2910164 genotypes were collected and the
frequency distribution of the CC, CG and GG genotypes was found to
be 30, 51 and 16, respectively (Fig.
1B). The mRNA expression level of miR-146a was also measured in
the 82 patients whose tumor specimens were available and its
association with the genetic variant was evaluated. As shown in
Fig. 1B, the expression of
pre-miR-146a in individuals carrying the rs2910164 GG genotype was
comparable with GC, and the expression in the GG and GC genotype
groups was significantly higher compared with CC carriers
(P<0.001). The effect of the rs2910164 C allele on the
expression level of miR-146a represented a recessive pattern in the
high-grade group. The expression pattern of iNOS was also examined
using western blot analysis in the collected 82 RCC patients with
different genotypes. The expression of miR-146a was higher and iNOS
was lower in the GG/GC group compared with in the CC group
(Fig. 1C).
To investigate the mechanism underlying the tumor
suppressive effect of miR-146a on RCC, iNOS was computationally
identified as a potential target gene of miR-146a, which was
subsequently confirmed using a luciferase assay, as shown in
Fig. 1D–G. Initially, two
potential seed sequences were identified in the 3′-UTR of iNOS as
the possible binding sites of miR-146a (Fig. 1D and F). By introducing mutations
and comparing the luciferase activities with the wild type
(Fig. 1E and F), the actual
binding site of miR-146a in the 3′-UTR of iNOS was located.
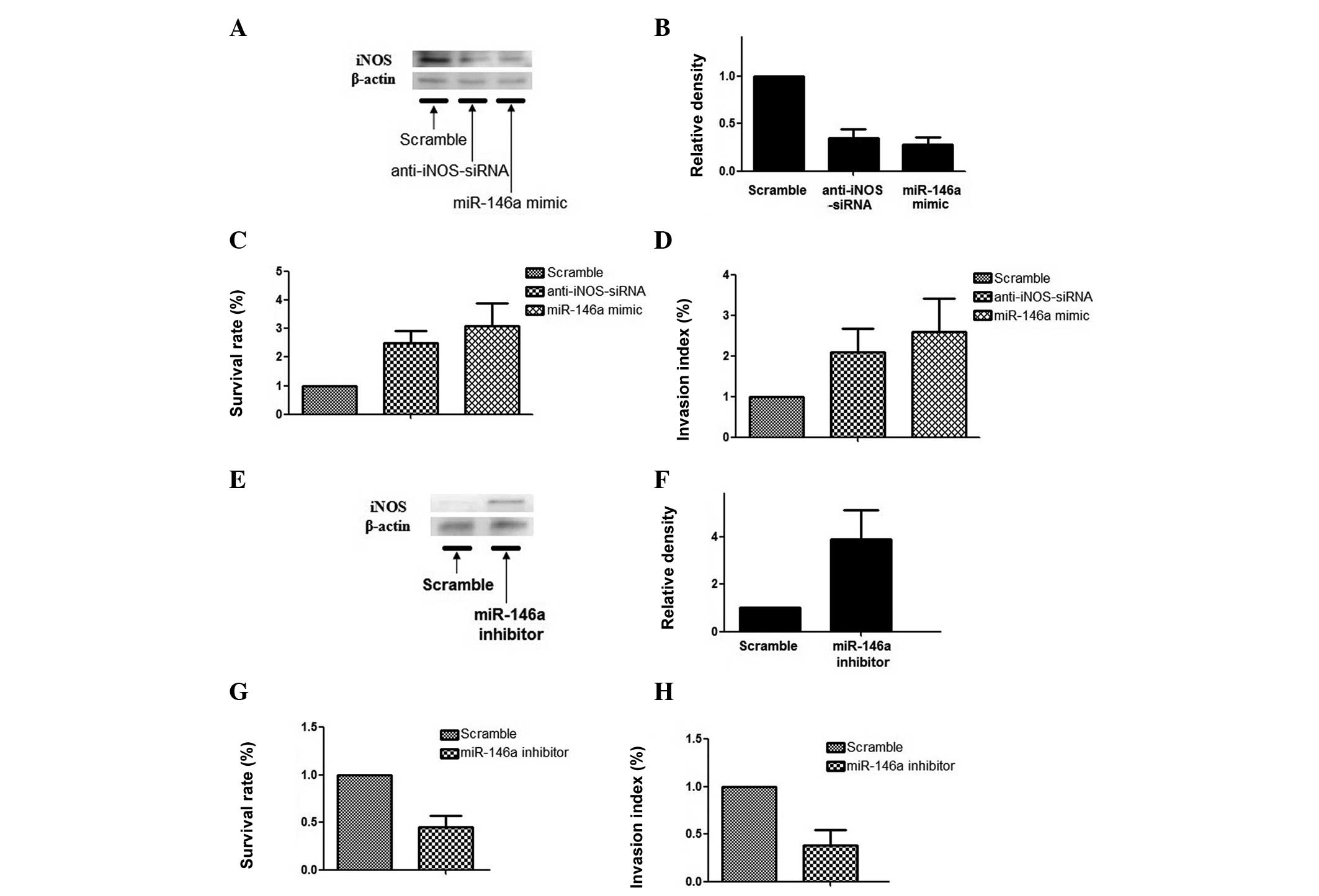
Furthermore, the effect of miR-146a on cell activities, including
proliferation and invasion, and the expression level of iNOS were
analyzed by qPCR and western blot analysis. The results
demonstrated that overexpression of miR-146a caused significant
downregulation of iNOS (Fig. 2A and
B) and introduction of miR-146a promoted the growth and
invasion of RCC cells (Fig. 2C and
D). In addition, miR-146a was downregulated by transfection of
miR-146a inhibitors. The results demonstrated that downregulation
of miR-146a consistently caused significant upregulation of iNOS in
RCC cells (Fig. 2E and F) and
downregulation of miR-146a suppressed the growth and invasion of
RCC cells (Fig. 2G and F).
Discussion
In the present study, the association between a
common polymorphism in miR-146a (rs2910164) and the susceptibility
and prognosis of RCC was investigated in a Chinese population
composed of 421 RCC patients and 432 control subjects. Considering
low frequencies of the GG genotype and a comparable level of
expression for heterozygotes (GC) and homozygotes (GG), suggesting
a recessive model, GC and GG genotypes were combined as a recessive
genetic model in the following association analysis. The present
study found that the rs2910164 GG and GC genotypes were associated
with an increased risk of RCC only in senior subjects (>57 years
old). Furthermore, the GC and GG genotypes were associated with a
poorer survival rate among patients with RCC compared with the CC
genotype.
Accumulative evidence has indicated that miRNAs,
which act as tumor suppressors or oncogenes, are involved in
numerous important biological processes, including differentiation,
proliferation, development and apoptosis (28,29).
Polymorphisms in miRNAs have been repeatedly reported to be
associated with various types of tumor, including breast cancer,
gastric cancer, colorectal cancer and lung cancer (30–33).
Jazdzewski et al demonstrated that the C allele of rs2910164
in pre-miR-146a altered the processing and maturation of the miRNA,
causing a 1.9-fold reduction of mature miR-146a compared with the G
allele (11). The SNP was located
at position +60 relative to the first nucleotide on the passenger
strand of pre-miR146a, and the C allele was predicted to lead to
mispairing within the mature hairpin. Consistently, the mRNA
expression level of miR-146a was measured in the 82 patients whose
tumor specimens were available and it was found that the expression
of pre-miR-146a in those who carried the rs2910164 GG genotype was
comparable with GC, and GG and GC genotype groups had a
significantly higher expression compared with CC carriers
(P<0.001).
It has been proposed that the rs2910164 genotype
contributes to carcinogenesis or tumor suppression by undermining
its interactions with key target genes (11). The same group also reported that
the C allele compromised its ability to bind with HeLa cell nuclear
protein and also caused inefficient inhibition of the miR146a
target genes, including tumor necrosis receptor-associated factor 6
(TRAF6) and interleukin-1 receptor-associated kinase 1
(IRAK1). Jazdewskiet al reported that the GC genotype
was associated with an increased risk for papillary thyroid cancer
compared with the GC/CC genotypes (11). Permuth-Wey et al reported
that the rs2910164 CC/GC genotypes are associated with an increased
risk of glioma, particularly among older individuals, and the C
allele was associated with a poorer survival rate among patients
with glioblastoma (34). Xu et
al found that the rs2910164 CC genotype was associated with a
decreased risk for prostate cancer (35). Xu et al found that the GG
genotype was associated with an increased risk of hepatocellular
carcinoma (36). Guo et al
demonstrated that the GG genotype was associated with increased
esophageal squamous cell carcinoma risk, particularly among smokers
(37). C allele carriers were
reported to be susceptible to gastric cancer in a Japanese
population (38), while the C
allele was found to be protective against gastric cancer in a
Chinese population (24).
Inconsistent results of the association between the rs2910164
miR-146a genotype and cancer risk may represent distinct
heterogeneous etiology in different tumor types or differences in
the ethnicity of investigated populations. The observation that
miR-146a was oncogenic in certain types of cancer, but acted as a
tumor suppressor in others indicated the miR-146a may target
different genes in different types of cancer. As it has been
reported that the expression of miR-146a was significantly elevated
in RCC (19), it is plausible to
hypothesize that miR-146a targets a tumor suppressor in the
development of RCC.
NOS are a family of enzymes catalyzing the
production of NO from L-arginine. The inducible isoform, iNOS, is
involved in the regulation of cell differentiation, proliferation
and immune responses (20–22). Several lines of investigation have
demonstrated a significantly decreased expression of iNOS in RCC
compared with normal kidney tissue (19,23,24).
Similarly, the NOS activity was higher in non-malignant kidney
tissue than in RCC tissue and was inversely correlated with tumor
grade. Furthermore, cytokine treatment induced a marked increase in
NOS activity and NO exerted cytostatic effects on cultured proximal
tubular cells and RCC cell lines (25). Consistently, interferon γ-induced
iNOS can inhibit the proliferation of murine RCC cells (26) and the infection of human renal
cancer cells by retroviruses harboring the murine iNOS gene can
induce the production of high levels of NO, which is associated
with autocytotoxicity, suppression of tumorigenicity and abrogation
of metastasis (24). In the
present study, iNOS was identified as a validated target of
miR-146a and introduction of miR-146a promoted the growth and
invasion of RCC cells. Overexpression of miR-146a simultaneously
caused significant downregulation of iNOS. In addition, miR-146a
was downregulated by transfection of miR-146a inhibitors and the
results demonstrated that down-regulation of miR-146a suppressed
the growth and invasion of RCC cells, and downregulation of
miR-146a consistently caused significant upregulation of iNOS in
RCC cells. The expression pattern of miR-146a and iNOS was also
examined in 82 tissue samples collected from RCC patients, which
was further compared between the GG/GC than CC group. The results
revealed that the expression level of miR-146a was higher and the
expression of iNOS was lower in the GG/GC than in the CC group,
suggesting the tumor suppressive effect of miR-146a in RCC was
mediated by its inhibitory effect on the expression of iNOS.
Although our findings suggest that the pre-miR-146a
rs2910164 polymorphism genotypes were associated with a higher risk
of RCC, the complex nature of RCC also suggests that this disorder
is likely to exhibit genetic characteristics similar to those of
other complex disorders heterogeneously affected by variants at
multiple gene loci. Further, larger and preferably prospective
studies are warranted to validate the role of this polymorphism in
the development of RCC. Since polymorphisms often vary in different
ethnic populations, additional investigations are also required to
verify the association of this polymorphism with RCC risk in
diverse ethnic groups.
References
|
1
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer Statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calle EE and Kaaks R: Overweight, obesity
and cancer: Epidemiological evidence and proposed mechanisms. Nat
Rev Cancer. 4:579–591. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Setiawan VW, Stram DO, Nomura AM, Kolonel
LN and Henderson BE: Risk factors for renal cell cancer: The
multiethnic cohort. Am J Epidemiol. 166:932–940. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeegers MP, Tan FE, Dorant E and van den
Brandt PA: The impact of characteristics of cigarette smoking on
urinary tract cancer risk: A meta-analysis of epidemiologic
studies. Cancer. 89:630–639. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee Y, Jeon K, Lee JT, Kim S and Kim VN:
MicroRNA maturation: Stepwise processing and subcellular
localization. EMBO J. 21:4663–4670. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Q, Jie Z, Ye S, Li Z, Han Z, Wu J,
Yang C and Jiang Y: Genetic variations in miR-27a gene decrease
mature miR-27a level and reduce gastric cancer susceptibility.
Oncogene. 33:193–202. 2014. View Article : Google Scholar
|
|
11
|
Jazdzewski K, Murray EL, Franssila K,
Jarzab B, Schoenberg DR and de la Chapelle A: Common SNP in
pre-miR-146a decreases mature miR expression and predisposes to
papillary thyroid carcinoma. Proc Natl Acad Sci USA. 105:7269–7274.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Turner JD, Williamson R, Almefty KK,
Nakaji P, Porter R, Tse V and Kalani MY: The many roles of
microRNAs in brain tumor biology. Neurosurg Focus. 28:E32010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ,
Park JY, Jeon HS, Park JS and Choi GS: A miR-146a polymorphism
(rs2910164) predicts risk of and survival from colorectal cancer.
Anticancer Res. 33:3233–3239. 2013.PubMed/NCBI
|
|
15
|
Chen L, Zhang R, Li P, Liu Y, Qin K, Fa
ZQ, Liu YJ, Ke YQ and Jiang XD: P53-induced microRNA-107 inhibits
proliferation of glioma cells and downregulates the expression of
CDK6 and Notch-2. Neurosci Lett. 534:327–332. 2013. View Article : Google Scholar
|
|
16
|
Zhang X, Chen T, Zhang J, Mao Q, Li S,
Xiong W, Qiu Y, Xie Q and Ge J: Notch1 promotes glioma cell
migration and invasion by stimulating β-catenin and NF-κB signaling
via AKT activation. Cancer Sci. 103:181–190. 2012. View Article : Google Scholar
|
|
17
|
Mei J, Bachoo R and Zhang CL:
MicroRNA-146a inhibits glioma development by targeting Notch1. Mol
Cell Biol. 31:3584–3592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ha E, Bang JH, Son JN, Cho HC and Mun KC:
Carbamylated albumin stimulates microRNA-146, which is increased in
human renal cell carcinoma. Mol Med Rep. 3:275–279. 2010.
|
|
19
|
Perske C, Lahat N, Sheffy Levin S,
Bitterman H, Hemmerlein B and Rahat MA: Loss of inducible nitric
oxide synthase expression in the mouse renal cell carcinoma cell
line RENCA is mediated by microRNA miR-146a. Am J Pathol.
177:2046–2054. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
MacMillan-Crow LA, Crow JP, Kerby JD,
Beckman JS and Thompson JA: Nitration and inactivation of manganese
superoxide dismutase in chronic rejection of human renal
allografts. Proc Natl Acad Sci USA. 93:11853–11858. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Williams MS, Noguchi S, Henkart PA and
Osawa Y: Nitric oxide synthase plays a signaling role in
TCR-triggered apoptotic death. J Immunol. 161:6526–6531.
1998.PubMed/NCBI
|
|
22
|
Brito C, Naviliat M, Tiscornia AC,
Vuillier F, Gualco G, Dighiero G, Radi R and Cayota AM:
Peroxynitrite inhibits T lymphocyte activation and proliferation by
promoting impairment of tyrosine phosphorylation and
peroxynitrite-driven apoptotic death. J Immunol. 162:3356–3366.
1999.PubMed/NCBI
|
|
23
|
Renaudin K, Denis MG, Karam G, Vallette G,
Buzelin F, Laboisse CL and Jarry A: Loss of NOS1 expression in
high-grade renal cell carcinoma associated with a shift of NO
signalling. Br J Cancer. 90:2364–2369. 2004.PubMed/NCBI
|
|
24
|
Juang SH, Xie K, Xu L, Shi Q, Wang Y,
Yoneda J and Fidler IJ: Suppression of tumorigenicity and
metastasis of human renal carcinoma cells by infection with
retroviral vectors harboring the murine inducible nitric oxide
synthase gene. Hum Gene Ther. 9:845–854. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jansson OT, Morcos E, Brundin L,
Bergerheim US, Adolfsson J and Wiklund NP: Nitric oxide synthase
activity in human renal cell carcinoma. J Urol. 160:556–560. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tate DJ Jr, Patterson JR, Velasco-Gonzalez
C, Carroll EN, Trinh J, Edwards D, Aiyar A, Finkel-Jimenez B and
Zea AH: Interferon-gamma-induced nitric oxide inhibits the
proliferation of murine renal cell carcinoma cells. Int J Biol Sci.
8:1109–1120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ficarra V, Martignoni G, Maffei N,
Brunelli M, Novara G, Zanolla L, Pea M and Artibani W: Original and
reviewed nuclear grading according to the Fuhrman system: A
multivariate analysis of 388 patients with conventional renal cell
carcinoma. Cancer. 103:68–75. 2005. View Article : Google Scholar
|
|
28
|
Bartels CL and Tsongalis GJ: MicroRNAs:
Novel biomarkers for human cancer. Clin Chem. 55:623–631. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harfe BD: MicroRNAs in vertebrate
development. Curr Opin Genet Dev. 15:410–415. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao LB, Bai P, Pan XM, Jia J, Li LJ, Liang
WB, Tang M, Zhang LS, Wei YG and Zhang L: The association between
two polymorphisms in pre-miRNAs and breast cancer risk: A
meta-analysis. Breast Cancer Res Treat. 125:571–574. 2011.
View Article : Google Scholar
|
|
31
|
Peng S, Kuang Z, Sheng C, Zhang Y, Xu H
and Cheng Q: Association of microRNA-196a-2 gene polymorphism with
gastric cancer risk in a Chinese population. Dig Dis Sci.
55:2288–2293. 2010. View Article : Google Scholar
|
|
32
|
Zhu L, Chu H, Gu D, Ma L, Shi D, Zhong D,
Tong N, Zhang Z and Wang M: A functional polymorphism in
miRNA-196a2 is associated with colorectal cancer risk in a Chinese
population. DNA Cell Biol. 31:350–354. 2012. View Article : Google Scholar
|
|
33
|
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L,
Zeng Y, Miao R, Jin G, Ma H, et al: Genetic variants of miRNA
sequences and non-small cell lung cancer survival. J Clin Invest.
118:2600–2608. 2008.PubMed/NCBI
|
|
34
|
Permuth-Wey J, Thompson RC, Burton Nabors
L, Olson JJ, Browning JE, Madden MH, Ann Chen Y and Egan KM: A
functional polymorphism in the pre-miR-146a gene is associated with
risk and prognosis in adult glioma. J Neurooncol. 105:639–646.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu B, Feng NH, Li PC, Tao J, Wu D, Zhang
ZD, Tong N, Wang JF, Song NH, Zhang W, et al: A functional
polymorphism in Pre-miR-146a gene is associated with prostate
cancer risk and mature miR-146a expression in vivo. Prostate.
70:467–472. 2010. View Article : Google Scholar
|
|
36
|
Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge
YY, Yang JR, Su H and Zhuang SM: A functional polymorphism in the
miR-146a gene is associated with the risk for hepatocellular
carcinoma. Carcinogenesis. 29:2126–2131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo H, Wang K, Xiong G, Hu H, Wang D, Xu
X, Guan X, Yang K and Bai Y: A functional variant in microRNA-146a
is associated with risk of esophageal squamous cell carcinoma in
Chinese Han. FAM Cancer. 9:599–603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Okubo M, Tahara T, Shibata T, Yamashita H,
Nakamura M, Yoshioka D, Yonemura J, Kamiya Y, Ishizuka T, Nakagawa
Y, et al: Association study of common genetic variants in
pre-microRNAs in patients with ulcerative colitis. J Clin Immunol.
31:69–73. 2011. View Article : Google Scholar
|