Introduction
Cervical cancer is the second most common cancer in
women worldwide with the fifth highest mortality rate (1,2).
Squamous cell carcinomas are the most common type, accounting for
80–85% of all cervical cancers (3). Infection with human papilloma virus
is the greatest risk factor for cervical cancer (4), followed by smoking (5). The five-year relative survival rate
for the earliest stage of invasive cervical cancer is 92%; however,
the prognosis is significantly lower when metastasis is present,
suggesting the importance of early diagnosis.
Studies have identified pathways associated with the
pathogenesis of cervical cancer, particularly the molecular
mechanisms underlying its invasiveness. Wnt signaling was reported
to be involved in the pathogenesis of cervical cancer (6). Tumor necrosis factor-α (TNF-α) is a
pro-inflammatory cytokine, which has been implicated in several
cancers. Duarte et al (7)
reported that G-308A TNF-α polymorphism is associated with an
increased risk of invasive cervical cancer. Chan et al
(8) indicated that overexpression
of forkhead box M1 transcription factor is associated with cervical
cancer progression and pathogenesis. Murphy et al (9) performed an immunocytochemical
analysis to reveal that p16INK4A, CDC6 and MCM5 are predictive
biomarkers in cervical pre-invasive neoplasia and cervical cancer.
Microarray technology is also widely adopted in the discovery of
crucial genes. Song et al (10) identified several candidate genes
associated with invasion of cervical cancer via microarray analysis
of normal cervix, in situ carcinoma and invasive cervical
cancer tissues. Zhai et al (11) identified genes contributing to the
invasive properties of cervical carcinoma cells. However, as these
findings have not resulted in an improved outcome for patients with
cervical carcinoma, additional study is required.
The present study, analyzed gene expression profiles
of high-grade squamous intraepithelial lesions (HSIL) and invasive
cervical squamous cell carcinomas (CSCC) with currently available
bioinformatic tools, attempting to identify crucial genes in the
pathogenesis of CSCC as well as potential biomarkers for diagnosis
or prognosis.
Materials and methods
Microarray data
A gene expression data set [accession no. GSE7803
(11)], including 10 normal
squamous cervical epitheilial, 7 HSIL and 21 invasive CSCC samples,
was downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Gene expression
levels were determined using the Affymetrix Human Genome U133A
Array (no. HG-U133A; Affymetrix Inc., Santa Clara, CA, USA), and
probe annotations were acquired.
Screening of DEGs
The 22,283 probes were mapped into 20,967 genes. A
log2 transformation was applied on the gene expression
levels (12). Analysis of
differentially expressed genes (DEGs) was performed for
pre-invasive cervical squamous cell carcinomas vs. normal control
and invasive cervical squamous cell carcinomas vs. normal control
groups using the Limma package (13) in R. Multiple testing correction
according to the Benjamini-Hochberg (BH) method (14) was applied to the P-values and the
false discovery rate (FDR) was calculated. FDR<0.05 was set as
the cut-off value to screen out significant DEGs.
To identify genes associated with the invasiveness
of CSCC, DEGs in HSIL were compared with those in invasive
CSCC.
Cluster analysis
Two-way cluster analysis was performed using the
expression levels of the DEGs with package pheatmap in R (15). An Euclidean distance was adopted in
the analysis.
Pathway enrichment analysis
Pathway enrichment analysis was performed for the
DEGs using KOBAS (16). The
statistical method is based on cumulative hypergeometric
distribution and P<0.05 was set as the threshold to filter out
significantly over-represented biological pathways.
Construction of a protein-protein
interaction (PPI) network
Proteins are involved in complex interaction
networks to fulfil certain biological functions. Therefore,
revealing the PPI is a useful method to identify molecular
mechanisms. A PPI network was constructed for the DEGs of invasive
CSCC using String (17), which was
then visualized by Cytoscape (18).
Functional enrichment analysis
Functional enrichment analysis was performed for the
DEGs in the PPI network using the Database for Annotation,
Visualization and Integration Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (19) online tool. The statistical method
is based on hypergeometric distribution. FDR<0.05 was set as the
cut-off value.
Prediction of relevant small
molecules
Connectivity map (Cmap) was designed to link gene
patterns associated with disease to corresponding patterns produced
by drug candidates (20,21). Relevant small molecules were
predicted using the DEGs and those with |score| >0.9 were
retained.
Results
Differentially expressed genes
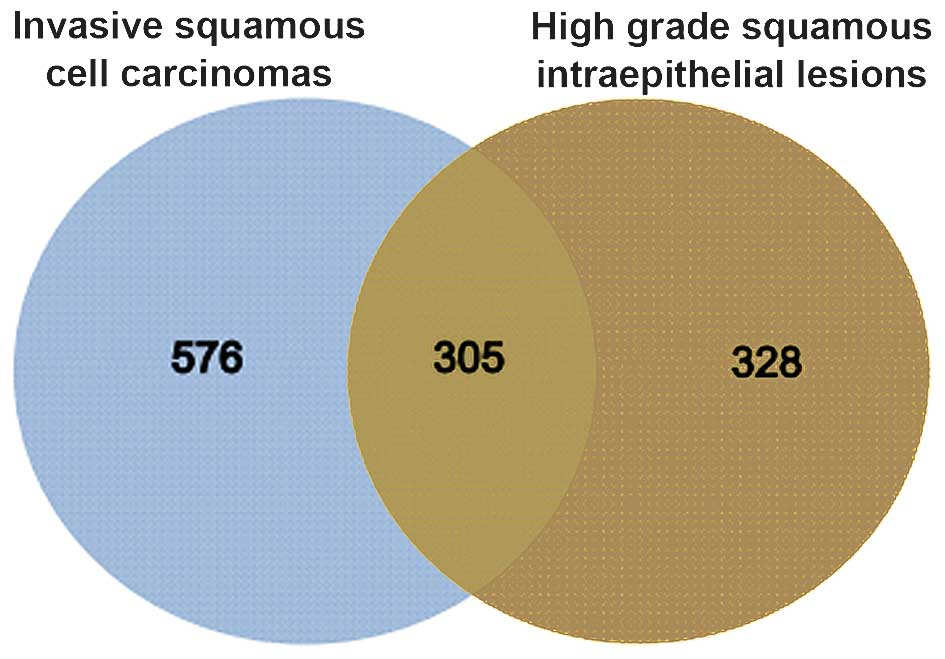
Compared with normal controls, 633 and 881 DEGs were
identified in HSIL and invasive CSCC, respectively.
The two groups of DEGs were compared and 305 genes
were found to be common between HSIL and invasive CSCC (Fig. 1).
Cluster analysis
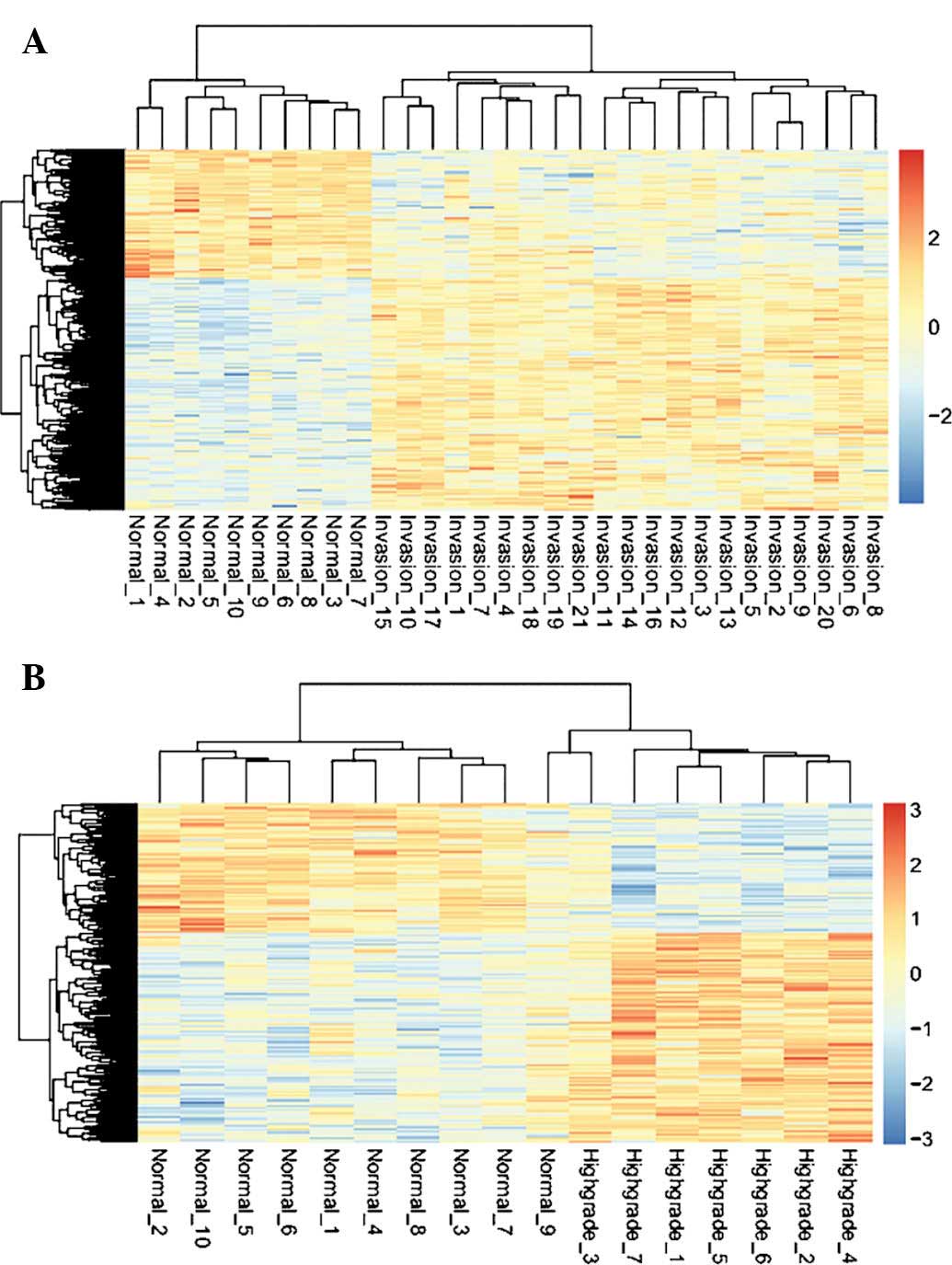
To verify the reliability of the DEG results,
two-way cluster analysis was performed with unique DEGs of HSIL and
invasive CSCC (Fig. 2). HSIL as
well as invasive CSCC were clearly separated from normal controls,
confirming the reliability of the DEG analysis.
Pathway enrichment analysis
Pathway enrichment analysis was performed for the
unique DEGs and common DEGs of HSIL and invasive CSCC using KOBAS
(Table I). the mitogen-activated
protein kinase (MAPK) signaling pathway was significantly enriched
in the unique DEGs of HSIL, while the cell cycle was
overrepresented in the unique DEGs of invasive CSCC.
 | Table ISignificantly enriched pathways in the
three groups of DEGs. |
Table I
Significantly enriched pathways in the
three groups of DEGs.
A, Unique DEGs in
high grade squamous intraepithelial lesions
|
|---|
| ID | Pathway
description | P-value |
|---|
| hsa04010 | MAPK signaling
pathway | 0.004436 |
| hsa00512 | O-Glycan
biosynthesis | 0.005334 |
| hsa05200 | Pathways in
cancer | 0.011241 |
| hsa00531 | Glycosaminoglycan
degradation | 0.013071 |
| hsa05221 | Acute myeloid
leukemia | 0.049570 |
B, Common DEGs
|
|---|
| hsa00590 | Arachidonic acid
metabolism |
8.98×10−4 |
| hsa05120 | Epithelial cell
signaling in Helicobacter pylori infection | 0.012220 |
| hsa00591 | Linoleic acid
metabolism | 0.018788 |
| hsa03030 | DNA replication | 0.036427 |
| hsa04115 | p53 signaling
pathway | 0.049630 |
C, Unique DEGs in
invasive squamous cell carcinomas
|
|---|
| hsa04110 | Cell cycle |
1.23×10−10 |
| hsa03030 | DNA replication |
1.01×10−5 |
| hsa04115 | p53 signaling
pathway |
8.46×10−5 |
| hsa04114 | Oocyte meiosis |
5.11×10−4 |
| hsa03440 | Homologous
recombination |
7.91×10−4 |
| hsa05200 | Pathways in
cancer | 0.003796 |
| hsa05215 | Prostate
cancer | 0.010327 |
| hsa03410 | Base excision
repair | 0.013273 |
| hsa03430 | Mismatch
repair | 0.013275 |
| hsa04610 | Complement and
coagulation cascades | 0.022293 |
| hsa03420 | Nucleotide excision
repair | 0.033021 |
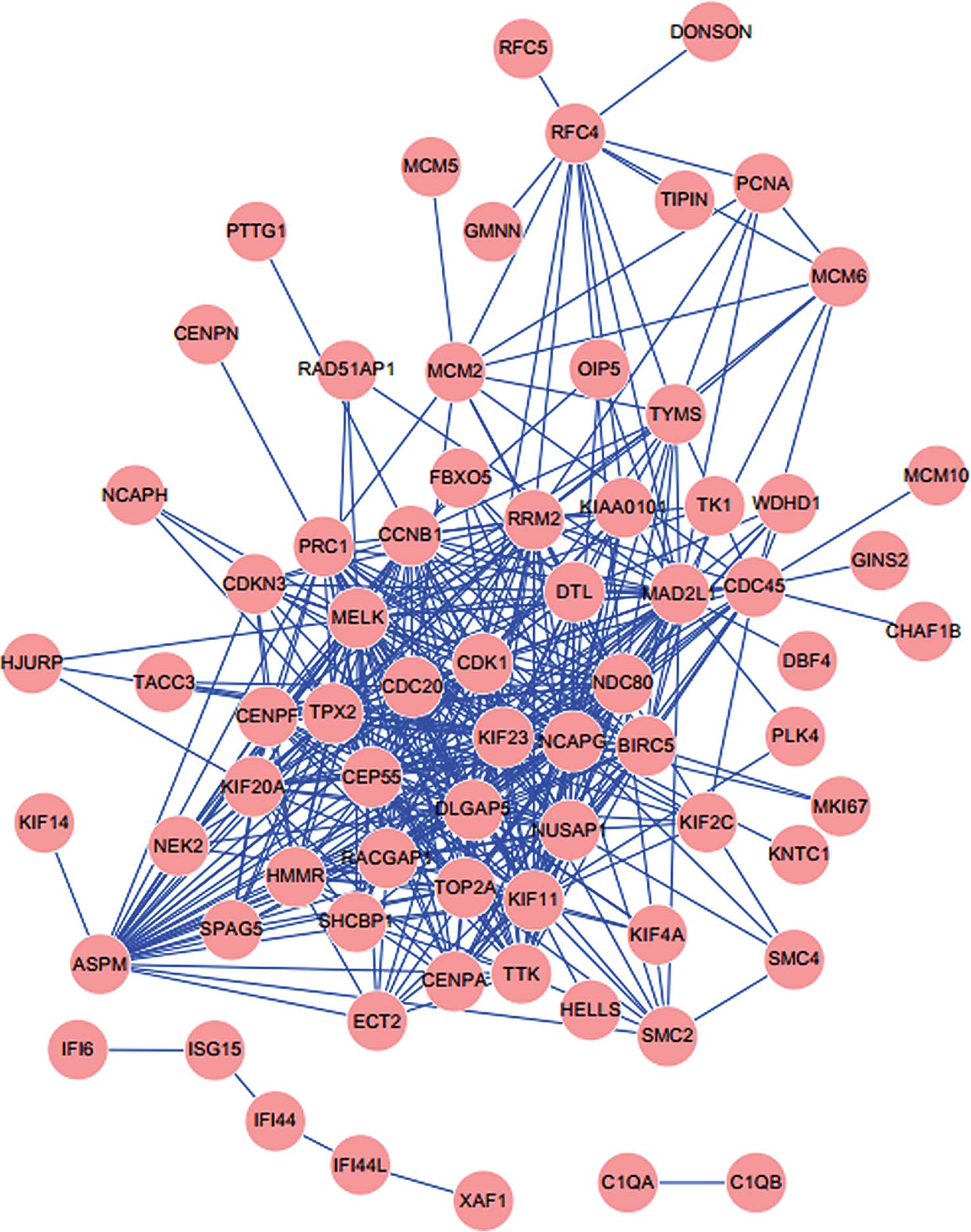
PPI network of DEGs in invasive CSCC
A PPI network was constructed for the DEGs in
invasive CSCC (Fig. 3). The
network consisted of 72 upregulated genes and 434 edges.
Functional enrichment analysis
Functional enrichment analysis was performed for the
genes in the PPI network using the DAVID online tool. The top 10
gene ontology (GO) terms are listed in Table II. All of these terms were
associated with the cell cycle, which was in accordance with the
results of the pathway enrichment analysis. A total of 41 DEGs were
involved in the cell cycle, including DBF4, TTK, PTTG1, CDC45,
CDK1, CDC20, MCM2, MCM6, CCNB1 and MAD2L1.
 | Table IISignificantly enriched GO terms in
the genes from the network. |
Table II
Significantly enriched GO terms in
the genes from the network.
| GO term | Function | Count | P-value | FDR |
|---|
| 0007049 | Cell cycle | 41 |
1.35×10−33 |
1.96×10−30 |
| 0000279 | M phase | 31 |
1.37×10−31 |
1.99×10−28 |
| 0000278 | Mitotic cell
cycle | 32 |
1.43×10−31 |
2.07×10−28 |
| 0022403 | Cell cycle
phase | 33 |
1.49×10−31 |
2.16×10−28 |
| 0007067 | Mitosis | 27 |
3.88×10−30 |
5.63×10−27 |
| 0000280 | Nuclear
division | 27 |
3.88×10−30 |
5.63×10−27 |
| 0022402 | Cell cycle
process | 35 |
4.74×10−30 |
6.88×10−27 |
| 0000087 | M phase of mitotic
cell cycle | 27 |
6.31×10−30 |
9.15×10−27 |
| 0048285 | Organelle
fission | 27 |
1.14×10−29 |
1.66×10−26 |
| 0051301 | Cell division | 27 |
9.56×10−27 |
1.39×10−23 |
Relevant small molecules
A total of six relevant small molecules were
predicted by Cmap with |score| >0.9 (Table III). Piperlongumine was the most
negatively correlated molecule. Previous studies have indicated
that piperlongumine has anti-tumor activity (22,23).
 | Table IIISmall molecules associated with the
pathology of cervical squamous cell carcinomas. |
Table III
Small molecules associated with the
pathology of cervical squamous cell carcinomas.
| Cmap name | Enrichment | P-value |
|---|
| Piperlongumine | −0.927 | 0.01099 |
| GW-8510 | −0.915 | 0.00010 |
| Alsterpaullone | −0.911 | 0.00128 |
| Quinostatin | −0.901 | 0.01976 |
| Prestwick-692 | 0.943 | <0.00010 |
| Isoflupredone | 0.959 | 0.00010 |
Discussion
In the present study, a comparative analysis of gene
expression profiles was performed between HSIL, invasive CSCC and
normal controls. A total of 633 and 881 DEGs were identified in
HSIL and invasive CSCC, respectively. Comparison of the two groups
of DEGs showed that the HSIL and CSCC groups had 305 DEGs in
common. Cluster analysis results verified the confidence of the
DEGs. Pathway enrichment analysis revealed that the MAPK signaling
pathway was significantly enriched in the unique DEGs of HSIL,
while the cell cycle was overrepresented among the unique DEGs of
invasive CSCC. The MAPK pathway can be activated by diverse
extracellular and intracellular stimuli and regulates a variety of
cellular activities, including proliferation, differentiation,
survival and death. Deregulation of MAPK pathways has been
implicated in numerous human diseases, including cancer (24,25).
Dysregulation of the cell cycle is the most common feature of
cancer (20,26) and the analysis of the present study
revealed that it was most significantly enriched in invasive
CSCC.
To further investigate the molecular mechanisms
underlying invasive CSCC, the PPI network was constructed, which
included 72 upregulated genes and 434 edges. Functional enrichment
analysis revealed that the cell cycle and GO terms associated with
the cell cycle were enriched in the genes from the network. This
finding was consistent with the results of the pathway enrichment
analysis. Several of the genes identified were key genes or
potential targets in invasive CSCC. NEK2 is a
serine/threonine-protein kinase that is involved in mitotic
regulation. Upregulation of NEK2 is observed in cell lines derived
from breast cancer (27) and
cervical cancer (28). Hayward and
Fry (28) suggested that NEK2
contributes to chromosome instability and may be a target for
chemotherapeutic intervention. DBF4 is involved in cell adhesion
and migration, possibly through its regulation of the arrangement
of the actin cytoskeleton (29).
The overexpression of DBF4 has been reported in numerous cancer
types (30), and the present study
revealed that it was upregulated in invasive CSCC and may
contribute to the metastasis of CSCC. PTTG1 has transforming
activity in vitro and tumorigenic activity in vivo,
and is highly expressed in various tumor types. Depletion of PTTG1
has anti-proliferative effects in multiple tumor types (31). It also increases cell motility and
promotes lymph node metastasis in esophageal squamous cell
carcinoma (32). Hence, the
present study speculated that PTTG1 may have a crucial role in the
proliferation and mobility of cervical cancer cells. Elevated
expression levels of MCM2 and MCM6 has been reported in cervical
neoplasia (33), suggesting that
these genes may be implicated in the development of CSCC.
Furthermore, the present study predicted associated
small molecules using the expression levels of DEGs in invasive
CSCC by using Cmap. Piperlongumine was the most negatively
correlated molecule, which is a bioactive compound isolated from
long peppers that shows selective toxicity towards a variety of
cancer cell types (34). The
cytotoxicity of piperlongumine has been attributed to increases in
reactive oxygen species in cancer cells. Jarvius et al
(35) reported that it induces
inhibition of the ubiquitin-proteasome system in cancer cells.
Ginzburg et al (36)
further reported that piperlongumine inhibits nuclear factor-κB
activity and attenuates aggressive growth characteristics of
prostate cancer cells. Piperlongumine may therefore be suitable for
controlling invasive CSCC. This result may be useful for the
development of drugs for invasive CSCC.
In conclusion, the present study identified a number
of DEGs in HSIL and invasive CSCC, which may provide direction for
future studies. Potential biomarkers and associated small molecules
for CSCC were revealed, which may contribute to the development of
novel diagnostic markers and therapeutics for CSCC.
References
|
1
|
Armstrong EP: Prophylaxis of cervical
cancer and related cervical disease: A review of the
cost-effectiveness of vaccination against oncogenic HPV types. J
Manag Care Pharm. 16:217–230. 2010.PubMed/NCBI
|
|
2
|
Steward BW and Wild CP: World Cancer
Report. World Health Organization; Geneva: 2014
|
|
3
|
Chaturvedi AK: Beyond cervical cancer:
Burden of other HPV-related cancers among men and women. J Adolesc
Health. 46(4 Suppl): S20–S26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schiffman M, Castle PE, Jeronimo J,
Rodriguez AC and Wacholder S: Human papillomavirus and cervical
cancer. Lancet. 370:890–907. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gadducci A, Barsotti C, Cosio S, Domenici
L and Riccardo Genazzani A: Smoking habit, immune suppression, oral
contraceptive use and hormone replacement therapy use and cervical
carcinogenesis: A review of the literature. Gynecol Endocrinol.
27:597–604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uren A, Fallen S, Yuan H, Usubütün A,
Küçükali T, Schlegel R and Toretsky JA: Activation of the canonical
wnt pathway during genital keratinocyte transformation: A model for
cervical cancer progression. Cancer Res. 65:6199–6206. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duarte I, Santos A, Sousa H, Catarino R,
Pinto D, Matos A, Pereira D, Moutinho J, Canedo P, Machado JC and
Medeiros R: G-308A TNF-alpha polymorphism is associated with an
increased risk of invasive cervical cancer. Biochem Biophys Res
Commun. 334:588–592. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chan D, Yu S, Chiu PM, Yao KM, Liu VW,
Cheung AN and Ngan HY: Over-expression of FOXM1 transcription
factor is associated with cervical cancer progression and
pathogenesis. J Pathol. 215:245–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murphy N, Ring M, Heffron CC, King B,
Killalea AG, Hughes C, Martin CM, McGuinness E, Sheils O and
O'Leary JJ: p16INK4A, CDC6 and MCM5: Predictive biomarkers in
cervical preinvasive neoplasia and cervical cancer. J Clin Pathol.
58:525–534. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song JY, Lee JK, Lee NW, Jung HH, Kim SH
and Lee KW: Microarray analysis of normal cervix, carcinoma in situ
and invasive cervical cancer: Identification of candidate genes in
pathogenesis of invasion in cervical cancer. Int J Gynecol Cancer.
18:1051–1059. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ,
Trimble CL, Fearon ER and Cho KR: Gene expression analysis of
preinvasive and invasive cervical squamous cell carcinomas
identifies HOXC10 as a key mediator of invasion. Cancer Res.
67:10163–10172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujita A, Sato JR, Rodrigues LO, Ferreira
CE and Sogayar MC: Evaluating different methods of microarray data
normalization. BMC bioinformatics. 7:4692006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
14
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. Journal of the Royal Statistical Society. Series
B (Methodological). 289–300. 1995.
|
|
15
|
Szekely GJ and Rizzo ML: Hierarchical
clustering via joint between-within distances: Extending Ward's
minimum variance method. Journal of Classification. 22:151–183.
2005. View Article : Google Scholar
|
|
16
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39(Web Server Issue): W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database Issue): D561–D568. 2011. View Article : Google Scholar :
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar :
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
20
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Catania A, Urban S, Yan E, Hao C, Barron G
and Allalunis-Turner J: Expression and localization of
cyclin-dependent kinase 5 in apoptotic human glioma cells. Neuro
Oncol. 3:89–98. 2001.PubMed/NCBI
|
|
22
|
Kong EH, Kim YJ, Kim YJ, Cho HJ, Yu SN,
Kim KY, Chang JH and Ahn SC: Piplartine induces caspase-mediated
apoptosis in PC-3 human prostate cancer cells. Oncol Rep.
20:785–792. 2008.PubMed/NCBI
|
|
23
|
Bezerra DP, Militão GC, de Castro FO,
Pessoa C, de Moraes MO, Silveira ER, Lima MA, Elmiro FJ and
Costa-Lotufo LV: Piplartine induces inhibition of leukemia cell
proliferation triggering both apoptosis and necrosis pathways.
Toxicology In Vitro. 21:1–8. 2007. View Article : Google Scholar
|
|
24
|
Kim EK and Choi EJ: Pathological roles of
MAPK signaling pathways in human diseases. Biochim Biophys Acta.
1802:396–405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kastan MB and Bartek J: Cell-cycle
checkpoints and cancer. Nature. 432:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hayward DG, Clarke RB, Faragher AJ, Pillai
MR, Hagan IM and Fry AM: The centrosomal kinase Nek2 displays
elevated levels of protein expression in human breast cancer.
Cancer Res. 64:7370–7376. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hayward DG and Fry AM: Nek2 kinase in
chromosome instability and cancer. Cancer Lett. 237:155–166. 2006.
View Article : Google Scholar
|
|
29
|
Chen Y, Lu B, Yang Q, Fearns C, Yates JR
III and Lee JD: Combined integrin phosphoproteomic analyses and
small interfering RNA-based functional screening identify key
regulators for cancer cell adhesion and migration. Cancer Res.
69:3713–3720. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bonte D, Lindvall C, Liu H, Dykema K,
Furge K and Weinreich M: Cdc7-Dbf4 kinase overexpression in
multiple cancers and tumor cell lines is correlated with p53
inactivation. Neoplasia. 10:920–931. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cho-Rok J, Yoo J, Jang YJ, Kim S, Chu IS,
Yeom YI, Choi JY and Im DS: Adenovirus-mediated transfer of siRNA
against PTTG1 inhibits liver cancer cell growth in vitro and in
vivo. Hepatology. 43:1042–1052. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ito T, Shimada Y, Kan T, David S, Cheng Y,
Mori Y, Agarwal R, Paun B, Jin Z, Olaru A, et al: Pituitary
tumor-transforming 1 increases cell motility and promotes lymph
node metastasis in esophageal squamous cell carcinoma. Cancer Res.
68:3214–3224. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Malinowski DP: Multiple biomarkers in
molecular oncology. I. Molecular diagnostics applications in
cervical cancer detection. Expert Rev Mol Diagn. 7:117–131. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu JM, Pan F, Li L, Liu QR, Chen Y, Xiong
XX, Cheng K, Yu SB, Shi Z, Yu AC and Chen XQ: Piperlongumine
selectively kills glioblastoma multiforme cells via reactive oxygen
species accumulation dependent JNK and p38 activation. Biochem
Biophys Res Commun. 437:87–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jarvius M, Fryknäs M, D'Arcy P, Sun C,
Rickardson L, Gullbo J, Haglund C, Nygren P, Linder S and Larsson
R: Piperlongumine induces inhibition of the ubiquitin-proteasome
system in cancer cells. Biochem Biophys Res Commun. 431:117–123.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ginzburg S, Golovine KV, Makhov PB, Uzzo
RG, Kutikov A and Kolenko VM: Piperlongumine inhibits NF-κB
activity and attenuates aggressive growth characteristics of
prostate cancer cells. Prostate. 74:177–186. 2014. View Article : Google Scholar :
|