Introduction
According to the World Health Organization, the
number of individuals diagnosed with cancer annually has reached
>1.2 million individuals, and breast cancer is responsible for
3% of female fatalities (1,2).
Although a number of identified molecules are involved in the way
breast cancers progress and metastasize, the mechanisms of breast
cancer remain to be identified (3,4).
Surgery is mainly used as a primary treatment, and chemotherapy,
radiotherapy and endocrine therapy are then directed at eliminating
the residual tumor cells, thus reducing the recurrence and
metastasis risk. However, there remain cases of relapse or
metastases in certain patients. At this point, few molecules
exhibit high efficiency in predicting chemotherapy sensitivity and
postoperative distant metastasis for breast cancer.
DEK, a non-histone nuclear phosphoprotein initially
identified as a putative proto-oncogene, has recently been found to
be associated with the regulation of hematopoiesis (5). Studies have demonstrated that not
only is it associated with chromatin reconstruction and gene
transcription, but it also contributes to cell apoptosis (5–7).
Thus, there is a direct correlation between high expression levels
of the human DEK and numerous types of human malignancy (5). Following investigation of the DEK
expression level in chronic lymphocytic leukemia, Wang et al
(6) observed a marked increase of
DEK mRNA expression in patients with chronic lymphocytic leukemia,
which may be useful to assess the prognosis in patients with
chronic lymphocytic leukemia. At present, the DEK expression status
and the clinical implication in breast cancer remain unclear. Thus,
the present study aimed to investigate the expression status of DEK
in breast cancer and the clinical implications in order to aid in
the development of breast cancer management.
Patients and methods
Patients and tissue specimens
Patients (n=628) with histologically confirmed
breast cancer who underwent radical surgery between January 2001
and January 2010 in Harbin Medical University (Nangang, China) were
enrolled in the present study. Samples were obtained for
immunohistochemical staining as well as prognostic analysis. The
mean age of the patients was 47.28±9.43 years (range, 27–78 years).
Patients underwent curative surgery, the resected specimens were
pathologically examined and >10 lymph nodes were pathologically
examined following surgery. Complete medical records including the
ER, PR, Her2, p53 and Ki67 status were available. The study
protocol was approved by Harbin Medical University. Patients were
informed of the details of the study and agreed to participate.
Western blot analysis
For western blot analysis, cells were lysed with
buffer (0.1% SDS, 50 mmol/l Tris-HCl, pH 7.6; 1% NP-40, 150 mmol/l
NaCl, 2 mg/ml aprotinin, 2 mg/ml leupeptin and 7 mg/ml PMSF). The
protein concentrations were determined using the bicinchoninic acid
Protein Assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA).
Proteins (30 µg) were separated on 10% SDS-PAGE gels (Varsal
Instruments, Beijing, China) and transferred to a polyvinylidene
difluoride membrane (Varsal Instruments). After blocking, the
membrane was incubated with an anti-DEK antibody (cat. no.
ab166624; 1:1,000; Abcam, Cambridge, MA, USA) at 4°C overnight.
After washing, the membrane was incubated with a secondary antibody
(cat. no. ZB-2301; Beijing Zhongshan Goldenbridge Biotechnology
Co., Ltd., Beijing, China) at a dilution 1:3,000 at room
temperature for 1 h. Proteins were detected with an enhanced
chemiluminescent kit (Varsal Instruments) and anti-β-actin antibody
(cat. no. SAB5500001; 1:1,000; Sigma-Aldrich, St. Louis, MO, USA)
was used as loading control. Densitometry was performed using
Gel-pro Analyzer 4.0 (Media Cybernetics, Silver Spring, MD,
USA).
Immunohistochemistry procedures
Thin slices of tumor tissue from all cases were
fixed in 4% formaldehyde solution (pH 7.0) for periods not
exceeding 24 h. Paraffin embedding was conducted, and 4
µm-thick sections were cut and placed on glass slides coated
with 3-aminopropyl triethoxysilane (Seebio Biotech Inc., Shanghai,
China) for immunohistochemistry. Tissue samples were stained with
hematoxylin and eosin to determine histological type and grade of
tumors.
Briefly, breast tumor tissues were cut at a
thickness of 4 µm using a cryostat. The sections were
mounted on microscope slides, air-dried and then fixed in a mixture
of 50% acetone and 50% methanol. The sections were then de-waxed
with xylene, gradually hydrated with gradient alcohol, and washed
with phosphate-buffered saline (PBS). Sections were incubated for
60 min with the primary antibody. Following washing with PBS,
sections were incubated for 30 min with the secondary biotinylated
antibody (Multilink Swine anti-goat/mouse/rabbit immunoglobulin;
Dako Inc., Carpinteria, CA, USA). Following washing, Avidin Biotin
Complex (1:1,000 dilution, Vector Laboratories Ltd., Burlingame,
CA, USA) was then applied to the sections for 30–60 min at room
temperature. The immunoreactive products were visualized by
catalysis of 3, 3-diaminobenzidine (DAB) by horseradish peroxidase
in the presence of H2O2, following extensive
washings. Sections were then counterstained in Gill's hematoxylin
and dehydrated in ascending grades of methanol prior to clearing in
xylene, and mounting under a coverslip. The sections were observed
under an Olympus CX31 microscope (Olympus, Tokyo, Japan).
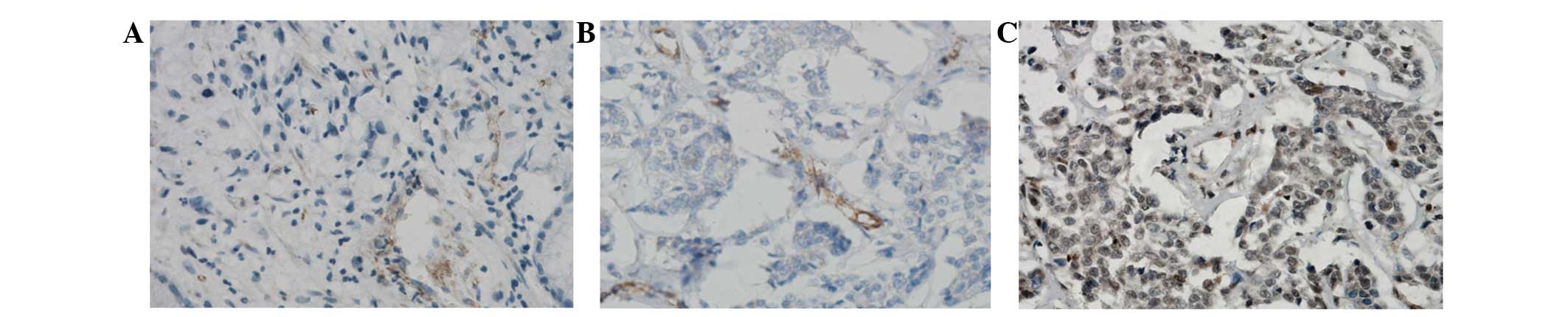
To score DEK as immunopositive staining, the
positive cells are shown as a yellow to brown color in the nucleus
and/or cytoplasm. DEK expression was classified semi-quantitatively
according to the following criteria: −, <1% of neoplastic cells
discretely expressed DEK; +, ≥1 of morphologically unequivocal
neoplastic cells discretely expressed DEK.
Statistical analysis
All data were analyzed with SPSS statistics software
(Version 13.0, SPSS Inc., Chicago, IL, USA). Correlations between
DEK and other parameters were investigated using the χ2
test, Fisher's exact test or independent t-tests. The Kaplan-Meier
method was adopted to analyze disease-specific survival, while the
log-rank test was used to analyze survival differences.
Multivariate analysis was performed using the Cox proportional
hazards model selected in forward stepwise. P<0.05 was
considered to indicate a statistically significant difference.
Results
Association between DEK expression and
clinico-pathological characteristics of breast cancer
Immunohistochemical examination showed that DEK was
located in the nucleus and/or cytoplasm of breast cancer cells. It
was observed that expression of DEK protein was significantly
higher in breast cancer tissues compared with paracancerous tissue
(61.94% vs. 6.53%; Fig. 1).
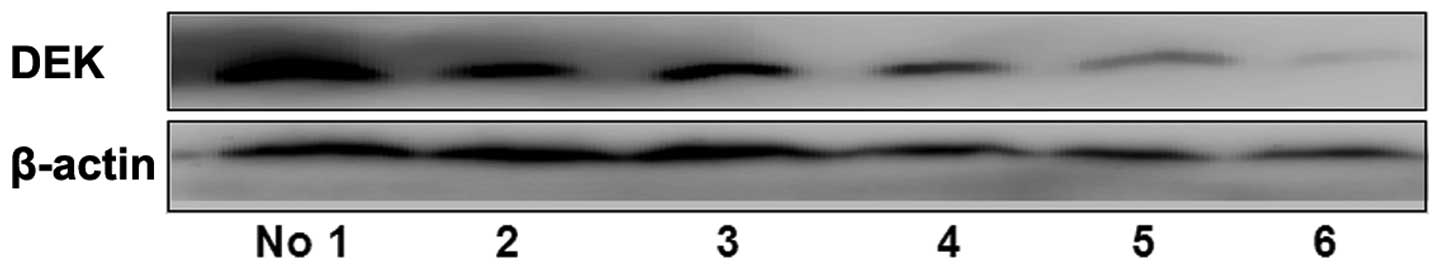
Western blot analysis showed that DEK protein was significantly
highly expressed in breast cancer tissues with lymph node
metastasis compared with those without (P=0.001; Fig. 2). After universal analysis, DEK was
observed to be correlated with age, tumor size, histological type,
lymph node metastasis and distant metastasis (P=0.024, 0.001,
0.001, 0.001 and 0.001, respectively; Table I).
 | Table ICorrelation between DEK expression and
clinico-pathological features of breast cancer (n=628). |
Table I
Correlation between DEK expression and
clinico-pathological features of breast cancer (n=628).
| Variable | n | DEK− | DEK+ | χ2 | P-value |
|---|
| Age, years | | | | 5.093 | 0.024 |
| <40 | 129 | 38 | 91 | | |
| >40 | 499 | 201 | 298 | | |
| Tumor size | | | | 97.559 | 0.001 |
| T1 | 126 | 86 | 40 | | |
| T2 | 463 | 105 | 358 | | |
| T3 | 27 | 6 | 21 | | |
| T4 | 12 | 2 | 10 | | |
| Histological
grade | | | | 46.489 | 0.001 |
| I | 51 | 38 | 13 | | |
| II | 415 | 165 | 250 | | |
| III | 162 | 36 | 126 | | |
| Metastatic nodes | | | | 40.896 | 0.001 |
| Negative | 304 | 158 | 146 | | |
| Positive | 324 | 91 | 243 | | |
| Distant
metastasis | | | | 26.714 | 0.001 |
| Negative | 379 | 175 | 204 | | |
| Positive | 249 | 64 | 185 | | |
| Triple-negative
breast cancer | | | | 0.203 | 0.653 |
| Yes | 140 | 51 | 89 | | |
| No | 488 | 188 | 300 | | |
Association between DEK expression and
the post-operative recurrence and chemotherapeutic resistance
Patients with high expression of DEK were shown to
have a significantly increased distant metastasis rate.
Furthermore, 185 (74.30%) of 249 breast cancers with distant
metastasis exhibited DEK expression compared with 204 (53.83%) of
379 cases of non-distant metastasis (P=0.001).
The factors associated with post-operative distant
metastasis with multiple analyses were also investigated. Age,
tumor size, histological type, triple negative subtype and DEK
expression were found to be associated with post-operative distant
metastasis in breast cancer (Table
II). In addition, the present study investigated the
correlation between DEK expression and chemotherapeutic sensitivity
in 107 patients with breast cancer who underwent neoadjuvant
chemotherapy. DEK expression was expressed in 78.57, 67.86, 46.30
and 18.18% of patients with complete response (CR), partial
response (PR), stable disease (SD) and progressive disease (PD)
(P=0.006; Table III).
 | Table IIMultivariate analysis of the factors
associated with post-operative distant metastasis. |
Table II
Multivariate analysis of the factors
associated with post-operative distant metastasis.
| Characteristic | Exp (B) | 95% CI for Exp
(B) | P-value |
|---|
| Age | 2.847 | 1.302–4.116 | 0.010 |
| Tumor size | 1.641 | 1.386–2.140 | 0.032 |
| Histological
type | 2.056 | 1.749–3.203 | 0.020 |
| Triple-negative
breast cancer | 3.821 | 2.542–5.075 | 0.001 |
| DEK | 3.425 | 2.582–4.169 | 0.001 |
| Constant | 0.002 | | |
 | Table IIICorrelations between DEK expression
and chemotherapeutic resistance in breast cancers [n=107; n
(%)]. |
Table III
Correlations between DEK expression
and chemotherapeutic resistance in breast cancers [n=107; n
(%)].
| Response | n | DEK− | DEK+ |
χ2-value | P-value |
|---|
| CR | 11 | 9 | 2 (18.18) | 12.489 | 0.006 |
| PR | 54 | 29 | 25 (46.30) | | |
| SD | 28 | 9 | 19 (67.86) | | |
| PD | 14 | 3 | 11 (78.57) | | |
Prognostic analysis
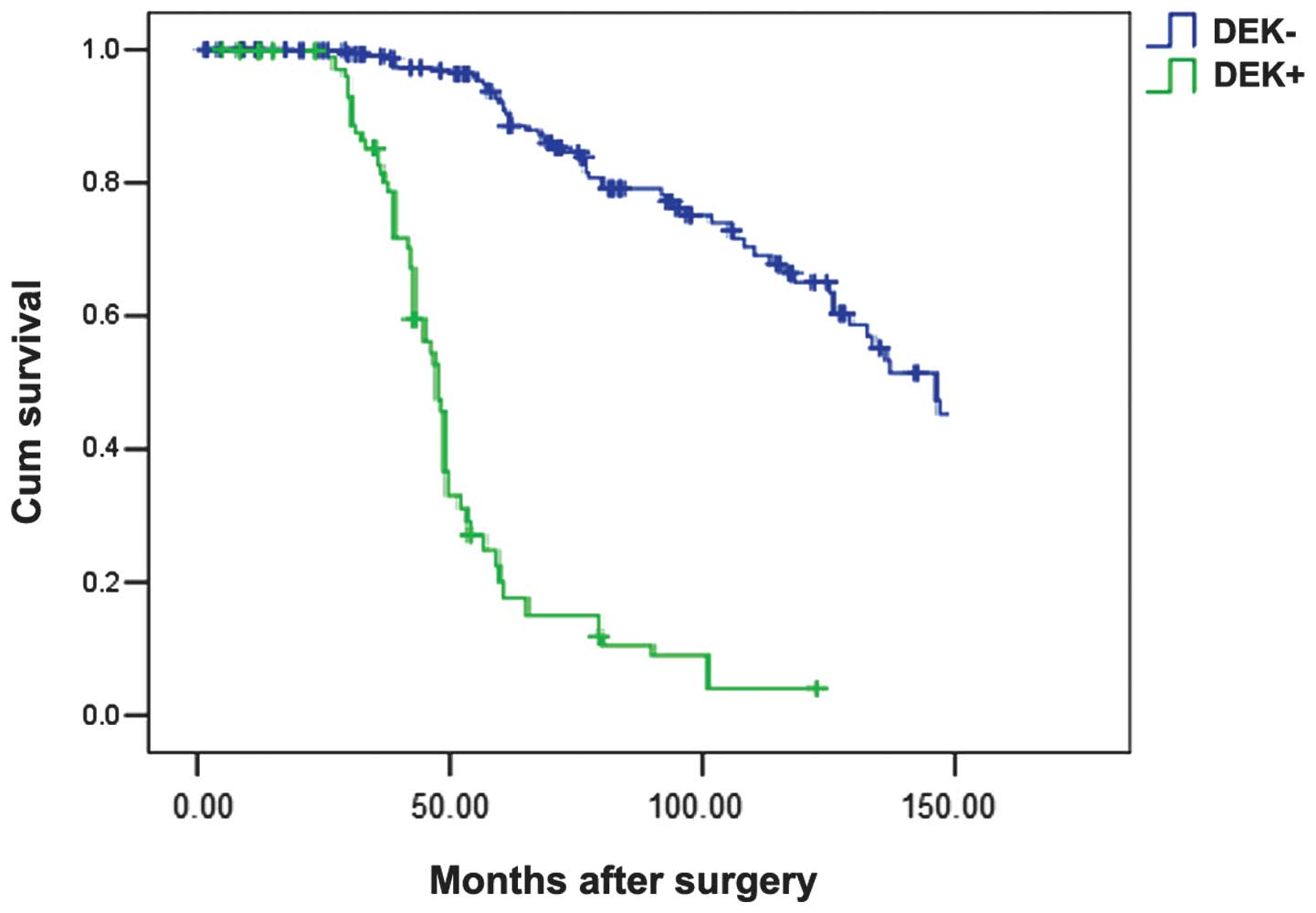
Furthermore, DEK along with age, histological type,
lymph node metastasis, affected survival rate in triple-negative
breast cancer. Patients with triple-negative breast cancer and high
DEK expression exhibited a poorer disease-specific survival
compared with those with none or low expressed DEK protein
(P=0.001; Fig. 3). In the Cox
regression test, DEK protein was detected as an independent
prognostic factor (P=0.001; Table
IV).
 | Table IVCox model regression analysis of the
breast cancer prognostic factors. |
Table IV
Cox model regression analysis of the
breast cancer prognostic factors.
| Variable | OR | 95% CI for OR | P-value |
|---|
| Age | 1.402 | 1.063–2.614 | 0.004 |
| Tumor size | 1.871 | 1.365–3.163 | 0.001 |
| Histological
type | 1.329 | 1.163–1.981 | 0.002 |
| Lymph node
metastasis | 2.132 | 1.655–2.806 | 0.001 |
| Triple-negative
breast cancer | 3.284 | 2.749–4.157 | 0.001 |
| DEK | 2.776 | 1.923–3.260 | 0.001 |
Discussion
DEK is a chromatin-associated oncogene whose
expression has been linked to cancer through multiple mechanisms,
including β-catenin activity. Recently, Privette reported that DEK
is a downstream target of Ron receptor activation in murine and
human models (7). The absence of
DEK in the MMTV-Ron mouse model led to a significant delay in tumor
development, characterized by decreased cell proliferation,
diminished metastasis and a decline in the number of cells
expressing breast cancer stem cell markers. Overexpression of DEK
was sufficient to promote cellular growth and invasion in cell
lines established from MMTV-Ron mouse models (7). In another recent study based on head
and neck squamous cell carcinoma (HNSCC), Adams et al
(8) reported that DEK is required
for optimal proliferation of E7-transgenic epidermal cells and for
the growth of HNSCC tumors. Notably, DEK protein is universally
upregulated in HPV-positive and –negative human HNSCC tumors
relative to adjacent normal tissue. Furthermore, DEK knockdown
inhibited the proliferation of HPV-positive and -negative HNSCC
cells, establishing a functional role for DEK in human disease
(8). DEK is also found to be
related to the poor prognosis of gastric cancer and colorectal
cancer (9,10). Thus, DEK may exhibit potential as a
breast cancer treatment target. At present, the expression status
of DEK protein in breast cancer and its correlation with the
biological behavior of breast cancer remains unclear. Furthermore,
studies addressing the association between DEK and chemotherapy
sensitivity and prognosis of breast cancer are limited.
In the present study, the correlation between DEK
expression and the biological behavior and clinico-pathological
characteristics of breast cancer was investigated. DEK protein
expression was observed to be significantly higher in cancerous
tissues than adjacent-tumor tissues. Furthermore, DEK protein was
found to be related to tumor size, histological type, lymph node
metastasis and post-operative distant metastasis in the 628 breast
cancers. Following further investigation of the association between
DEK expression and chemotherapeutic sensitivity, it was found that
DEK expression was significantly correlated with poor chemotherapy
response in breast neoadjuvant chemotherapy.
After survival analysis, cases with high-level DEK
expression were significantly more likely to develop post-operative
distant metastasis and exhibit poor postoperative disease specific
survival. Cox regression analysis showed DEK protein was detected
as an independent prognostic factor. The outcomes suggested that
DEK expression has been shown to be associated with poor breast
cancer prognosis. DEK may be involved in breast cancer oncogenesis
and may be a potential biomarker for the metastasis and
chemotherapy resistance of breast cancer.
References
|
1
|
Gaffan J, Dacre J and Jones A: Educating
undergraduate medical students about oncology: a literature review.
J Clin Oncol. 24:1932–1939. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dowling EC, Klabunde C, Patnick J and
Ballard-Barbash R: For the international cancer screening network
(ICSN): Breast and cervical cancer screening programme
implementation in 16 countries. J Med Screen. 17:139–146. 2010.
View Article : Google Scholar
|
|
3
|
Dilaveri CA, Mac Bride MB, Sandhu NP, Neal
L, Ghosh K and Wahner-Roedler DL: Breast manifestations of systemic
diseases. Int J Womens Health. 4:35–43. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Magee JA, Piskounova E and Morrison SJ:
Cancer stem cells: impact, heterogeneity and uncertainty. Cancer
Cell. 21:283–296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Broxmeyer HE, Mor-Vaknin N, Kappes F,
Legendre M, Saha AK, Ou X, O'Leary H, Capitano M, Cooper S and
Markovitz DM: Concise review: role of DEK in stem/progenitor cell
biology. Stem Cells. 31:1447–1453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang DM, Liu L, Fan L, Zou ZJ, Zhang LN,
Yang S, Li JY and Xu W: Expression level of DEK in chronic
lymphocytic leukemia is regulated by fludarabine and Nutlin-3
depending on p53 status. Cancer Biol Ther. 13:1522–1528. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Privette Vinnedge LM, Benight NM, Wagh PK,
Pease NA, Nashu MA, Serrano-Lopez J, Adams AK, Cancelas JA, Waltz
SE and Wells SI: The DEK oncogene promotes cellular proliferation
through paracrine Wnt signaling in Ron receptor-positive breast
cancers. Oncogene. 34:2325–2336. 2015. View Article : Google Scholar
|
|
8
|
Adams AK, Hallenbeck GE, Casper KA, Patil
YJ, Wilson KM, Kimple RJ, Lambert PF, Witte DP, Xiao W, Gillison
ML, Wikenheiser-Brokamp KA, et al: DEK promotes HPV-positive and
-negative head and neck cancer cell proliferation. Oncogene.
34:868–877. 2015. View Article : Google Scholar
|
|
9
|
Piao J, Shang Y, Liu S, Piao Y, Cui X, Li
Y and Lin Z: High expression of DEK predicts poor prognosis of
gastric adenocarcinoma. Diagn Pathol. 9:672014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin L, Piao J, Gao W, Piao Y, Jin G, Ma Y,
Li J and Lin Z: DEK over expression as an independent biomarker for
poor prognosis in colorectal cancer. BMC Cancer. 13:3662013.
View Article : Google Scholar : PubMed/NCBI
|