Introduction
Colon cancer is a common type of cancer worldwide
and the treatment of colon cancer is currently problematic.
Although there have been marked advances in diagnostic techniques,
as well as improvements in treatment modalities, the prognosis of
colon cancer remains poor due to its high recurrence (1–3).
Numerous reports show that various signaling pathways are activated
during colon cancer progression (1–3);
however, the mechanism remains unclear and the identification of
novel molecular targets for colon cancer is required.
MicroRNAs (miRNAs) are small RNAs, which negatively
regulate gene expression in various types of cell (4). The target genes of miRNAs can be
downregulated at a post-transcriptional level. Furthermore, miRNAs
acting as tumor inhibitors of onco-miRNAs locate in
cancer-associated genomic regions. Their function in the cells
depends on their direct target genes (5–7). The
primary causes of colon cancer patient mortality are the poor
prognosis and metastases. Therefore, it is considered to be
critical to establish a novel diagnostic marker and continue to
investigate the underlying molecular mechanism of miRNAs for
therapeutic application. Emerging evidence demonstrates that a
group of miRNAs is involved in the regulation of basic cellular
processes in colon cancer, such as cell proliferation,
differentiation, apoptosis and metastasis. Previous reports
revealed that miR-34a, miR-21, miR-155, miR-499a, miR-99a, miR-101,
as well as other miRNAs are involved in colon cancer progression
(8–11). Furthermore, an miRNA profile
reported that miR-34a is downregulated in colon cancer (9–11).
miR-34a is a known tumor suppressor in various types
of cancer, including lung, breast, prostate and liver cancer, as
well as in colon cancer (12–18).
However, the role of miR-34a in colon cancer requires further
investigation. Thus, the aim of the present study was to
investigate the underlying effect of miR-34a in colon cancer.
miR-34a expression was analyzed in human colon cancer tissues and
cells and, using bioinformatics, platelet-derived growth factor
receptor α (PDGFRA) was determined to be a target gene of miR-34a.
Furthermore, miR-34a was identified to be a tumor suppressor, which
negatively regulates the PDGFRA signaling pathway in the
progression of colon cancer. The roles of miR-34a in cell
proliferation, migration and invasion were also evaluated.
Materials and methods
Colon cancer samples
Primary colon cancer specimens and normal biopsies
were obtained from The First Affiliated Hospital of Dalian Medical
University (Dalian, China). A total of 176 colon cancer specimens
and normal biopsies were obtained from patients undergoing radical
surgery for colon cancer at The First Affiliated Hospital (Dalian
Medical University, Dalian, China). There were 95 male and 86
female patients (mean age, 57.2-years old). Colon cancer and
healthy tissues were confirmed by hematoxylin and eosin staining in
the pathology department of The First Affiliated Hospital. The
study was approved by the Ethics Committee of The First Affiliated
Hospital (Dalian Medical University, Dalian, China) Analyses of
tissue samples were performed according to the instructions laid
out by the Ethics Committee of the Hospital. All of the patients
provided written informed consent.
Cell lines and culture
Colon cancer cells, HT-29, SW620, Lovo and Colo205,
were originally obtained from the American Type Culture Collection.
The cells were cultured in Dulbecco's modified Eagle's medium
(Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (Gibco Life Technologies, Carlsbad, CA,
USA), 100 U/ml of penicillin and 100 µg/ml of streptomycin
(Sigma-Aldrich, St. Louis, MO, USA) at 37°C in a humidified
atmosphere of 5% CO2. Normal colon epithelial cells,
NCM460 and FHC, were cultured according to the manufacturer's
instructions.
Luciferase assay
A psiCHECK™-2 Dual-Luciferase miRNA target
expression vector was used for the 3′-untranslated region (UTR)
luciferase assays (Sangon Biotech, Co., Ltd., Shanghai, China). The
target genes of miR-34a were selected on the basis of the following
miRNA target database: http://www.microrna.org/microrna/home.do. The 3′-UTR
sequence of the PDGFRA gene was amplified using polymerase chain
reaction (PCR) and cloned into the psiCHECK™-2 vector to produce a
wild-type reporter. The following primer sequences were used to
amplify the wild-type 3′-UTRs of PDGFRA: Forward,
5′-TCTAGACCGGCCTGAGAAACACTATTTGTG-3′ and reverse,
5′-TCTAGAACATGAACAGGGGCATTCGTAATACA-3′. The mutant reporter
construct was generated using the Site-Directed Gene Mutagenesis
kit (Beyotime Institute of Biotechnology, Shanghai, China)
according to the manufacturer's instructions. There are two binding
sites in the PDGFRA 3′-UTR. For one mutant 3′-UTR of PDGFRA, the
following primer sequences were used: Forward,
5′-ACTGCCAAAACATTTATGACAAGCTGTATCGCCTCG-3′ and reverse,
5′-CGAGGCGATACAGCTTGTCATAAATGTTTTGGCAGT-3′. For the other mutant
3′-UTR of PDGFRA, the following primer sequences were used:
Forward, 5′-ACTGCCAAAACATTTATGACAAGCTGTATGGTCGTTTATATTT-3′ and
reverse, 5′-AAATATAAACGACCATACAGCTTGTCATAAATGTTTTGGCAGT-3′. For the
luciferase assay, a total of 5,000 cells were transfected with
miR-34a mimics and the psiCHECK™-2 Dual-Luciferase miRNA target
expression vectors containing the wild-type or the mutant target
sequences using Lipofectamine 2000 (Invitrogen Life Technologies).
Data are presented as relative firefly luciferase activity
normalized to Renilla luciferase activity from the same
construct.
Lentiviral infection
The sequence of mature miR-34a was synthesized,
amplified and cloned into a GV232-Puro Vector by Genechem Co., Ltd.
(Shanghai, China). DNA sequencing was used to identify the correct
sequences. Lentivirus mediating miR-34a or its control was produced
in 293T cells using Lipofectamine 2000 reagent according to the
manufacturer's instructions. For transduction, the colon cancer
cells were infected with miR-34a or its control and subsequently
selected using 1 µg/ml puromycin (Sigma-Aldrich).
Lentivirus-mediated silencing of miR-34a was verified by
reverse-transcription quantitative (RT-q)PCR analysis.
RNA extraction, isolation and
RT-qPCR
Total RNA was extracted from the cells and the
tissues using TRIzol reagent (Invitrogen Life Technologies). RNA (2
µg) was reverse transcribed to cDNA using a SYBR®
Green PCR kit (Takara Biotechnology Co., Ltd., Shiga, Japan). To
measure miRNA expression, qPCR was performed for DNA amplification
(ABI-Prism; Applied Biosystems Life Technologies, Foster City, CA,
USA). The cycling conditions were 10 min of polymerase activation
at 95°C, followed by 40 cycles at 95°C for 15 sec and 60°C for 60
sec. The relative expression levels were calculated by comparing
the cycle threshold values of the samples with those of the
references, all data were normalized to glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) or the U6 sRNA control.
MTT assay
An MTT assay was conducted to detect cell viability.
The colon cancer cells were seeded into 96-well plates following
transfection with miRNA or the plasmids. At day 1, 2, 3, 4 and 5,
20 µl MTT solution (Sigma-Aldrich) was added to each well (5
mg/ml; 0.5% MTT) and the cells continued to culture for 4 h. The
supernatant was discarded, 150 µl dimethyl sulfoxide (Sangon
Biotech, Co., Ltd.) was added to each well and the culture plate
was agitated at 50–75 × g for 10 min to ensure crystals were
completely dissolved. An ELISA reader was used to measure the
absorbance at 570 nm. The MTT assay was performed three times.
Colony forming assay
The colon cancer cells were transfected with miRNA
or DNA plasmids and collected following transfection for 24 h. The
transfected cells were seeded into 6-well plates with 200 cells per
well, cultured for three days and the nonadherent cells were
removed. After 10–14 days, the colonies were dyed with 0.1% crystal
violet (Sigma-Aldrich) and counted.
Western blot analysis
Cells were lysed using radioimmunoprecipitation
assay buffer (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) with
1X protease inhibitor cocktail and total protein was extracted. The
concentration of protein was determined using the Bradford assay
(Bio-Rad Laboratories, Inc., Philadelphia, PA, USA). SDS-PAGE
(12.5%) was conducted to separate the proteins and the separated
proteins were transferred to polyvinylidene difluoride membranes
(Merck Millipore, Bedford, MA, USA) at 55 V for 4 h at 4°C. The
membranes were blocked in 5% fat-free milk, incubated in the
primary antibodies and then incubated with secondary antibodies for
1 h at room temperature. PDGFRA (cat. no. 3164), p-Akt (Ser473;
cat. no. 4060), Akt (cat. no. 9272) and p21 (cat. no. 2946) primary
antibodies were purchased from Cell Signaling Technology, Inc.
(Danvers, MA, USA). The dilutions of the above antibodies were all
1:1,000. GAPDH antibody was purchased from Santa Cruz
Biotechnology, Inc. (1:5,000). Protein bands were visualized on
X-ray film (Sigma-Aldrich) using an enhanced chemiluminescence
detection system.
Wound healing assays
Cell migration was assessed by measuring the
movement of cells into an acellular area, which was scraped using a
200-µl pipette tube; the spread of wound closure was
observed after 48 h. Images were obtained to assess the level of
migration in each group of transfected cells. Migration was
quantified by counting the total number of cells that migrated
toward the original wound field.
Migration and invasion assay
Colon cancer cells were trans-fected with miRNA or
DNA plasmids and collected following transfection for 48 h. For the
migration assay, 1×105 colon cancer cells were plated in
the top chamber of the non-coated membrane (24-well insert; pore
size, 8 µm; Corning Costar) and allowed to migrate toward
serum-containing medium in the lower chamber. Cells were fixed with
methanol after a 24-h incubation and stained with 0.1% crystal
violet (2 mg/ml). The number of cells invading through the membrane
was counted under a light microscope (three random fields per
well). For the invasion assay, the steps were the same as the
migration assay, however, the membrane was coated with Matrigel (BD
Biosciences, San Jose, CA, USA). The number of cells invading
through the membrane was counted under a light microscope
(magnification, ×40; three random fields per well).
Statistical analysis
Statistical analysis was conducted using SPSS 15.0
(SPSS, Inc., Chicago, IL, USA). Student's t-test was used to
analyze the results, all experiments were performed in triplicate
and the data are expressed as means ± standard deviation. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-34a expression is correlated with
clinicopathological features of colon cancer
Firstly, miR-34a expression was evaluated in 176
primary colon cancer tissue samples and their adjacent non-tumor
tissues using RT-qPCR. The colon cancer TNM staging was according
to the American Joint Committee on Cancer and International Union
Against Cancer (19,20). miR-34a expression in colon cancer
tissues was observed to be markedly lower when compared with that
of their adjacent non-tumor tissues. The analyzed data revealed
that the low expression level of miR-34a in colon cancer tissues
was significantly associated with lymph node metastasis and a poor,
advanced pT stage, as well as invasion into lymphatic vessels
(Table I).
 | Table IAssociation between miRNA expression
and clinicopathological features. |
Table I
Association between miRNA expression
and clinicopathological features.
| Characteristic | miRNA expression
| P-value |
|---|
| Low | High |
|---|
| Age (years) | | | 0.054 |
| ≥60 | 52 (59.1) | 36 (40.9) | |
| <60 | 56 (63.6) | 32 (36.4) | |
| Gender | | | 0.204 |
| Male | 54 (60.0) | 40 (44.4) | |
| Female | 36 (40.0) | 50 (55.6) | |
| Tumor size (cm) | | | 0.280 |
| ≥6 | 40 (45.5) | 28 (31.8) | |
| <6 | 50 (55.6) | 58 (67.4) | |
| Histology grade | | | 0.016 |
| Well | 10 (11.1) | 0 (0) | |
| Moderate | 54 (60.0) | 44 (48.9) | |
| Poor | 26 (28.9) | 46 (51.1) | |
| LNM | | | 0.515 |
| No | 38 (42.2) | 30 (33.3) | |
| Yes | 52 (57.8) | 60 (66.7) | |
| TNM stage | | | 0.012 |
| I | 1 (2.3) | 54 (60.0) | |
| II | 10 (11.4) | 28 (31.1) | |
| III | 40 (45.5) | 6 (6.8) | |
| IV | 39 (40.9) | 2 (2.2) | |
PDGFRA is a novel target gene of miR-34a
in colon cancer cells
miR-34a exerts a suppressive role in various types
of cancer, however, its target gene has not been fully elucidated.
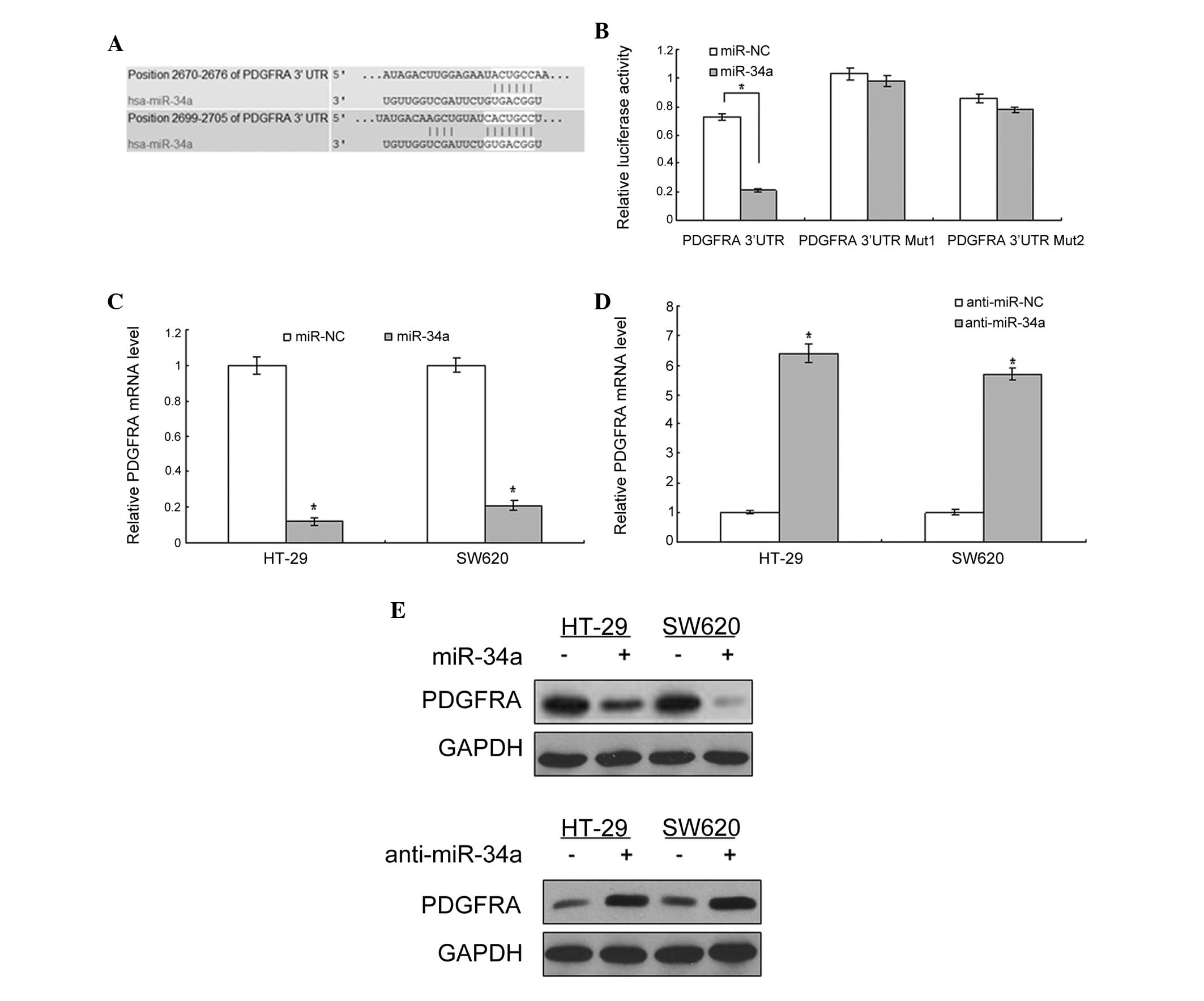
Bioinformatic analysis using TargetScan (http://www.targetscan.org/vert_61/) revealed that
PDGFRA was directly suppressed by miR-34a (Fig. 1A). As shown in Fig. 1B, the luciferase activity of PDGFRA
in 293T cells was significantly lower than in the control cells
(P<0.01). The luciferase activity of the cells with the mutated
3′-UTR of PDGFRA was not changed significantly when compared with
the control. Subsequently, whether miR-34a regulates endogenous
PDGFRA expression in the cells was examined. In HT-29 and SW620
cells, compared with the control, endogenous PDGFRA mRNA expression
(Fig. 1C and D) and protein levels
(Fig. 1E) were downregulated in
the cells with LV-miR-34a infection and upregulated with
anti-miR-34a infection.
PDGFRA expression is negatively
associated with miR-34a expression in colon cancer tissues
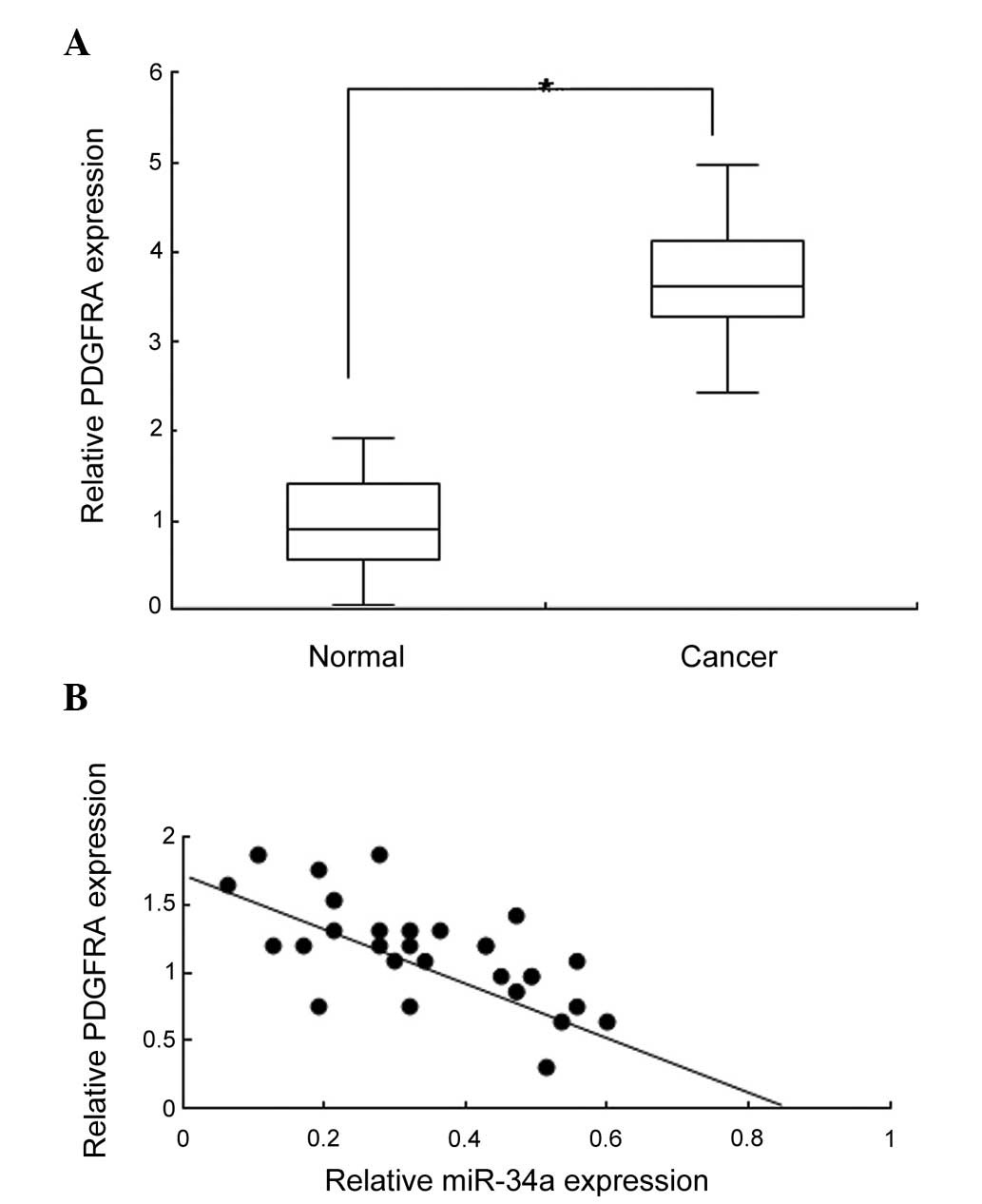
To identify the association between the expression
of PDGFRA and miR-34a in colon cancer tissue, the expression of
PDGFRA in 26 pairs of colon cancer tissue and normal control
samples was detected using qRT-qPCR. PDGFRA expression was observed
to increase markedly in the cancer tissues when compared with the
normal tissues (Fig. 2A;
P<0.05). Analysis of the data demonstrated that PDGFRA was
negatively associated with miR-34a expression in the majority of
tissues (Fig. 2B). These data
further indicated that PDGFRA was a target of miR-34a in colon
cancer.
PDGFRA increases human colon cancer cell
proliferation and metastasis
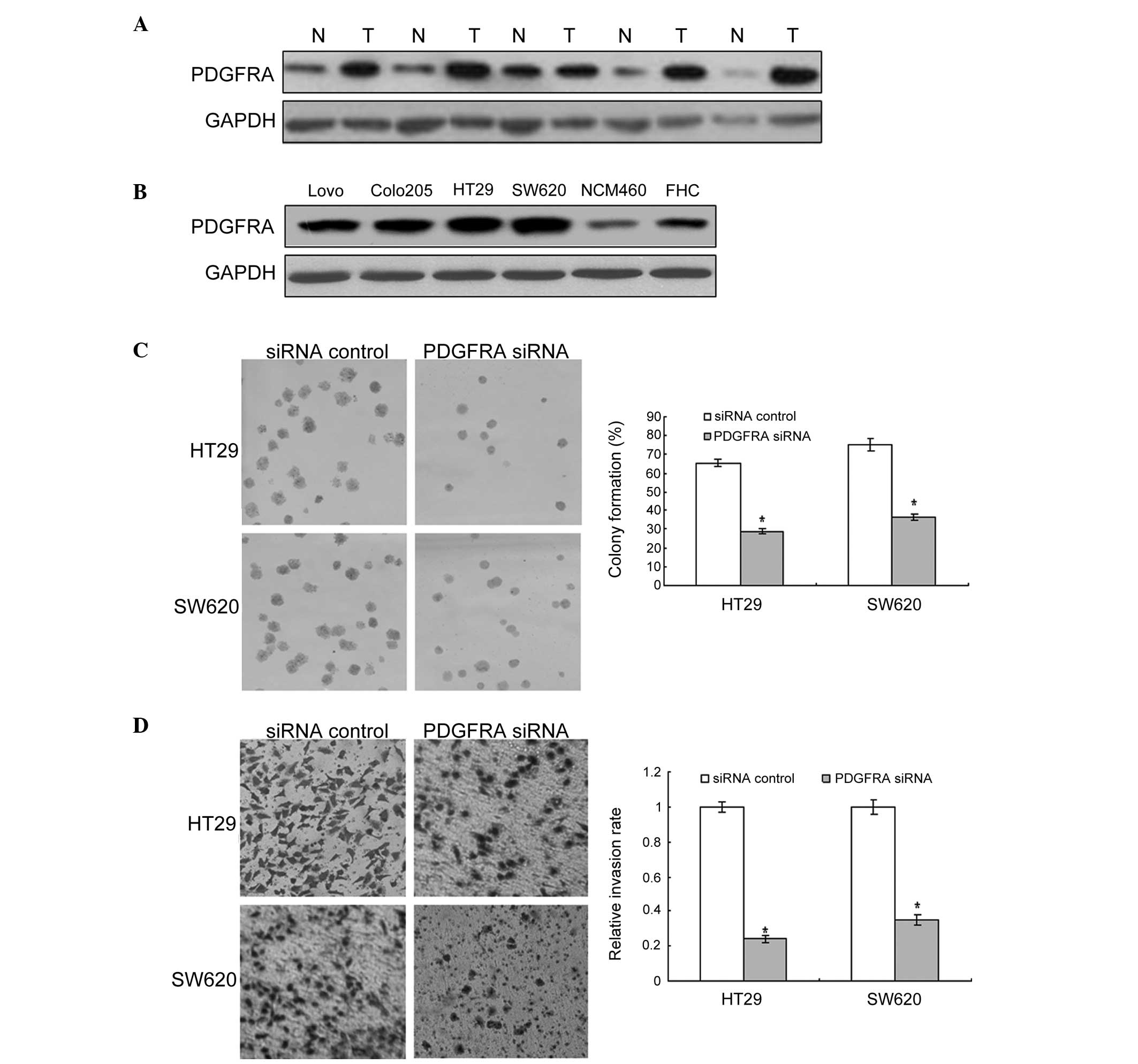
Previous reports demonstrated that PDGFRA is
overexpressed in colon cancer, however, its role has not been
determined. To investigate the possible role of PDGFRA in colon
cancer cells, the PDGFRA protein was examined in colon cancer
cells. As shown in Fig. 3A, PDGFRA
expression in colon cancer tissue samples was higher than that in
adjacent normal tissue samples. Similarly, PDGFRA was markedly
higher in human colon cancer cells than in the normal colon cells
(Fig. 3B). These results provide
initial evidence that PDGFRA may exert a tumor-suppressing role in
the development of human colon cancer. Furthermore, it was noted
that cell proliferation and metastasis were decreased in colon
cancer cells exhibiting PDGFRA inhibition (Fig. 3C and D).
miR-34a inhibits colon cancer progression
by targeting PDGFRA and its signaling pathway
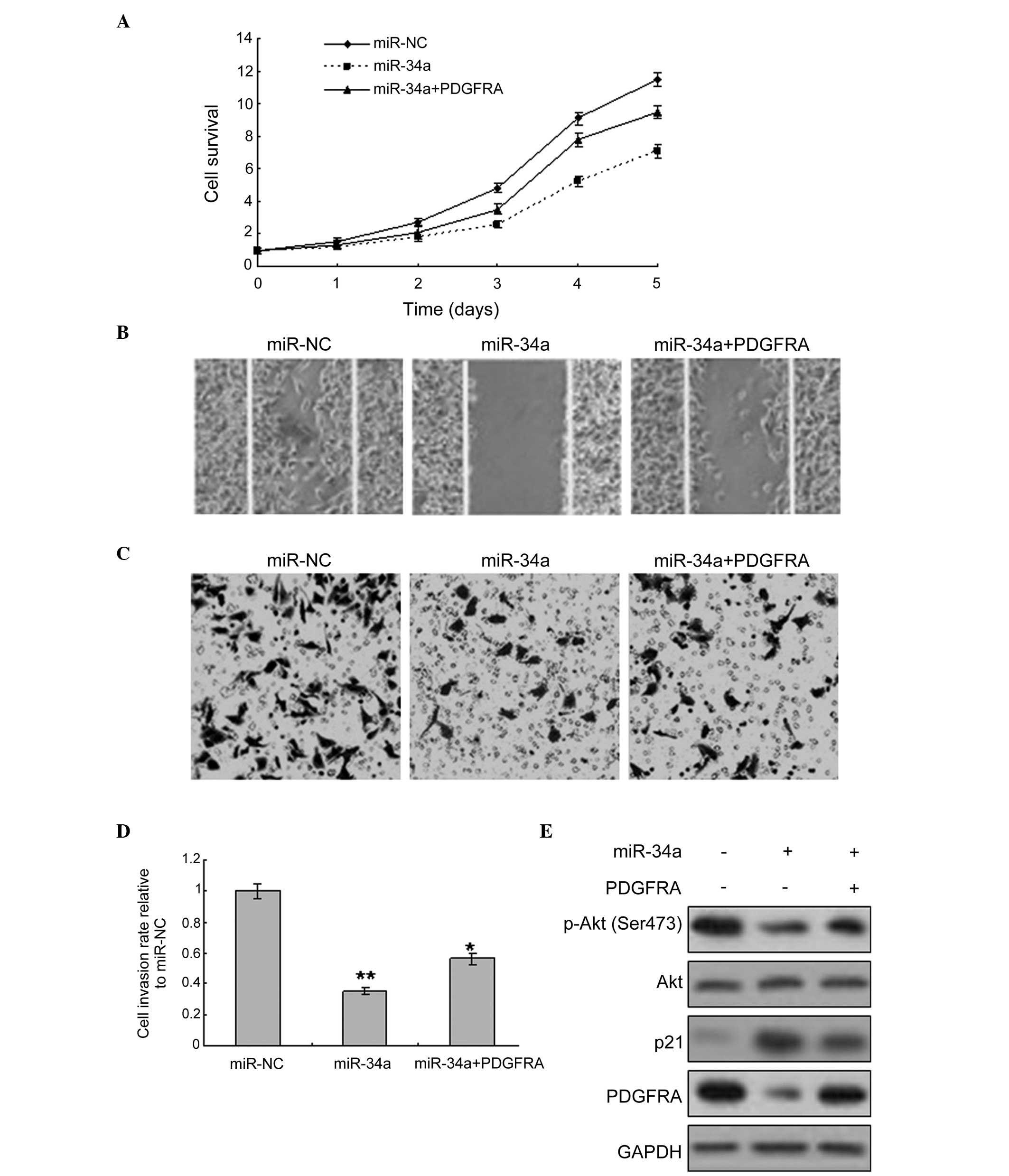
The above-mentioned data indicate that PDGFRA is a
target gene of miR-34a. In the present study, elucidating whether
miR-34a regulates biological behavior, such as proliferation,
apoptosis and metastasis was investigated. PDGFRA overexpression
was used to assess whether miR-34a inhibited HT-29 cell metastasis.
The result revealed that miR-34a expression in the cells inhibited
proliferation of HT-29 cells with or without PDGFRA overexpression
(Fig. 4A). The migration and
invasion of the cells increased with PDGFRA expression, and when
the cells were infected with LV-miR-34a, migration and invasion
were decreased (Fig. 4B-D). As the
PDGFRA-mediated signaling pathway is an important pathway involved
in cancer development, various proteins and downstream proteins in
the signaling pathway, such as Akt, were detected and observed to
be decreased by miR-34a overexpression (Fig. 4E).
Discussion
Previous reports demonstrated that miR-34a is
involved in colon cancer progression, including inducing apoptosis,
and inhibiting cell proliferation and metastasis. However, the role
of miR-34a requires further research. In the present study, miR-34a
was found to be downregulated in colon cell lines and tissues, and
has an important role in metastasis by targeting PDGFRA.
Firstly, the results from the clinical data
indicated that miR-34a is associated with the colon cancer stage
and metastasis, which indicates that miR-34a is involved in colon
cancer cell metastasis. In HT-29 and SW620 colon cancer cells,
restoration of miR-34a expression promoted cell growth, and
decreased cell migration and invasion (Fig. 3). This indicated that a reduced
expression level of miR-34a in colon cancer cells may facilitate
cell invasion and metastasis of colon cancer. In addition, PDGFRA
expression was observed to be negatively correlated with miR-34a
expression in colon cancer cells and tissues.
Using bioinformatic prediction, PDGFRA was
identified to be the direct target of miR-34a. Furthermore,
overexpression of miR-34a decreased the expression levels of PDGFRA
protein and mRNA, indicating that PDGFRA is a downstream gene of
miR-34a in colon cancer cells. However, miR-34a in colon cancer may
target other genes. It is well known that there are molecules
influencing miR-34a, which are involved in colon cancer progression
and growth, as well as other functions (12,13).
PDGFRA is overexpressed in various malignancies and
is crucial in promoting cell proliferation and metastasis (21–25).
In the present study, miR-34a expression was observed to be
negatively correlated with PDGFRA. Furthermore, the present results
indicate that miR-34a downregulates PDGFRA expression by directly
targeting the PDGFRA 3′-UTR. Therefore, it may be possible to use
miRNA to target PDGFRA during cancer therapy. However, expression
of miRNA or its target genes is influenced by multiple factors,
such as epigenetic modification, including DNA methylation, histone
modification, chromosome deficiency and transcriptional regulation
(4,5). Thus, the underlying mechanisms of the
repression of miRNA require further investigation.
In conclusion, the present study verified that the
deregulated expression of miR-34a is associated with a poor
prognosis and an aggressive phenotype of colon cancer. Furthermore,
the findings indicate that miR-34a is significantly involved in the
regulation of colonic malignant behavior, including cell
proliferation and invasion, by directly targeting PDGFRA.
Therefore, miR-34a may serve as a potential prognostic biomarker
and as a therapeutic molecule for the treatment of colon
cancer.
Acknowledgments
The present study was supported by grants from the
Joint Funds of the Natural Science Foundation of Liaoning Province
(grant no. 2013023024), the National Natural Science Foundation of
China (grant no. 81172052) and the Yingcai Program of Dalian
Medical University.
References
|
1
|
Weitz J, Koch M, Debus J, Höhler T, Galle
PR and Büchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dyzmann-Sroka A and Malicki J: Cancer
incidence and mortality in the greater Poland region-analysis of
the year 2010 and future trends. Rep Pract Oncol Radiother.
19:296–300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Swiderska M, Choromańska B, Dąbrowska E,
Konarzewska-Duchnowska E, Choromańska K, Szczurko G, Myśliwiec P,
Dadan J, Ladny JR and Zwierz K: The diagnostics of colorectal
cancer. Contemp Oncol (Pozn). 18:1–6. 2014.
|
|
4
|
Dalmay T: MicroRNAs and cancer. J Intern
Med. 263:366–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Negrini M, Ferracin M, Sabbioni S and
Croce CM: MicroRNAs in human cancer: From research to therapy. J
Cell Sci. 120:1833–1840. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho WC: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tokarz P and Blasiak J: The role of
microRNA in metastatic colorectal cancer and its significance in
cancer prognosis and treatment. Acta Biochim Pol. 59:467–74.
2012.PubMed/NCBI
|
|
10
|
Liu C and Tang DG: MicroRNA regulation of
cancer stem cells. Cancer Res. 71:5950–5954. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lai X, Wolkenhauer O and Vera J: Modeling
miRNA regulation in cancer signaling systems: miR-34a regulation of
the p53/Sirt1 signaling module. Methods Mol Biol. 880:87–108. 2012.
View Article : Google Scholar
|
|
12
|
Chen F and Hu SJ: Effect of microRNA-34a
in cell cycle, differentiation and apoptosis: A review. J Biochem
Mol Toxicol. 26:79–86. 2012. View Article : Google Scholar
|
|
13
|
Hermeking H: The miR-34 family in cancer
and apoptosis. Cell Death Differ. 17:193–199. 2010. View Article : Google Scholar
|
|
14
|
Ma Y, Bao-Han W, Lv X, Su Y, Zhao X, Yin
Y, Zhang X, Zhou Z, MacNaughton WK and Wang H: MicroRNA-34a
mediates the autocrine signaling of PAR2-activating proteinase and
its role in colonic cancer cell proliferation. PLoS One.
8:e723832013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bu P, Chen KY, Chen JH, Wang L, Walters J,
Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, et al: A
microRNA miR-34a-regulated bimodal switch targets Notch in colon
cancer stem cells. Cell Stem Cell. 12:602–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Winton DJ: miR-34a sets the 'sweet spot'
for notch in colorectal cancer stem cells. Cell Stem Cell.
12:499–501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rudy DR and Zdon MJ: Update on colorectal
cancer. Am Fam Physician. 61:1759–1770. 1773–1774. 2000.PubMed/NCBI
|
|
20
|
Jass JR and Morson BC: Reporting
colorectal cancer. J Clin Pathol. 40:1016–1023. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu Y, Wang Y, Guan B, Rao Q, Wang J, Ma
H, Zhang Z and Zhou X: C-kit and PDGFRA gene mutations in triple
negative breast cancer. Int J Clin Exp Pathol. 7:4280–4285.
2014.PubMed/NCBI
|
|
22
|
Stock AM, Hahn SA, Troost G, Niggemann B,
Zänker KS and Entschladen F: Induction of pancreatic cancer cell
migration by an autocrine epidermal growth factor receptor
activation. Exp Cell Res. 326:307–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Demoulin JB and Essaghir A: PDGF receptor
signaling networks in normal and cancer cells. Cytokine Growth
Factor Rev. 25:273–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hayes BJ, Riehle KJ, Shimizu-Albergine M,
Bauer RL, Hudkins KL, Johansson F, Yeh MM, Mahoney WM Jr, Yeung RS,
Campbell JS, et al: Activation of platelet-derived growth factor
receptor alpha contributes to liver fibrosis. PLoS One.
9:e929252014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roh JW, Huang J, Hu W, Yang X, Jennings
NB, Sehgal V, Sohn BH, Han HD, Lee SJ, Thanapprapasr D, et al:
Biologic effects of platelet-derived growth factor receptor α
blockade in uterine cancer. Clin Cancer Res. 20:2740–2750. 2014.
View Article : Google Scholar : PubMed/NCBI
|