Introduction
Acute myeloid leukemia (AML) is a hematopoietic stem
cell disorder characterized by the clonal proliferation of myeloid
precursors with inhibition of differentiation, leading to the
accumulation of immature cells at various stages and reduction in
the production of normal hematopoietic components: Erythrocytes,
platelets and mature granulocytes (1). AML is the most common lethal
hematological malignancy in children and young adults, and
represents 3% of all cancer cases, and accounts for ~257,000 cases
of cancer-associated mortality worldwide annually (2). With conventional intensive
chemotherapy, the prognosis for patients with AML remains poor,
with an overall long-term survival of <30% in patients younger
than 60 years old, and only 10–20% for older patients (3). Previous studies have demonstrated
that therapeutic strategies targeting tumor cell growth and
survival signaling pathways may provide a novel strategy to
optimize AML therapy (4–6). Although numerous molecules have been
identified as potential targets, only a few have a pivotal role in
tumor cell proliferation and survival (7). Therefore, the identification of novel
therapeutic targets and the development of novel therapeutic
strategies that may effectively regulate cellular function appear
to be of central importance.
Homeobox (HOX) genes comprise a large family of
homeodomain-containing transcription factors, present in four
separate clusters (A–D), which are key regulators of embryonic
development, hematopoietic differentiation and leukemogenesis
(8). Previous studies demonstrated
that increased HOXA9 and Meis homeobox 1 expression were strongly
associated with cytogenetically normal-AML (9,10).
HOXA5 is a member of the HOX gene family, which is known to have
important roles in embryonic development and in the regulation of
the differentiation process of adult cells. In addition, HOXA5 has
been implicated in the differentiation of epithelial and
hematopoietic cells (11).
Overexpression of HOXA5 in hematopoietic progenitors results in
increased granulocytic/monocytic differentiation, but reduced
erythroid/megakaryocytic differentiation, which suggests that HOXA5
functions as an important regulator of hematopoietic lineage
determination and maturation (12).
Studies of homeobox genes in leukemic cells provided
evidence that aberrant expression may have an important role during
leukemogenesis (13,14). However, whether HOXA5 regulates the
proliferation and apoptosis of leukemia cells, as well as the
nature of the underlying mechanism, remain unknown. The aim of the
present study was to investigate the potential role of HOXA5 in the
development of leukemia.
Materials and methods
Cell culture
Human U937 AML cells were purchased from the
Shanghai Institutes for Biological Sciences of the Chinese Academy
of Sciences (Shanghai, China) and cultured in RPMI-1640 medium (GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 15%
fetal bovine serum (GE Healthcare Life Sciences) and 1% antibiotics
(100 IU/ml penicillin and 100 µg/ml streptomycin; GE
Healthcare Life Sciences), at 37°C in a humidified atmosphere
containing 5% CO2.
GFP assays
Following transfection, GFP expression in U937 cells
was observed under a fluorescent microscope (Olympus DP71; Olympus,
Tokyo, Japan) and then the percentage of GFP-positive cells was
estimated by flow cytometry (FACS FC500; Beckman Coulter, Brea CA,
USA).
In vitro transfection with shRNA
Three pairs of shRNA sequences targeting HOXA5,
termed HOXA5 shRNA-1, -2, -3, and one scramble sequence that served
as a control were designed and synthesized by Shanghai GenePharma
Co., Ltd. (Shanghai, China). Their target sequences are shown in
Table I. The U937 cells in
logarithmic growth phase were seeded into a six-well plate at a
density of 5×105 cells/well and transfected using
HiPerFect (Qiagen, Inc., Valencia, CA, USA) according to the
manufacturer's instructions. After 48 h, transfection efficiency
was examined under the fluorescence microscope. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analyses were performed to determine inhibitory
efficacy.
 | Table ICandidate HOXA5 shRNAs and their
target sequences. |
Table I
Candidate HOXA5 shRNAs and their
target sequences.
| Recombinant | Target sequence
(5′-3′) |
|---|
| HOXA5 shRNA-1 |
GGACTACCAGTTGCATAATTA |
| HOXA5 shRNA-2 |
GCTTTCTGTTCATCTCTTTGT |
| HOXA5 shRNA-3 |
GCAGAAGGAGGATTGAAATAG |
RT-qPCR
Total RNA was isolated using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA),
according to the manufacturer's instructions. A total of 2
µg total RNA was reverse transcribed into cDNA. The reverse
transcription reaction was performed using a PrimeScript™ RT
reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Otsu,
Japan) and RT-qPCR was performed using a SYBR Green reaction kit
(Takara Biotechnology Co., Ltd.) on an ABI PRISM 7500 real-time PCR
system (Applied Biosystems, Foster City, CA, USA). The reaction
system of the PCR was composed of SYBR green reagent, forward
primer, reverse primer, template cDNA and nuclease-free distilled
water. The PCR conditions were as follows: 95°C for 30 sec, 40
cycles of 95°C for 5 sec, and 60°C for 34 sec. The PCR primer
sequences for the RT-qPCR were as follows: Forward:
5′-TGCACCACCACCTGCTTAGC-3′ and reverse: 5′-GGCATGGACTGTGGTCATGAG-3′
for human GAPDH; and forward: 5′-AGCCACAAATCAAGGACACA-3′ and
reverse: 5′-GCTCGCTCACGGAACTATG-3′ for HOXA5. qPCR for each gene of
each cDNA sample was assayed in triplicate. The results were
calculated using the 2−∆∆Ct method using the following
equations: ΔCt = Ct(target gene) − Ct(actin); ΔΔCt =
ΔCt(HOXA5 shRNA-treated cells) − ΔCt(untreated
control).
Western blot analysis
The cells were washed twice with cold
phosphate-buffered saline (PBS) and harvested in 100 µl cell
lysis buffer (Nanjing KeyGEN Biotech Co., Ltd., Nanjing, China)
containing protease inhibitors (Nanjing KeyGEN Biotech Co., Ltd.).
A total of 50 µg extracted proteins were separated on 10%
SDS-PAGE (Beyotime Institute of Biotechnology, Shanghai, China),
and then transferred electrophoretically onto a poly-vinylidene
difluoride membrane (EMD Millipore, Bedford, MA, USA). The
membranes were blocked with 5% skimmed milk for 2 h at room
temperature, and washed three times with Tris-buffered saline with
Tween-20 [TBST; 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.1%
Tween-20], and then incubated overnight at 4°C with specific
antibodies. The primary antibodies used were as follows: Rabbit
monoclonal anti-HOXA5 (ImmunoWay Biotechnology, Newark, DE, USA;
1:1,000 dilution; cat. no. YT2211), rabbit polyclonal anti-survivin
(Wuhan Boster Biological Technology, Ltd., Wuhan, China; 1:300
dilution; cat. no. BA4055-2), rabbit polyclonal anti-caspase-3
(Wuhan Boster Biological Technology, Ltd.; 1:300 dilution; cat. no.
BA2885-2) antibodies, and rabbit polyclonal anti-β-actin (Beyotime
Institute of Biotechnology, Haimen, China; 1:1000 dilution; cat.
no. BA2305) antibody was used as a loading control. The following
day, after washing with TBST, the membrane was incubated in
horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G
(1:5,000; cat. no. ZB-5301; OriGene Technologies, Beijing, China)
for 1 h at room temperature. Finally, images were captured using a
FluorChem FC2 gel imaging system (Alpha Innotech, San Leandro, CA,
USA).
Cell proliferation assay
Cell proliferation was determined using a Cell
Counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). The cells were plated at a density of
1–2×104 cells/well in 96-well culture plates. Following
treatment, 10 µl CCK-8 solution was added to each well and
incubated for 1–4 h. The absorbance was then measured at 450 nm
using an ELISA reader (F-7000; Hitachi High-Technologies
Corporation, Tokyo, Japan).
Cell cycle analysis
Cell cycle analysis was conducted using a Cell Cycle
Detection kit (Nanjing KeyGEN Biotech Co., Ltd.) following the
manufacturer's instructions 48 h post-transfection. Briefly, the
cells were collected and fixed with 70% cold ethanol at 4°C
overnight. The DNA was stained with RNase and propidium iodide (PI)
for 30 min at room temperature, and then analyzed by flow cytometry
(FACS FC500; Beckman Coulter).
Cell apoptosis analysis
A total of 1×105 cells were collected 48
h post-transfection, washed twice with PBS, and then resuspended in
binding buffer (Nanjing KeyGEN Biotech Co., Ltd.). Cell suspension
was stained with Annexin V-fluorescein isothiocyanate and PI
(Nanjing KeyGEN Biotech Co., Ltd.) for 5–15 min at room
temperature, and analyzed by flow cytometry.
Statistical analysis
The data are expressed as the mean ± standard
deviation. The results were evaluated by one-way analysis of
variance using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
result.
Results
Efficacy of shRNA vectors in decreasing
HOXA5 expression in U937 cells
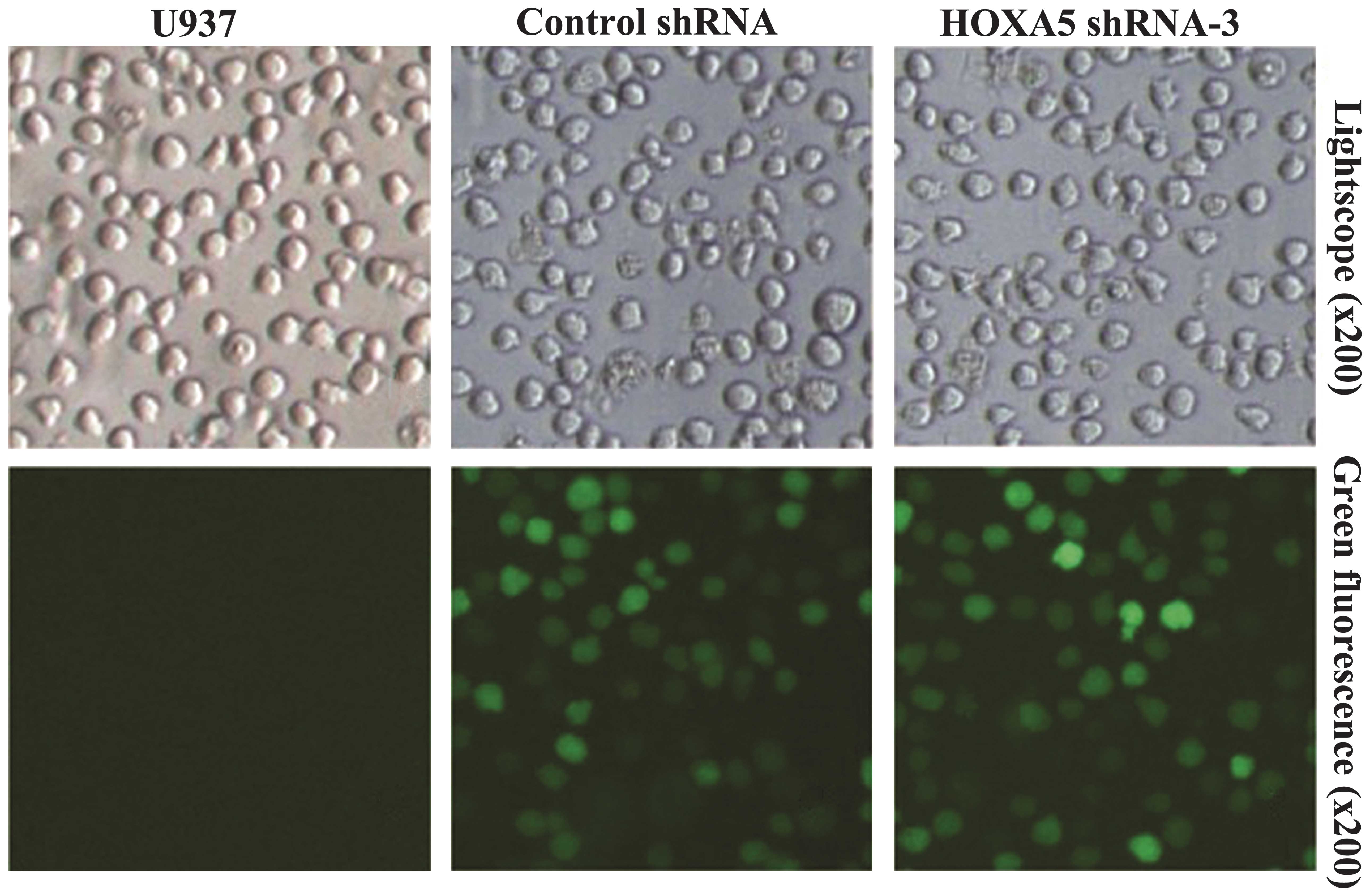
A green fluorescent protein (GFP)-containing vector,
pGPH1, was used to knock down HOXA5. Post-transfection, >70% of
cells were GFP-positive, indicating high transfection efficiency
(Fig. 1). To evaluate the effects
of shRNAs on gene silencing, three HOXA5 shRNAs targeting the HOXA5
gene (HOXA5 shRNA-1, 2 and 3) were investigated in U937 cells by
RT-qPCR and western blotting, with U937 cells alone serving as a
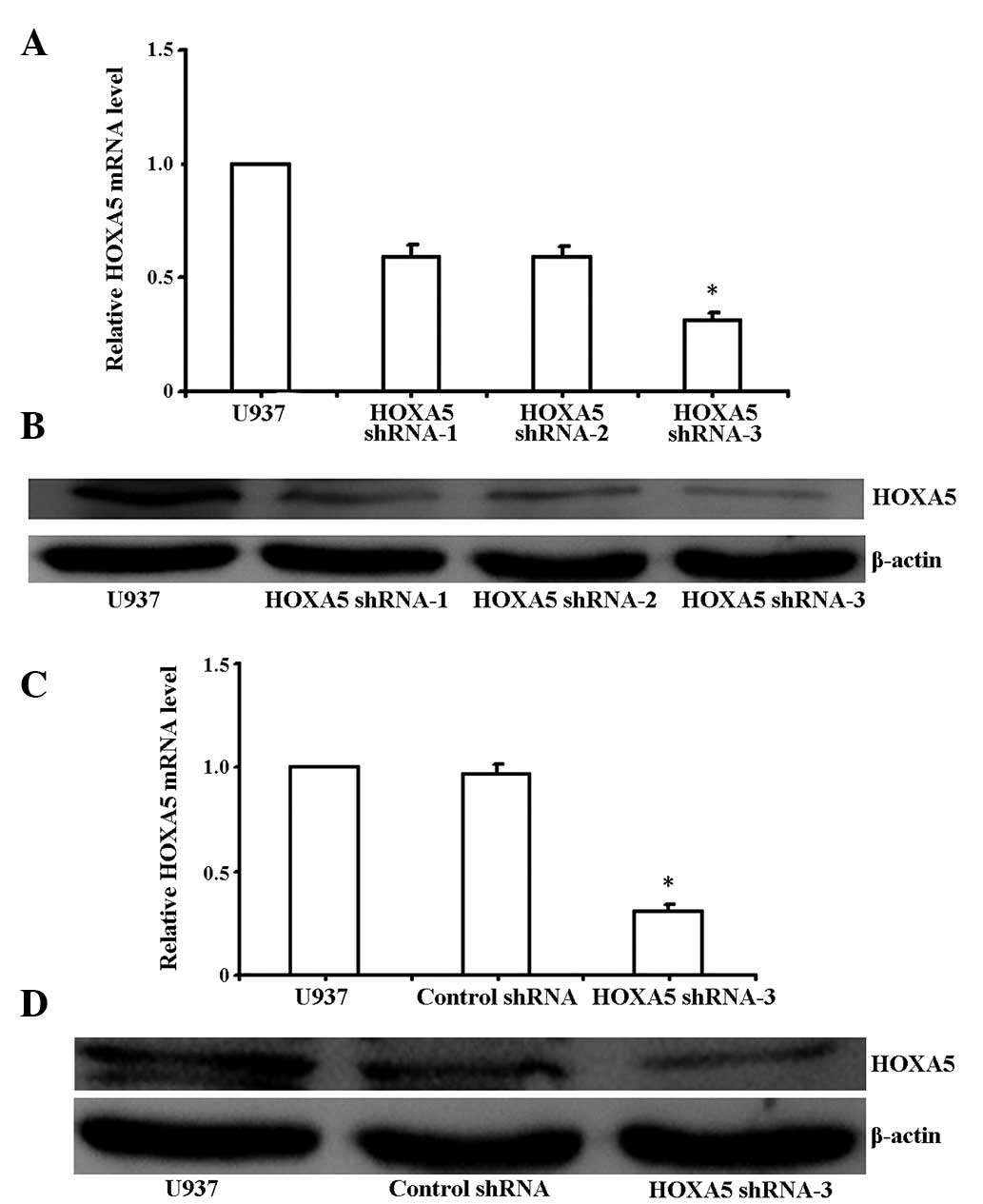
control. HOXA5 shRNA-3 was determined to be the most effective
compared with the other shRNAs. shRNA-3 induced a marked
downregulation of HOXA5 transcript in U937 cells after 48 h
(Fig. 2A and B; P<0.05).
RT-qPCR analysis revealed that HOXA5 mRNA expression levels in the
U937 cells transfected with HOXA5 shRNA-3 decreased by ~70%,
compared with those of the untransfected U937 cells, whereas the
control shRNA had no influence on HOXA5 mRNA expression levels in
the U937 cells (Fig. 2C;
P<0.05). Western blot analysis also showed a significant
decrease in HOXA5 expression in the U937 cell lines (Fig. 2D; P<0.05).
Knockdown of HOXA5 expression inhibits
cell proliferation as determined by a CCK-8 assay
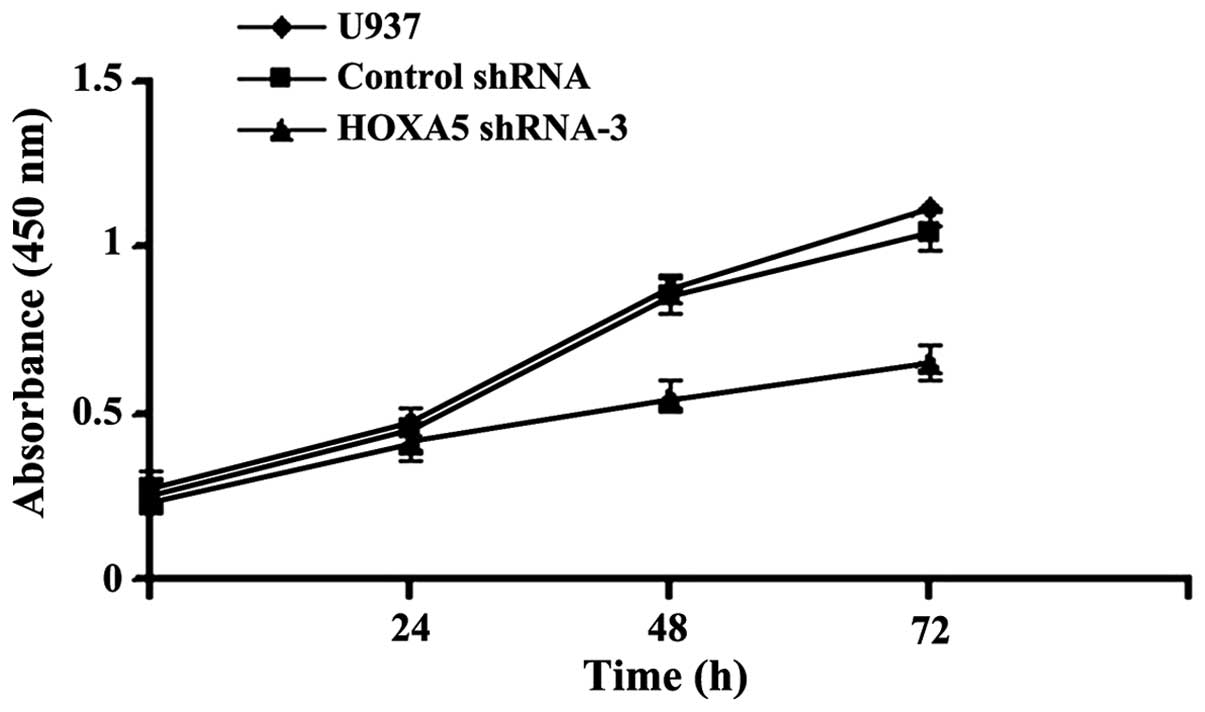
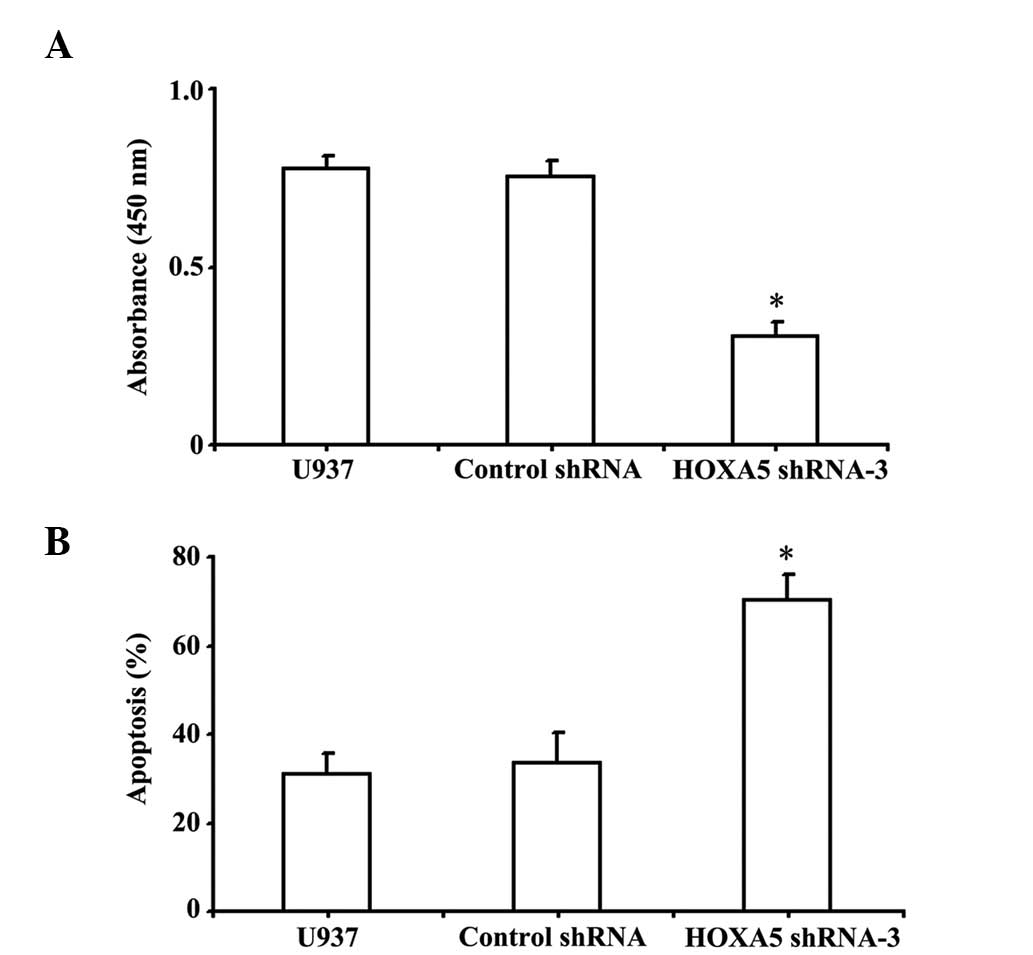
To determine whether downregulation of HOXA5
affected cell growth in vitro, the levels of cell
proliferation were assessed following HOXA5 RNAi by CCK-8 assay,
and the results are shown in Fig.
3. After 0, 24, 48 and 72 h transfection, the growth of HOXA5
shRNA-3 transfected cells decreased compared with cells transfected
with control shRNA and parental U937 (P<0.05), suggesting that
transfection with HOXA5 shRNA-3 inhibited cell proliferation.
Knockdown of HOXA5 expression induces
cell cycle G1 phase arrest in U937 cells in vitro
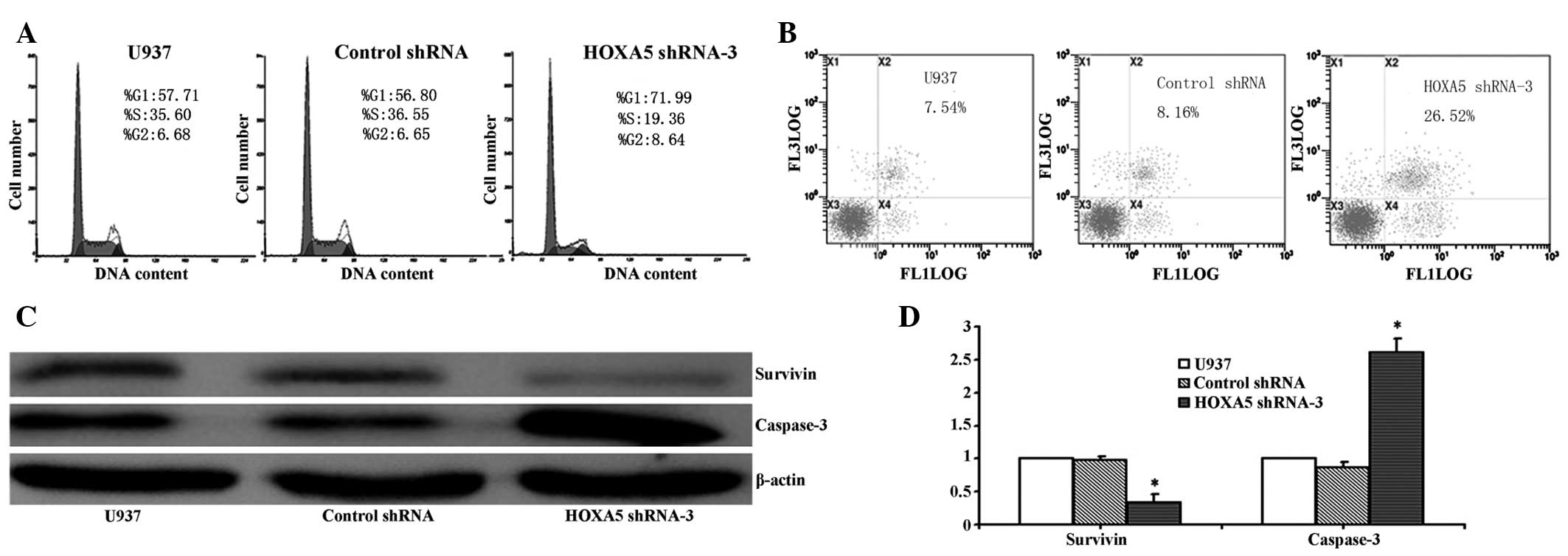
To evaluate the effects of HOXA5 knockdown on cell
cycle distribution in U937 cells, DNA content was measured by flow
cytometry in HOXA5 shRNA-3 transfected cells and control cells. As
shown in Fig. 4A, 48 h
post-transfection, interference of HOXA5 expression led to cell
cycle arrest in the G1 phase, compared with the controls
(P<0.05). In addition, the increase in the G1 phase
cell population was accompanied by a concomitant decrease in cells
in the S phase in the U937 cell lines with HOXA5 knockdown
(P<0.05), without any observable changes in the cell population
in the G2 phase. These data suggest that the induction
of G1 phase arrest accounts for the inhibitory effects
of HOXA5 knockdown on U937 cell growth.
Knockdown of HOXA5 induces U937 cell
apoptosis
The effects of HOXA5 knockdown on cell apoptosis
were also investigated using flow cytometric analysis. Apoptotic
rates were significantly higher in HOXA5 shRNA-3-transfected U937
cells. As shown in Fig. 4B, U937
cells transfected with HOXA5 shRNA-3 underwent increased apoptosis,
compared with control shRNA and parental U937 cells. Furthermore,
western blot analysis demonstrated that compared with control
cells, the protein expression levels of survivin were significantly
reduced, whereas the expression levels of caspase-3 were
significantly increased in HOXA5 shRNA-3-transfected U937 cells
(Fig. 4C; P<0.05). These
results suggest that inhibition of cell growth by HOXA5 shRNA-3
induces cell death in U937 cells via apoptotic events.
Knockdown of HOXA5 increases the
sensitivity of U937 cells to cytarabine
To assess whether downregulation of HOXA5 was able
to enhance the sensitivity of leukemic cells to cytarabine, a
common antitumor drug used in the treatment of AML, 48 h
post-transfection HOXA5 shRNA-3-transfected cells, control
shRNA-transfected cells, and parental U937 cells were incubated
with cytarabine for 24 h, and cell proliferation inhibition was
determined by a CCK-8 assay. As shown in Fig. 5A, compared with the controls, HOXA5
shRNA-3 exhibited significantly higher inhibitory levels on cell
proliferation, indicating enhanced sensitivity to cytarabine
(P<0.05). Furthermore, the present study investigated whether
knockdown of HOXA5 was able to enhance cytarabine-induced cell
apoptosis. HOXA5 shRNA-3 transfection combined with cytarabine
resulted in a significant increase in apoptotic cell death,
compared with cytarabine or HOXA5 shRNA-3 alone (Fig. 5B; P<0.05), suggesting that HOXA5
knockdown was able to enhance the sensitivity of U937 cells to
cytarabine.
Discussion
Deregulation of HOX genes influences tumorigenesis
and cancer cell biology via differentiation, apoptosis, receptor
signaling and other unknown mechanisms (15,16).
Aberrant expression of Hox has been observed in various types of
cancer, such as hematologic malignancies, breast carcinoma,
prostate cancer and lung cancer (17–21).
Previous studies have suggested that overexpression of individual
HOXA genes alone or in combination with MEIS1 results in leukemia
(22–24). HOXA5 is a member of the HOX gene
family and its overexpression in hematopoietic progenitor cells
results in increased granulocytic/monocytic differentiation, but
reduced erythroid/megakaryocytic differentiation (9). HOXA5 is critical for leukemic
transformation in CALM-AF10-mediated leukemia, and is overexpressed
in numerous leukemia cell lines (25,26).
These results suggested that HOXA5 is an important regulator of
hematopoietic lineage determination and maturation. Therefore, the
present study was designed to investigate the cellular functions of
HOXA5, in order to elucidate the mechanism underlying its
contribution to leukemia.
The present study used shRNA to reduce the
expression of HOXA5 in human U937 leukemia cell lines. The selected
shRNA-containing vector efficiently suppressed HOXA5 expression at
the mRNA and protein levels. The effect of HOXA5 knockdown on the
cellular functions of U937 cells was subsequently explored. Cell
proliferation of U937 cells transfected with HOXA5 shRNA-3 also
indicated that the cell growth of U937 cells decreased
significantly compared with control cells. Cell cycle analysis
demonstrated that the percentage of cells in the G1
phase was markedly increased, and those in the S phase was markedly
decreased in HOXA5 shRNA-3-transfected U937 cells. These results
suggested that knockdown of the HOXA5 gene was able to arrest cell
cycle progression, and inhibit cell proliferation. A flow
cytometric apoptosis assay demonstrated that HOXA5-specific shRNA
treatment resulted in a significant increase in the percentage of
cell apoptosis.
Apoptosis is regulated by a balance of pro-apoptotic
and anti-apoptotic genes, in which multiple genes and signaling
pathways are implicated. Abnormal expression and functional changes
of these genes are closely associated with tumorogenesis. Survivin
is a member of the inhibitor of apoptosis family. The survivin
protein is able to inhibit caspase activation via intrinsic and
extrinsic signaling pathways, thereby leading to negative
regulation of apoptosis or programmed cell death (27). These results have been demonstrated
by the disruption of survivin induction signaling pathways leading
to increased levels of apoptosis and a decrease in tumor growth
(28–32). Caspase-3 is an important enzyme in
apoptosis and the executor of apoptosis. The two signaling pathways
of cellular apoptosis, the intrinsic and extrinsic signaling
pathways, converge at caspase-3 to activate other caspases leading
to a proteolytic cascade and eventually apoptotic morphology.
Western blot analysis demonstrated that transfection with
HOXA5-targeted shRNA significantly decreased the expression levels
of survivin, and elevated the expression levels of caspase-3
compared with the controls, implying a role for HOXA5 as a
regulator of the expression of apoptosis-associated molecules.
Cytarabine is a DNA polymerase inhibitor widely used
for the treatment of several types of malignancies, including
leukemia. The present study demonstrated that HOXA5 knockdown
significantly enhances the cytotoxicity of cytarabine, and the
combination of cytarabine and HOXA5-targeted shRNA exerts greater
antiproliferative effects compared with cytarabine treatment alone,
demonstrating the role of HOXA5 in the chemoresistance of U937
cells. Therefore, these results suggested that suppression of HOXA5
by shRNA may sensitize leukemic cells to cytarabine. These findings
highlight the potential of combining conventional chemotherapeutics
with gene therapy as an attractive therapeutic strategy for
leukemia.
In conclusion, the results of the present study
demonstrated that HOXA5 knockdown with shRNA in U937 human leukemia
cells exerted antiproliferative effects, induced apoptosis and cell
cycle arrest, and enhanced the cytotoxicity of cytarabine. These
findings suggested that the shRNA-silencing of HOXA5 may be
considered as a promising therapeutic target for leukemia, and may
be used with conventional chemotherapeutics to increase therapeutic
efficacy.
Acknowledgments
The authors of the present study would like to thank
Dr Xie Shuyang and Dr Li Youjie for their technical assistance. The
present study was supported by funding from the NCET-10-0919
'Taishan Scholar' position of the National Natural Science
Foundation (grant nos. 31371321 and 81200601); the Shandong Science
and Technology Committee (grant no. 2010GSF10264); and the
Foundation of Shandong Educational Committee of China (grant nos.
J10LC60 and J11LC01).
Abbreviations:
|
AML
|
acute myeloid leukemia
|
|
CCK-8
|
cell counting kit-8
|
|
GFP
|
green fluorescent protein
|
|
CN-AML
|
cytogenetically normal acute myeloid
leukemia
|
|
PBS
|
phosphate-buffered saline
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
SDS-PAGE
|
sodium dodecyl sulphate-polyacrylamide
gel electrophoresis
|
|
shRNA
|
short hairpin RNA
|
References
|
1
|
Passegué E, Jamieson CH, Ailles LE and
Weissman IL: Normal and leukemic hematopoiesis: Are leukemias a
stem cell disorder or a reacquisition of stem cell characteristics?
Proc Natl Acad Sci USA. 100(Suppl 1): 11842–11849. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics. 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Estey E and Döhner H: Acute myeloid
leukaemia. Lancet. 368:1894–1907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bali P, George P, Cohen P, Tao J, Guo F,
Sigua C, Vishvanath A, Scuto A, Annavarapu S, Fiskus W, et al:
Superior activity of the combination of histone deacetylase
inhibitor LAQ824 and the FLT-3 kinase inhibitor PKC412 against
human acute myelogenous leukemia cells with mutant FLT-3. Clin
Cancer Res. 10:4991–4997. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Christiansen DH, Andersen MK, Desta F and
Pedersen-Bjergaard J: Mutations of genes in the receptor tyrosine
kinase (RTK)/RAS-BRAF signal transduction pathway in
therapy-related myelodysplasia and acute myeloid leukemia.
Leukemia. 19:2232–2240. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Plass C, Oakes C, Blum W and Marcucci G:
Epigenetics in acute myeloid leukemia. Semin Oncol. 35:378–387.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yao K, Xing H, Yang W, Liao A, Wu B, Li Y,
Zhang R and Liu Z: Knockdown of RLIP76 expression by RNA
interference inhibits proliferation, enhances apoptosis, and
increases chemosensitivity to daunorubicin in U937 leukemia cells.
Tumour Biol. 35:8023–8031. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cillo C, Cantile M, Faiella A and
Boncinelli E: Homeobox genes in normal and malignant cells. J Cell
Physiol. 188:161–169. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bullinger L, Döhner K, Bair E, Fröhling S,
Schlenk RF, Tibshirani R, Döhner H and Pollack JR: Use of
gene-expression profiling to identify prognostic subclasses in
adult acute myeloid leukemia. N Engl J Med. 350:1605–1616. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grubach L, Juhl-Christensen C, Rethmeier
A, Olesen LH, Aggerholm A, Hokland P and Ostergaard M: Gene
expression profiling of Polycomb: Hox and Meis genes in patients
with acute myeloid leukaemia. Eur J Haematol. 81:112–122. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thorsteinsdottir U, Sauvageau G and
Humphries RK: Hox homeobox genes as regulators of normal and
leukemic hematopoiesis. Hematol Oncol Clin North Am. 11:1221–1237.
1997. View Article : Google Scholar
|
|
12
|
Crooks GM, Fuller J, Petersen D, Izadi P,
Malik P, Pattengale PK, Kohn DB and Gasson JC: Constitutive HOXA5
expression inhibits erythropoiesis and increases myelopoiesis from
human hematopoietic progenitors. Blood. 94:519–528. 1999.PubMed/NCBI
|
|
13
|
Lawrence HJ and Largman C: Homeobox genes
in normal hematopoiesis and leukemia. Blood. 80:2445–2453.
1992.PubMed/NCBI
|
|
14
|
Celetti A, Barba P, Cillo C, Rotoli B,
Boncinelli E and Magli MC: Characteristic patterns of HOX gene
expression in different types of human leukemia. Int J Cancer.
237–244. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Drabkin HA, Parsy C, Ferguson K, Guilhot
F, Lacotte L, Roy L, Zeng C, Baron A, Hunger SP, Varella-Garcia M,
et al: Quantitative HOX expression in chromosomally defined subsets
of acute myelogenousleukemia. Leukemia. 16:186–195. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Golub TR, Slonim DK, Tamayo P, Huard C,
Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri
MA, Bloomfield CD and Lander ES: Molecular classification of
cancer: Class discovery and class prediction by gene expression
monitoring. Science. 286:531–537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shah N and Sukumar S: The hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Graham A, Papalopulu N and Krumlauf R: The
murine and drosophila homeobox gene complexes have common features
of organization and expression. Cell. 57:367–378. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Waltregny D, Alami Y, Clausse N, de Leval
J and Castronovo V: Overexpression of the homeobox gene HOXC8 in
human prostate cancer correlates with loss of tumor
differentiation. Prostate. 50:162–169. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Makiyama K, Hamada J, Takada M, Murakawa
K, Takahashi Y, Tada M, Tamoto E, Shindo G, Matsunaga A, Teramoto
K, et al: Aberrant expression of HOX genes in human invasive breast
carcinoma. Oncol Rep. 13:673–679. 2005.PubMed/NCBI
|
|
21
|
Abe M, Hamada J, Takahashi O, Takahashi Y,
Tada M, Miyamoto M, Morikawa T, Kondo S and Moriuchi T: Disordered
expression of HOX genes in human non-small cell lung cancer. Oncol
Rep. 15:797–802. 2006.PubMed/NCBI
|
|
22
|
Thorsteinsdottir U, Mamo A, Kroon E,
Jerome L, Bijl J, Lawrence HJ, Humphries K and Sauvageau G:
Overexpression of the myeloid leukemia-associated hoxa9 gene in
bone marrow cells induces stem cell expansion. Blood. 99:121–129.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bach C, Buhl S, Mueller D, García-Cuéllar
MP, Maethner E and Slany RK: Leukemogenic transformation by HOXA
cluster genes. Blood. 115:2910–2918. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mulgrew NM, Kettyle LM, Ramsey JM, Cull S,
Smyth LJ, Mervyn DM, Bijl JJ and Thompson A: c-Met inhibition in a
HOXA9/Meis1 model of CN-AML. Dev Dyn. 243:172–181. 2014. View Article : Google Scholar
|
|
25
|
Okada Y, Jiang Q, Lemieux M, Jeannotte L,
Su L and Zhang Y: Leukaemic transformation by CALM-AF10 involves
upregulation of hoxa5 by hDOT1 L. Nat Cell Biol. 8:1017–1024. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Quentmeier H, Dirks WG, Macleod RA,
Reinhardt J, Zaborski M and Drexler HG: Expression of HOX genes in
acute leukemia cell lines with and without MLL translocations. Leuk
Lymphoma. 45:567–574. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deveraux QL and Reed JC: IAP family
proteins-suppressors of apoptosis. Gen Dev. 13:239–252. 1999.
View Article : Google Scholar
|
|
28
|
Karami H, Baradaran B, Esfahani A, Estiar
MA, Naghavi-Behzad M, Sakhinia M and Sakhinia E: siRNA-mediated
silencing of survivin inhibits proliferation and enhances etoposide
chemosensitivity in acute myeloid leukemia cells. Asian Pac J
Cancer Prev. 14:7719–7724. 2013. View Article : Google Scholar
|
|
29
|
Riedl SJ and Shi Y: Molecular mechanisms
of caspase regulation during apoptosis. Nat Rev Mol Cell Biol.
5:897–907. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sommer KW, Schamberger CJ, Schmidt GE,
Sasgary S and Cerni C: Inhibitor of apoptosis protein (IAP)
survivin is upregulated by oncogenic c-H-Ras. Oncogene.
22:4266–4280. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamanaka K, Nakata M, Kaneko N, Fushiki H,
Kita A, Nakahara T, Koutoku H and Sasamata M: YM155, a selective
survivin suppressant, inhibits tumor spread and prolongs survival
in a spontaneous metastatic model of human triple negative breast
cancer. Int J Oncol. 39:569–575. 2011.PubMed/NCBI
|
|
32
|
Blanc-Brude OP, Mesri M, Wall NR, Plescia
J, Dohi T and Altieri DC: Therapeutic targeting of the survivin
pathway in cancer: Initiation of mitochondrial apoptosis and
suppression of tumor-associated angiogenesis. Clin Cancer Res.
9:2683–2692. 2003.PubMed/NCBI
|