Introduction
Adult stem cells are self-renewing, pluripotent
cells, which can cross lineage boundaries to transdifferentiate
into different types of tissues (1,2).
Since the late 1990s, the number of reports regarding the
transdifferentiation of adult stem cells has increased (3). Our previous study demonstrated the
differentiation of mouse Thy-1(+) Lin(−) bone marrow cells into
hepatocyte-like cells (4). It was
further demonstrated that the transplantation of autologous bone
marrow-derived liver stem cells in patients with hepatitis B
virus-induced liver cirrhosis exhibit improved liver function and
structure (5).
The amino-terminal domains of histones are
susceptible to enzymatic modifications, including acetylation,
methylation, phosphorylation, ubiquitination and SUMOylation
(6,7). Epigenetic modification of histones
may result in altered chromatin conformation and affect the binding
of transcription factors to chromatin. Histone acetylation can
induce a reduction in DNA binding to histones and the relaxation of
chromatin, providing binding sites for proteins, which are crucial
for transcriptional activation (7). Histone methylation refers to the
addition of between one and three methyl groups to the nitrogen
atoms of lysine, arginine or histidine (8). Histone methylation regulates
transcription through providing specific binding sites, which are
attractive to effector protein complexes, promoting the recruitment
and stabilization of regulatory proteins (9). It has been reported that the
acetylation and methylation of histone H3 is important in the
regulation of transcription (10,11).
The epigenetic histone modification of embryonic
stem cells during differentiation into hepatocytes, or
hepatocyte-like cells, has been reported (11-13).
However, histone modification during the transdifferentiation of
adult stem cells into hepatocyte, or hepatocyte-like cells, has not
been reported. Furthermore, the dynamics of epigenetic
modifications during transdifferentiation remain to be fully
elucidated. It has been previously demonstrated that the histone
deacetylase inhibitor, valproic acid, induces the hepatic
differentiation of mouse bone marrow stromal stem cells in
vitro and in vivo, suggesting histone acetylation during
the transdifferentiation of adult stem cells to hepatocytes
(14). The present study aimed to
examine he epigenetic modifications of histone H3 during the
transdifferentiation of Thy-1(+) Lin(−) bone marrow cells into
hepatocytes in vitro, to provide insight into the role of
histone modification in the transdifferentiation of bone marrow
stem cells.
Materials and methods
Experimental animals
A total of 20 rats were used for this study. The
animals were kept in collective cages (four animals per group) at a
controlled temperature of 25±1°C and a 12/12 h light/dark cycle of
with access to food and water ad libitum. The bone marrow
cell donors in the present study comprised 5-6-week-old male inbred
Wistar rats, weighing 90-100 g, which were purchased from the
Experimental Animal Center of Sun Yet-sen University (Guangzhou,
China). The animals were maintained in a pathogen-free environment.
All experimental procedures conformed to the European Convention on
Animal Care (15). The present
study was approved by the Institutional Animal Care and Use
Committee of Southern Medical University (Guangzhou, China).
Preparation of the Thy-1(+) Lin(−) bone
marrow cells
The rats were sacrificed by cervical dislocation.
The femurs and tibias of lower limbs of the rats were collected
under sterile conditions, and the marrow cavity was exposed by
cutting the spongious end of each bone. The marrow cavity was
flushed with DMEM/MCDB201 (Beijing Solarbio Science and Technology
Co., Ltd., Beijing, China) three times to collect the bone marrow
cells. The red blood cells (RBCs) were removed using RBC lysis
buffer (Beijing Solarbio Science and Technology Co., Ltd.). The
RBC-free bone marrow cells were filtered through a 400-mesh sieve
(30 µm opening); and the cells were counted using a
hemocytometer (Beyotime Institute of Biotechnology, Shanghai,
China). The cell suspension density was adjusted to 100 M cells/ml.
A two-step immuno-magnetic cell sorting method was used to obtain
the Thy-1(+) Lin(−) bone marrow cells (16). First, the Lin(−) cells were
separated from the bone marrow cell suspension (negative
selection), following which the Thy-1(+) Lin(−) cells were
separated (positive selection). The lineage cell depletion kit and
immunomagnetic beads were purchased from Miltenyi Biotec GmbH
(Bergish Gladbach, Germany), and used, according to the
manufacturer's instructions. Overall, the viability of the obtained
Thy-1(+) Lin(−) cells was >95%, which was assessed using trypan
blue staining. Briefly, the cell suspension was mixed with 0.4%
trypan blue solution (Beyotime Institute of Biotechnology) at a 1:1
ratio. After 1–2 min incubation at room temperature, the mixture
was loaded onto one chamber of hemocytometer and the squares of the
chamber were observed under a BX51 light microscope (Olympus,
Tokyo, Japan). The viable/live (clear) and non-viable/dead (blue)
cells were counted and the viability was calculated using the
following formula: (number of live cells counted / total number of
cells counted) × 100. Cell detection by flow cytometry (EPICS
Altra; Beckman Coulter, Brea, CA, USA) was conducted within 1 h.
The number of Thy-1(+) Lin(−) bone marrow cells was determined by
counting the percentage of all cells. Flow cytometry revealed that
the cell purity was >98%.
Induction of the differentiation of
Thy-1(+) Lin(−) bone marrow cells into hepatocytes
The induction of differentiation of Thy-1(+) Lin(−)
bone marrow cells into hepatocytes has been previously described
(17). Briefly, the Thy-1(+)
Lin(−) bone marrow cells were seeded into 6-well plates coated with
25 ng/ml fibronectin (1×10 cells/ml; Sigma-Aldrich, St. Louis, MO,
USA) in DMEM/F-12 (Gibco Life Technologies, Carlsbad, CA, USA)
containing 10% fetal bovine serum (GE Healthcare Life Sciences,
Logan, UT, USA), 20 mM HEPES (Sigma-Aldrich), 10 nM dexamethasone
(Sigma-Aldrich), penicillin and streptomycin. Coverslips were
placed in the wells. Following cell adhesion, the medium was
replaced with DMEM/F-12 containing 2.5% cholestatic rat serum
(Gibco Life Technologies), 25 ng/ml hepatocyte growth factor
(PeproTech EC, Ltd., London, UK), 20 mM HEPES, 10 nM dexamethasone,
penicillin and streptomycin (Gibco Life Technologies). The cells
were induced for 14 days consecutively At 37°C, and the
medium was replaced once every 3 days. Subsequently, cell growth
and morphology were observed, and the cells were collected on days
0, 7 and 14 for analysis. Cell growth and morphology were observed
by a BX51 light microscope. Five visual fields were selected
randomly.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The mRNA expression levels of the octamer-binding
transcription factor 4 (OCT4), α-fetoprotein (AFP) and albumin
(ALB) genes during the differentiation of bone marrow stem cells
were detected using one-step RT-qPCR. Total RNA was extracted using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA).
The 1–3 µg mRNA was reverse transcribed using a QuantiTect
Reverse Transcription kit (Takara Bio, Inc., Otsu, Japan). The mRNA
expression levels the of OCT4, AFP and ALB genes were determined 0,
7 and 14 days following induction using a Takara One Step RNA PCR
kit (Takara Biotechnology, Co., Ltd., Dalian, China). The following
primers were used: ALB, forward 5′-atc ctg aac cgt ctg tgtgt-3′ and
reverse 5′-cag tta tcc ttg tcg gcagc-3′; AFP, forward 5′-gcg cat
cca ttt cct tcctt-3′ and reverse 5′-tct aaa cac cca tcg ccagt-3′;
OCT4, forward 5′-agt ttg cca agc tgc tgaaa-3′and reverse
5′-cactcgaaccacatccctct-3′; β-actin, forward
5′-gagaaattgtgcgtgacatca-3′ and reverse 5′-cctgaacctctcattgcca-3′.
All primers were purchased from Sangon Biotech (Beijing, China).
Each qPCR reaction was performed using a final volume of 50
µl, containing 24 µl nuclease-free water, 5 µl
10X One Step RNA PCR buffer, 1 µl of each 0.4 µM
forward and reverse primer, 10 µl 5 mM MgCl2, 5
µl 1 mM deoxynucleoside triphosphate mixture, 1 µl
0.8 U/µl RNase inhibitor, 0.1 U/µl AMV reverse
transcriptase (RTase) XL (Sigma-Aldrich) and AMV-Optimized Taq
(Sigma-Aldrich). Expression levels of different genes were
validated by RT-qPCR analysis using the ABI 7500 sequence detector
system according to the manufacturer's instructions (Applied
Biosystems, Foster City, CA, USA). The thermocy-cling conditions
were as follows: 50°C for 30 min for RT, 94°C for 2 min for RTase
denaturation, 94°C for 30 sec, 25-30 cycles of 65°C for 30 sec;
72°C for 10 min, 50°C for 15 min, 94°C for 2 min, 94°C for 30 sec
and 60°C for 30 sec (28 cycles). β-actin was used as a positive
control. Normal rat Thy-1(+) Lin(−) bone marrow cells, which had
not undergone induction, were used as a negative control. Each qPCR
reaction was repeated three times. Agarose gel electrophoresis was
used to verify RNA integrity, and the quantity and quality of RNA
samples were measured with the NanoDrop ND-1000 Spectrophotometer
(NanoDrop Technologies, Wilmington, DE, USA). The ΔΔCt method was
used for quantification.
Immunofluorescence staining
Histone modification, including dimethylation and
acetylation of H3 at lysine 9 (H3K9me2 and H3K9ac), dimethylation
and acetylation of H3 at lysine 14 (H3K14me2 and H3K14ac) and
dimethylation and acetylation of H3 at lysine 27 (H3K27me2 and
H3K27ac), during the differentiation of bone marrow stem cells was
determined using immunofluorescence staining. The cells were
collected on days 0, 7, and 14 following induction and seeded into
6-well plates (1×105 cells/well). Cells were incubated
for 30 min with the cytokeratin-18 (CK-18) primary antibody at
37°C. Following cell adhesion, the medium was discarded and the
cells were rinsed with phosphate-buffered saline (PBS).
Subsequently, the cells were fixed in paraformaldehyde
(Sigma-Aldrich) at room temperature for 20 min and rinsed with PBS
(three times, 10 min each). The cells were then blocked with PBS
containing 10% normal goat serum for 1 h. The cells were then
incubated with the following antibodies against histone
modification: Anti-histone H3 (dimethyl K9; rabbit monoclonal;
1:100 dilution; cat. no. ab32521; Abcam, Cambridge, MA, USA),
anti-histone H3 (dimethyl K14; rabbit monoclonal; 1:1,000 dilution;
cat. no. ab202416; Abcam), anti-histone H3 (dimethyl K27; rabbit
polyclonal; 1:100 dilution; cat. no. ab194690; Abcam), anti-histone
H3 (acetyl K9; rabbit polyclonal; 1:500 dilution; cat. no. ab61231;
Abcam), anti-histone H3 (acetyl K14; rabbit polyclonal; 1:100
dilution; cat. no. ab82501; Abcam) or anti-histone H3 (acetyl K27;
rabbit monoclonal; 1:200 dilution; cat. no. ab45173; Abcam) (Abcam,
Cambridge, MA, USA). The sample was incubated at 4°C overnight.
After incubation with the primary antibodies, cells were washed in
PBS three times and then incubated with their specific secondary
antibodies (anti-rabbit; cat. no. ZDR-5306; Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd., Beijing, China) for 1 h at
room temperature. For the assessment of CK-18, the primary antibody
used was anti-CK18 rabbit anti-rat antibody (cat. no. sc-24603;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and the samples
were incubated at 37°C for 30 min. The secondary antibody used for
detection was HRP-labeled anti-rabbit (cat. no. ZF-0513; Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd.). Following
incubation, the sample was rinsed with PBS containing 0.1% Tween-20
(three times, 10 min each). The cells were then incubated with
either fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit
IgG (1:1,000; cat. no. ZF-0511) or FITC-labeled goat anti-mouse IgG
(1:2,000; cat. no. ZF-0512) (Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd.) for the detection of histone modification
or the CK-18, respectively. The cells were incubated for 1 h or 30
min, respectively at 37°C. Following secondary antibody removal,
diamidino-2-phenylindole staining solution Invitrogen Life
Technologies) was added and the sample was incubated at room
temperature for 15 min. The sample was then rinsed with PBS
containing 0.1% Tween-20 (three times, 10 min each). The stained
cells were then observed under a fluorescent microscope (ECLIPSE
TE2000-U, Nikon Corporation, Tokyo, Japan), and images were
captured using a Universal Hood II gel imaging system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Western blot analysis
The histone modification expression levels of
H3K9me2, H3K14me2, H3 K27me2, H3K9ac, H3K14ac and H3 K27ac during
the differentiation of bone marrow stem cells were detected using
western blot analysis. Total protein was extracted from the cells
using a nuclear and cytoplasmic protein extraction kit (Beyotime
Institute of Biotechnology, Haimen, China), according to the
manufacturer's instructions. Protein concentrations were measured
using the Bradford method. The methods of the western blot assay
used were those previously described (18). Briefly, the blots were incubated
with the same corresponding first and secondary antibodies used for
the above-mentioned immuno-fluorescence staining. Subsequently, the
samples were detected using an enhanced chemiluminescence kit
(PerkinElmer, Inc, Waltham, MA, USA). Normal rat Thy-1(+) Lin(−)
bone marrow cells, which did not undergo induction, were used as a
negative control. The rats received an intravenous injection of
heparin and the liver was cannulated. The liver was perfused
through the portal vein, first with perfusion buffer for 10 min and
then with collagenase buffer for 12 min at a flow rate of 25
ml/min. Cells were separated from the digested liver using a 100
µm cell strainer (BD Biosciences, Franklin Lakes, NJ, USA)
to obtain a cellular suspension. The cell suspension was
centrifuged for 3 min at 200 × g. The upper layer was discarded and
10 ml of washing buffer was added. Rat hepatocytes, which were
freshly separated, were used as a positive control. β-actin was
used as a loading control. All analyses were repeated three times.
The intensity of bands was determined by using Quantity One
software (Bio-Rad Laboratories, Inc.), and the gray-scale value of
each target protein vs. that of β-actin was quantitatively
analyzed. All analyses were repeated three times.
Statistical analysis
Friedman one-way analysis of variance was used to
detect differences in the mRNA expression levels of the OCT4, AFP
and ALB genes; as well as the expression of the histone
modifications, including H3K9me2, H3K9ac, H3K14me2, H3K14ac,
H3K27me2 and H3K27ac, between groups. A Wilcoxon rank sum test was
used for pair-wise comparisons. Statistical analyses were performed
using SAS 9.2 (SAS Institute Inc., Cary, NC, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Differentiation of Thy-1(+) Lin(−) bone
marrow cells into hepatocytes in vitro
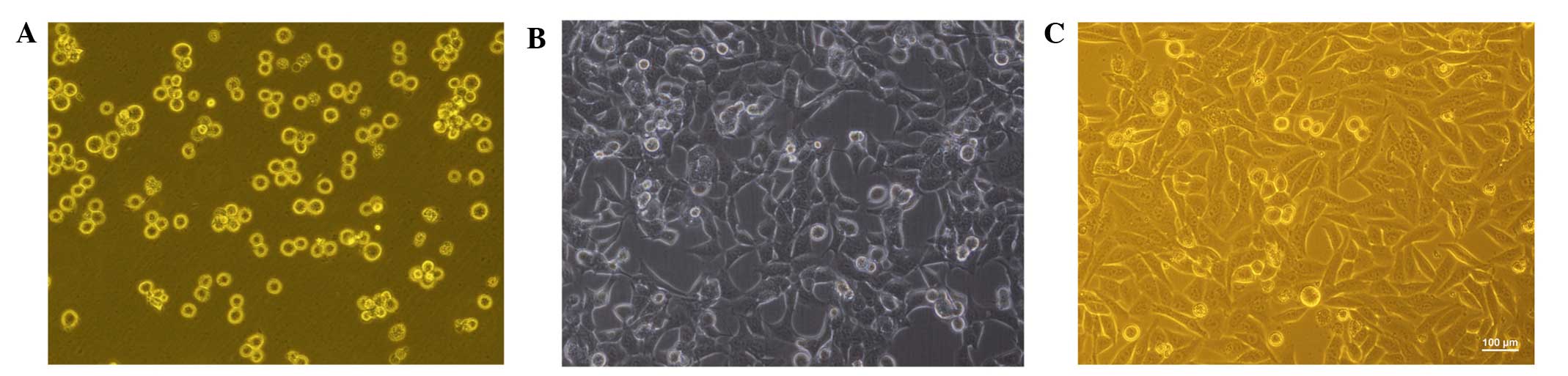
The freshly separated Thy-1(+) Lin(−) bone marrow
cells were uniform in size and they exhibited a lymphocyte-like
round shape. Prior to induction, the shape of the adherent cells
was variable, resembling spindle-, bipolar-, and fibroblast-like
cells. On day 7 post-induction, the morphology of the adherent
cells had diversified, and resembled short-spindle, oval- and
polygonal-like cells. On day 14 post-induction, the cell morphology
had changed further, with cells exhibiting an oval, round or
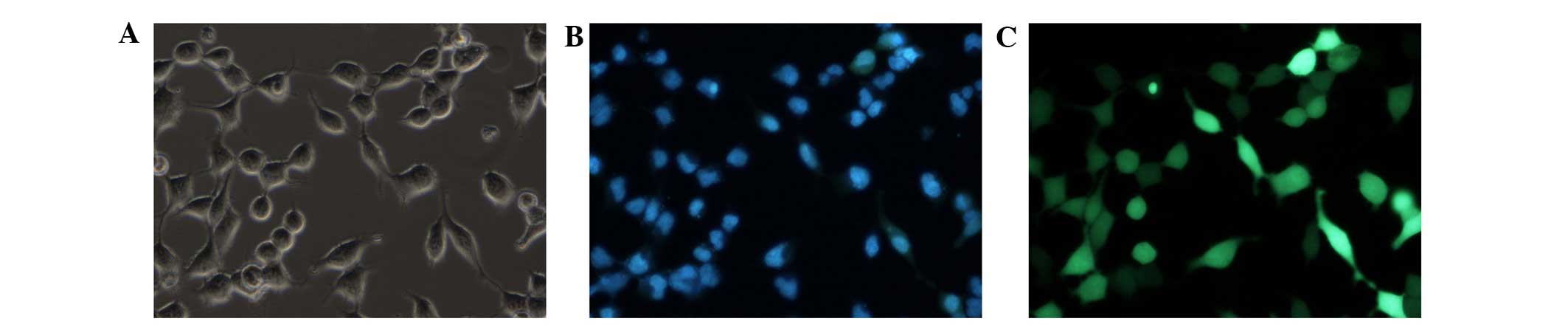
polygonal shape with an epithelioid shape (Fig. 1). On day 14 post-induction, the
bone marrow stem cells, which were derived from the hepatic lineage
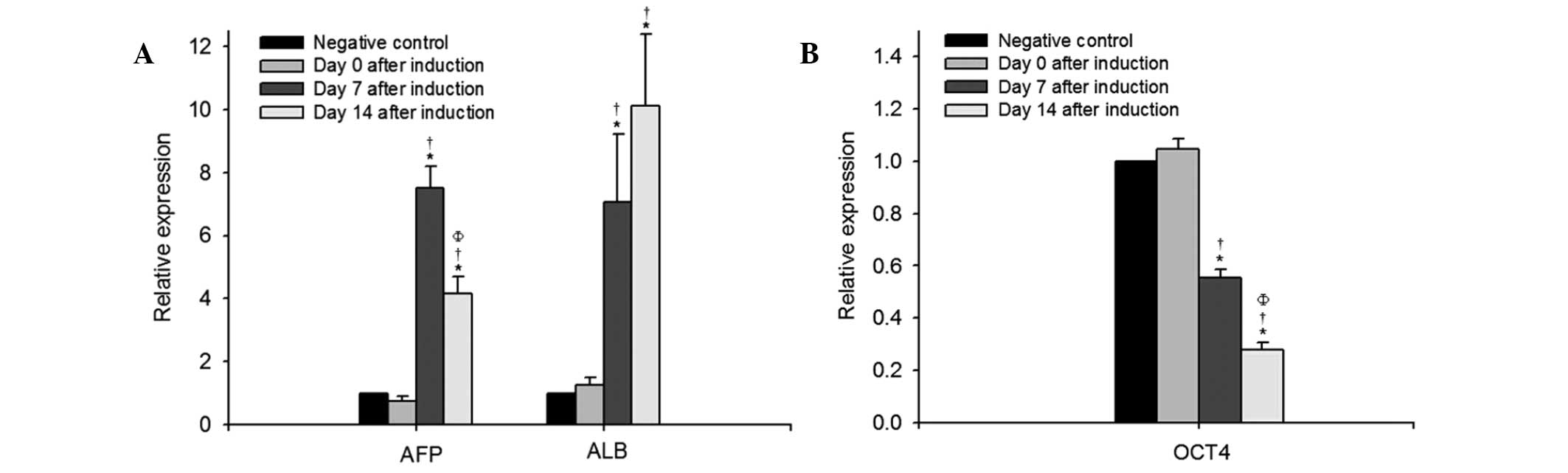
cells, expressed the hepatocyte-specific marker, CK-18 (Fig. 2). The results of the RT-qPCR
analysis demonstrated that the mRNA expression levels of
hepatocyte-specific AFP and ALB increased significantly on day 7
and day 14 following induction, compared with the negative controls
or those on day 0 (Fig. 3A).
Epigenetic modifications in Thy-1(+)
Lin(−) bone marrow cells during differentiation into
hepatocytes
The mRNA expression levels of the key pluripotency
factor, OCT4, in the bone marrow stem cell-derived hepatic lineage
cells was significantly reduced 7 days following induction,
compared with that on day 0, followed by a more marked reduction on
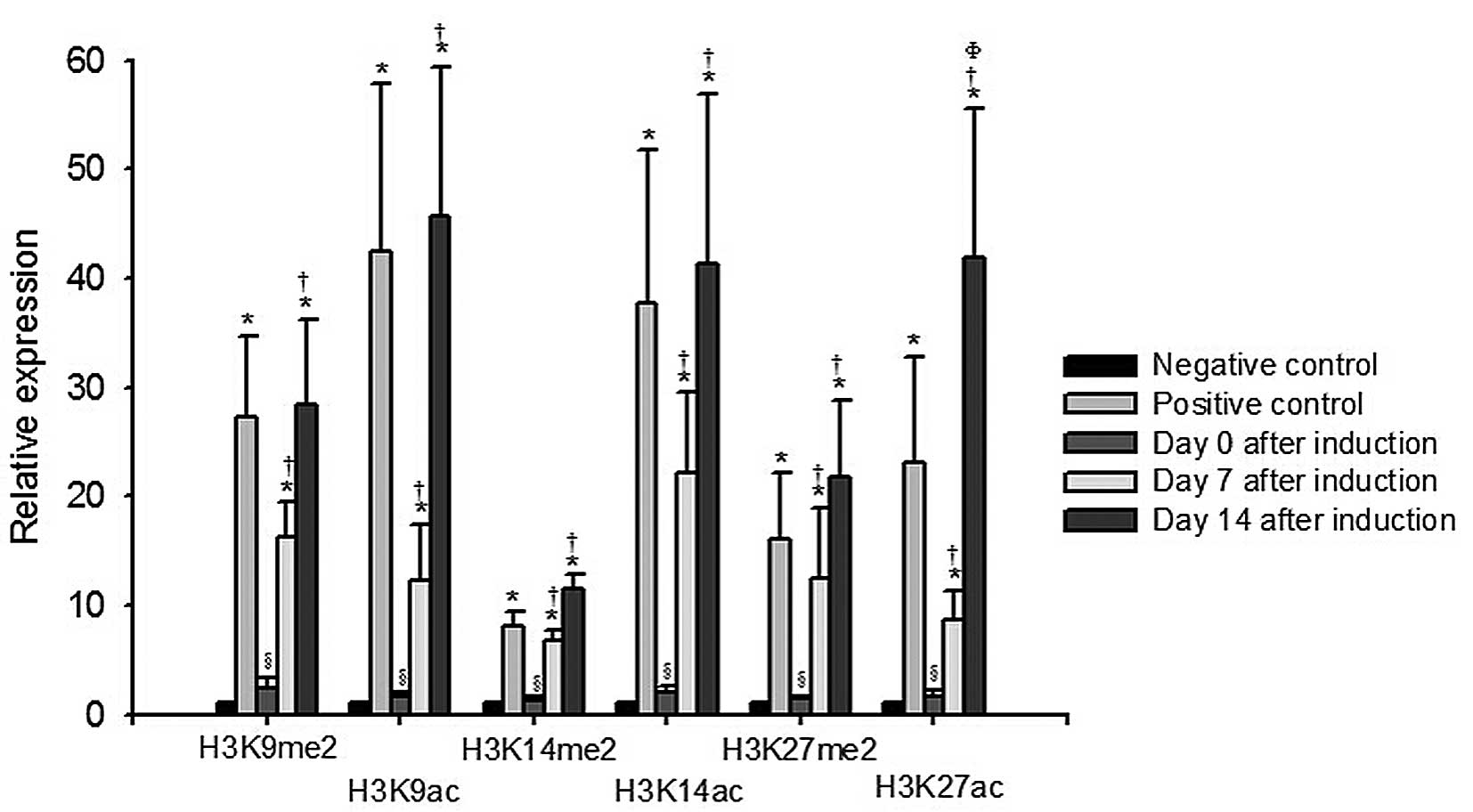
day 14 post- induction (P<0.05; Fig. 3B). Changes in histone modification,
indicated by the expression levels of H3K9me2, H3K9ac, H3K14me2,
H3K14ac, H3K27me2 and H3K27ac, are shown in Fig. 4. On days 7 and 14 following
induction, the levels of H3K9me2, H3K9ac, H3K14me2, H3K14ac,
H3K27me2 and H3K27ac increased significantly, compared with the
levels in the negative controls or those on day 0 (P<0.05). The
level of H3K27ac on day 14 post-induction was higher than that
observed on day 7 post-induction (P<0.05).
Discussion
The in vitro induction of the differentiation
of mesenchymal stem cells derived from bone marrow, adipose tissue,
skin, placenta, and umbilical cord into endodermal cells have been
reported (19-22). Epigenetic reprogramming is critical
in the differentiation of stem cells, and is essential for the
differentiation process, in which the cells overcome lineage
borders and attain the specialized functions of a new tissue
(23). In the present study, the
differentiation of Thy-1(+) Lin(−) bone marrow cells into
hepatocytes was induced in vitro. The bone marrow derived
epithelioid cells demonstrated morphological characteristics of
primary hepatocytes, and expressed the hepatocyte-specific marker
cytoskeletal protein, CK18, on day 14 post-induction, and the mRNA
expression of AFP and ALB were observed on days 7 and 14
post-induction, indicating a hepatic phenotype of the bone marrow
stem cell-derived epithelioid cells. In addition, histone
modifications, including significant increases in the levels of
H3K9me2, H3K9ac, H3K14me2, H3K14ac, H3K27me2 and H3K27ac were
observed on days 7 and 14 following induction, compared with their
levels on day 0 or in the negative control.
Embryonic stem cell investigations have progressed
by identifying three key transcription factors, Oct4, sex
determining region Y-box 2 (Sox2) and NANOG (24,25),
the interplay of which leads to the formation of regulatory
circuitry for the maintenance of the exclusive features of
embryonic stem cells (26). It is
now well recognized that Oct4, Sox2 and NANOG are essential for the
pluripotency of embryonic stem cells in vivo and in
vitro (27). Pluripotent stem
cells have been induced from somatic cells through the
overexpression of four transcription factors, including Oct4 and
Sox2 (28). Previous studies have
demonstrated the downregulation of Oct4 during the differentiation
of embryonic stem cells; with de novo DNA methylation of the
Oct4 regulatory region occurring during differentiation (29). Kim et al reported that the
transcription of Oct4 is elevated in pluripotent human embryonic
stem cells and reduces significantly during the differentiation of
embryonic stem cells into hepatocytes in vitro (12). In menstrual blood-derived stem
cells that can differentiate into functional hepatocyte-like cells
following induction, expression of the Oct 4 pluripotency marker
gene has also been detected (30).
The present study demonstrated that, during the
transdifferentiation of bone marrow stem cells into hepatocytes, a
similar decrease in the transcription of the Oct 4 pluripotency
marker gene was observed.
It has been suggested that histone tail
modifications, including acetylation, methylation,
phosphorylation/ubiquitination/sumoylation, ADP-ribosylation and
glycosylation can alter chromatin structure and subsequent gene
expression (31-33). In the present study, increases in
histone H3 modifications, including H3K9me2, H3K9ac,
H3K14me2, H3K14ac, H3K27me2 and H3K27ac were observed during the
transdifferentiation of bone marrow stem cells into hepatocytes
in vitro. To the best of our knowledge, histone modification
during the transdifferentiation of adult stem cells has not been
reported previously, and investigations on epigenetic modifications
during the transdifferentiation of adult stem cells have been
limited. It has been demonstrated that the chromatin remodeling
agent trichostatin A induces the transdifferentiation of human
mesenchymal stem cells into hepatocyte-like cells (34) and dimethylsulfoxide enhances the
transdifferentiation of human adipose tissue-derived stromal cells
into hepatic lineage cells (35).
DNA methyltransferase inhibitors have also been observed to induce
transdifferentiation (36-38). However, reports regarding
epigenetic modifications during the transdifferentiation of adult
stem cells remain minimal, and further investigations are
required.
There are previous reports describing epigenetic
modification during the differentiation of embryonic stem cells
(11-13). Well-acknowledged epigenetic
modifications associated with transcriptionally active chromatin
include DNA hypomethylation, the acetylation of histone H3 and H4
and the methylation of H3K4, H3K36 and H3K79 (13). Representative epigenetic
modifications associated with the repressive chromatin state
include the hypoacetylation of H3 and H4 and methylation of H3K9,
H4K20, H3K27 or H4K20 (11).
During the differentiation of embryonic stem cells into hepatocytes
in vitro, permissive histone modifications, including
H3K4me3 and H3K9ac within the promoter regions of the OCT4, SOX2,
and NANOG pluripotency marker genes and hepatocyte marker genes are
rich in embryonic stem cells prior to induction, and then decreased
rapidly in definitive endoderm cells and hepatocytes (12). By contrast, repressive histone
modifications, including H3K27me3, H3K9me2 and H3K9me3 in the
promoter regions of the pluripotency marker genes and hepatocyte
marker genes increase gradually during the differentiation of
embryonic stem cells into hepatocytes (12). These findings suggest that, during
the differentiation of embryonic stem cells, a combination of
specific histone modifications can modulate gene transcription.
However, further investigations are required to compare the histone
code (histone signature) during the differentiation of embryonic
stem cells in the trans-differentiation of adult stem cells into
hepatocytes.
In the present study, the hepatic characteristics of
bone marrow-derived hepatocyte-like cells at a morphological, RNA
and protein level were demonstrated. A limitation of the
investigation was the lack of functional assays available to
examine the glycogen uptake and urea synthesis of the bone
marrow-derived hepatocyte-like cells. Urea synthesis is one of the
unique functions of mature hepatocytes (39,40).
However, the in vivo transplantation of stem cell-derived
hepatocytes in immunodeficient animal models with liver injury is
essential for validating functional hepatic behavior (41). In our previous study, the same
induction method was used to induce the differentiation of bone
marrow stem cells into hepatocytes, and the hepatocytes generated
were transplanted ex vivo into animal models and patients
(42-44). In those investigations, the
function of the stem cell-derived hepatocytes had been supported
previously (4,5). The focus of the present study was to
investigate the levels of histone H3 modifications during the
differentiation of bone marrow stem cells into hepatocytes, and the
changes associated with these histone modification, with respect to
key lineage-specific genes during transdifferentiation, require
further examination. Future results may improve current
understanding of the intrinsic mechanism governing
transdifferentiation-inducing signals.
In conclusion, the present study demonstrated that,
during the transdifferentiation of Thy-1(+) Lin(−) bone marrow
cells into hepatocytes in vitro, the mRNA expression of the
key pluripotency factor, OCT4, was significantly reduced. This
change resembled that, which occurs during the differentiation of
embryonic stem cells into hepatocytes. The levels of histone H3
modifications, including H3K9me2, H3K9ac, H3K14me2, H3K14ac,
H3K27me2 and H3K27ac increased during the transdifferentiation of
the Thy-1(+) Lin(−) bone marrow cells into hepatocytes. Future
investigations are required to examine the changes in histone
modification, which are pertinent to key lineage-specific genes
during the transdifferentiation of adult stem cells. The results
may assist in understanding the similarity and discrepancy in the
epigenetic modifications during the transdifferentiation of adult
stem cells, and the differentiation of embryonic stem cells, into
hepatocytes.
Acknowledgments
This study was supported by the Sci-Tech Project of
Guangdong, China (grant. no. 2010B080701069).
References
|
1
|
Filip S, English D and Mokrý J: Issues in
stem cell plasticity. J Cell Mol Med. 8:572–577. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tang Y, He H, Cheng N, Song Y, Ding W,
Zhang Y, Zhang W, Zhang J, Peng H and Jiang H: PDGF, NT-3 and IGF-2
in combination induced transdifferentiation of muscle-derived stem
cells into Schwann cell-like cells. PLoS One. 9:e734022014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Snykers S, De Kock J, Rogiers V and
Vanhaecke T: In vitro differentiation of embryonic and adult stem
cells into hepatocytes: State of the art. Stem Cells. 27:577–605.
2009. View Article : Google Scholar :
|
|
4
|
Tang H, Liao CX, Zhou J, Jin HS, Tan YF,
Su J, Zhang CX and Zhang SH: Differentiation of transplanted mouse
c-Kit+lin-bone marrow cells into hepatocytes in vitro. Journal of
Southern Medical University. 26:567–569. 2006.In Chinese.
|
|
5
|
Liao X, AnCheng JY, Zhou QJ and Liao C:
Therapeutic effect of autologous bone marrow-derived liver stem
cells transplantation in hepatitis B virus-induced liver cirrhosis.
Hepatogastroenterology. 60:406–409. 2013.
|
|
6
|
Vaillant I and Paszkowski J: Role of
histone and DNA methylation in gene regulation. Curr Opin Plant
Biol. 10:528–533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Allis CD, Berger SL, Cote J, Dent S,
Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar
R, et al: New nomenclature for chromatin-modifying enzymes. Cell.
131:633–636. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miao F and Natarajan R: Mapping global
histone methylation patterns in the coding regions of human genes.
Mol Cell Biol. 25:4650–4661. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bannister AJ, Zegerman P, Partridge JF,
Miska EA, Thomas JO, Allshire RC and Kouzarides T: Selective
recognition of methylated lysine 9 on histone H3 by the HP1 chromo
domain. Nature. 410:120–124. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schones DE and Zhao K: Genome-wide
approaches to studying chromatin modifications. Nat Rev Genet.
9:179–191. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim H, Jang MJ, Kang MJ and Han YM:
Epigenetic signatures and temporal expression of lineage-specific
genes in hESCs during differentiation to hepatocytes in vitro. Hum
Mol Genet. 20:401–412. 2011. View Article : Google Scholar
|
|
13
|
Taverna SD, Li H, Ruthenburg AJ, Allis CD
and Patel DJ: How chromatin-binding modules interpret histone
modifications: Lessons from professional pocket pickers. Nat Struct
Mol Biol. 14:1025–1040. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Pan RL, Zhang XL, Shao JZ, Xiang
LX, Dong XJ and Zhang GR: Induction of hepatic differentiation of
mouse bone marrow stromal stem cells by the histone deacetylase
inhibitor VPA. J Cell Mol Med. 13:2582–2592. 2009. View Article : Google Scholar
|
|
15
|
Guittin P and Decelle T: Future
improvements and implementation of animal care practices within the
animal testing regulatory environment. ILAR J. 43(Suppl): S80–S84.
2002.PubMed/NCBI
|
|
16
|
Zhang CX, Liao SH, Zhang J, Sun J and
Zhou: Separation of bone marrow-derived liver stem cell subset
Thy-1.2 + Lin-cells using immunomagnetic beads. Shandong University
Transaction (Medical Sciences). 46:108–110. 2008.
|
|
17
|
Oh SH, Miyazaki M, Kouchi H, Inoue Y,
Sakaguchi M, Tsuji T, Shima N, Higashio K and Namba M: Hepatocyte
growth factor induces differentiation of adult rat bone marrow
cells into a hepatocyte lineage in vitro. Biochem Biophys Res
Commun. 279:500–504. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Elyaman W, Yardin C and Hugon J:
Involvement of glycogen synthase kinase-3beta and tau
phosphorylation in neuronal Golgi disassembly. J Neurochem.
81:870–880. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kang XQ, Zang WJ, Bao LJ, Li DL, Song TS,
Xu XL and Yu XJ: Fibroblast growth factor-4 and hepatocyte growth
factor induce differentiation of human umbilical cord blood-derived
mesen-chymal stem cells into hepatocytes. World J Gastroenterol.
11:7461–7465. 2005. View Article : Google Scholar
|
|
20
|
Lee KD, Kuo TK, Whang-Peng J, Chung YF,
Lin CT, Chou SH, Chen JR, Chen YP and Lee OK: In vitro hepatic
differentiation of human mesenchymal stem cells. Hepatology.
40:1275–1284. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kang XQ, Zang WJ, Song TS, Xu XL, Yu XJ,
Li DL, Meng KW, Wu SL and Zhao ZY: Rat bone marrow mesenchymal stem
cells differentiate into hepatocytes in vitro. World J
Gastroenterol. 11:3479–84. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ong SY, Dai H and Leong KW: Hepatic
differentiation potential of commercially available human
mesenchymal stem cells. Tissue Eng. 12:3477–3485. 2006. View Article : Google Scholar
|
|
23
|
Leu YW, Huang TH and Hsiao SH: Epigenetic
reprogramming of mesenchymal stem cells. Adv Exp Med Biol.
754:195–211. 2013. View Article : Google Scholar
|
|
24
|
Yamanaka Y and Ralston A: Early embryonic
cell fate decisions in the mouse. Adv Exp Med Biol. 695:1–13. 2010.
View Article : Google Scholar
|
|
25
|
Chui HC and Damasio AR: Progressive
dialysis encephalopathy (ʻdialysis dementia'). J Neurol.
222:145–157. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boyer LA, Lee TI, Cole MF, Johnstone SE,
Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG,
et al: Core transcriptional regulatory circuitry in human embryonic
stem cells. Cell. 122:947–956. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nichols J, Zevnik B, Anastassiadis K, Niwa
H, Klewe-Nebenius D, Chambers I, Schöler H and Smith A: Formation
of pluripotent stem cells in the mammalian embryo depends on the
POU transcription factor Oct4. Cell. 95:379–391. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Athanasiadou R, de Sousa D, Myant K,
Merusi C, Stancheva I and Bird A: Targeting of de novo DNA
methylation throughout the Oct-4 gene regulatory region in
differentiating embryonic stem cells. PLoS One. 5:e99372010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Khanjani S, Khanmohammadi M, Zarnani AH,
Talebi S, Edalatkhah H, Eghtesad S, Nikokar I and Kazemnejad S:
Efficient generation of functional hepatocyte-like cells from
menstrual blood-derived stem cells. J Tissue Eng Regen Med. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Turner BM: Reading signals on the
nucleosome with a new nomenclature for modified histones. Nat
Struct Mol Biol. 12:110–112. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Szyf M: DNA methylation and demethylation
as targets for anticancer therapy. Biochemistry (Mosc). 70:533–549.
2005. View Article : Google Scholar
|
|
33
|
Moggs JG, Goodman JI, Trosko JE and
Roberts RA: Epigenetics and cancer: Implications for drug discovery
and safety assessment. Toxicol Appl Pharmacol. 196:422–430. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Snykers S, Vanhaecke T, De Becker A,
Papeleu P, Vinken M, Van Riet I and Rogiers V: Chromatin remodeling
agent tricho-statin A: A key-factor in the hepatic differentiation
of human mesenchymal stem cells derived of adult bone marrow. BMC
Dev Biol. 7:242007. View Article : Google Scholar
|
|
35
|
Seo MJ, Suh SY, Bae YC and Jung JS:
Differentiation of human adipose stromal cells into hepatic lineage
in vitro and in vivo. Biochem Biophys Res Commun. 328:258–264.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Milhem M, Mahmud N, Lavelle D, Araki H,
DeSimone J, Saunthararajah Y and Hoffman R: Modification of
hematopoietic stem cell fate by 5aza 2′deoxycytidine and
trichostatin A. Blood. 103:4102–4110. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sgodda M, Aurich H, Kleist S, Aurich I,
König S, Dollinger MM, Fleig WE and Christ B: Hepatocyte
differentiation of mesenchymal stem cells from rat peritoneal
adipose tissue in vitro and in vivo. Exp Cell Res. 313:2875–2886.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yoshida Y, Shimomura T, Sakabe T, Ishii K,
Gonda K, Matsuoka S, Watanabe Y, Takubo K, Tsuchiya H, Hoshikawa Y,
et al: A role of Wnt/beta-catenin signals in hepatic fate
specification of human umbilical cord blood-derived mesenchymal
stem cells. Am J Physiol Gastrointest Liver Physiol.
293:G1089–G1098. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gómez-Lechón MJ, Donato MT, Castell JV and
Jover R: Human hepatocytes in primary culture: The choice to
investigate drug metabolism in man. Curr Drug Metab. 5:443–462.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kmieć Z: Cooperation of liver cells in
health and disease. Adv Anat Embryol Cell Biol. 161:III–XIII.
1–151. 2001.
|
|
41
|
Imamura T, Cui L, Teng R, Johkura K,
Okouchi Y, Asanuma K, Ogiwara N and Sasaki K: Embryonic stem
cell-derived embryoid bodies in three-dimensional culture system
form hepatocyte-like cells in vitro and in vivo. Tissue Eng.
10:1716–1724. 2004. View Article : Google Scholar
|
|
42
|
Liao CX, Tang H, Zhou J, Su J, Zhang SH
and Zhang CX: Sieving of hepatic stem cells derived from bone
marrow and their potential of differentiation. Chinese Journal of
Hepatobiliary Surgery. 9:614–616. 2007.In Chinese.
|
|
43
|
Yuan J, Liao CX, Qin AC, Liao XX, Huang
YP, Gong ZY and Liao H: Protocols for cloning human bone
marrow-derived hepatic stem cells in vitro. Nan Fang Yi Ke Da Xue
Xue Bao. 30:318–320. 2010.In Chinese. PubMed/NCBI
|
|
44
|
Qin AC, Liao CX, Wang Y, Yuan J, Huang YP,
Liao XX, Lai YQ and Gong ZY: Intrahepatic transplantation of in
vitro induced autologous bone marrow-derived liver stem cells in
patients with posthepatitic cirrhosis. Nan Fang Yi Ke Da Xue Xue
Bao. 30:529–531. 2010.In Chinese. PubMed/NCBI
|