Introduction
Type 2 diabetes (T2D) is one of the most common
chronic diseases worldwide, and it is estimated to affect >400
million individuals by 2030 (1).
The prevalence of T2D has markedly increased in the states of the
Cooperation Council for the Arab States of the Gulf that include
Bahrain, and these countries are now among the top 10 with the
highest rate of T2D worldwide (2).
A combination of genetic and
environmental/epigenetic factors may impair insulin sensitivity in
target tissues, as well as insulin secretion from pancreatic β
cells. When insulin secretion from the β cells is no longer able to
prevent insulin resistance, glucose levels rise (hyperglycemia),
leading to pre-diabetes at an early stage and T2D at a later stage
(3,4).
Substantial evidence has demonstrated that there is
a long and latent asymptomatic period during which T2D may be
detected, and thus T2D may remain undiagnosed for a number of years
(5,6). At present, there are no biomarkers
for predicting T2D, and the currently available blood glucose tests
serve as diagnostic rather than predictive tools. Since lifestyle
interventions are highly effective at delaying the onset and
progression of the T2D (7), the
development of diagnostic or prognostic biomarkers for the early
detection of T2D is clinically significant.
MicroRNAs (miRNAs) are endogenous small non-coding
RNAs that regulate gene expression by targeting mRNA for cleavage
or translational repression. miRNAs are involved in highly
regulated processes such as proliferation, differentiation,
apoptosis and metabolic processes (8). Specifically, miRNAs are important for
β cell function and the regulation of glucose stimulated insulin
secretion (9–11), and for controlling insulin
signaling in target tissues (12).
Consequently, aberrant expression of several miRNAs has been
implicated in the pathogenicity of diabetes (13).
miRNAs have been detected in the peripheral blood in
a notably stable form that is protected from endogenous RNase
activity (14). The biological
function of circulating miRNAs and the exact mechanism underlying
the entry of miRNAs into the blood remains elusive. It has been
reported that miRNAs may be packaged into microvesicles or
associated with apoptotic bodies and actively secreted into the
blood circulation, and subsequently target recipient cells in order
to exert specific molecular functions, thus mediating cell-to-cell
communication (15,16).
To date, changes in single miRNAs and in miRNA
signatures have been reported in the serum and plasma of patients
with various diseases, such as cancer (17,18)
and cardiovascular diseases (19,20).
Furthermore, a previous population-based cohort study identified a
set of plasma miRNAs that were differentially expressed in patients
with T2D and healthy controls (21), and among them, miR-15a was
demonstrated to be the most strongly associated with T2D.
In addition to the changes in miRNA expression
levels in the serum and plasma, recent blood-based miRNA studies
have reported altered miRNA expression in the peripheral whole
blood of patients with various types of cancers (22,23),
acute myocardial infarction (24),
and other human diseases (25),
suggesting a potential use of peripheral blood miRNAs as disease
biomarkers.
The present study examined the expression levels of
T2D-associated miR-15a in peripheral whole blood samples from
patients with T2D, pre-diabetes individuals with impaired fasting
glucose (IFG) and impaired glucose tolerance (IGT), and healthy
control subjects; and investigated the biomarker potential of
peripheral blood miR-15a for T2D and pre-diabetes prediction.
Subjects and methods
Study subjects and clinical data
A total of 70 subjects were included in the present
study: 24 patients with T2D (10 men and 14 women), 22 pre-diabetes
individuals with IFG and IGT (10 men and 12 women), and 24 healthy
control subjects (13 men and 11 women). Both the patients and
controls were selected from the King Abdullah University Medical
Centre (College of Medicine and Medical Sciences, Arabian Gulf
University, Kingdom of Bahrain). Individuals with malignant tumors,
cardiovascular disease, nephropathy or other chronic diseases which
could effect the miRNA expression levels were excluded from the
present study. The study was approved by the Medical Research and
Ethics Committee in the College of Medicine and Medical Sciences of
the Arabian Gulf University, Kingdom of Bahrain (Manama, Kingdom of
Bahrain). Written informed consent was obtained from all
subjects.
The presence of T2D in the patients was established
according to World Health Organization criteria (26): Fasting glucose (FG) levels ≥7.0
mmol/l, glucose levels ≥11.1 mmol/l (200 mg/dl) as determined by a
2 h oral glucose tolerance test (OGTT) when glycated hemoglobin
(HbA1c) levels >6.5%, or when the subjects had a clinical
diagnosis of the disease. Individuals with FG levels of 6.1–6.9
mmol/l (110–125 mg/dl) or 2 h OGTT glucose levels of 7.8–11.0
mmol/l (140–199 mg/dl) were designated as exhibiting pre-diabetes.
The healthy controls were defined as subjects with FG of 4.8–5.2
mmol/l (110 mg/dl), and 2 h OGTT glucose levels of <7.8 mmol/l
(140 mg/dl).
miRNA extraction
A total of 5 ml of peripheral blood were collected
from the subjects and placed into tubes containing
ethylenediaminetetraacetic acid (EDTA; BD Biosciences, Franklin
Lakes, NJ, USA). Aliquots (0.5 ml each) of blood were mixed with
1.3 ml RNAlater, an RNA stabilization reagent (Ambion Life
Technologies, Carlsbad, CA, USA) and stored at -80°C. Prior to RNA
extraction, the frozen samples were thawed at room temperature, and
total RNA including small RNA was isolated using the miRNeasy kit
(Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's
instructions. Concentration and purity of the RNA was quantified by
measuring the absorbance at 260 nm (A260) and 280 nm (A280) using a
NanoDrop ND-100 spectrophotometer (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was performed using the TaqMan®
MicroRNA Reverse Transcription kit and Biosystems real-time PCR
detection system (Applied Biosystems Life Technologies, Foster
City, CA, USA). cDNA was synthesized from total RNA (20 ng) using
specific stem-loop RT primers and reagents from the
TaqMan® MicroRNA Reverse Transcription kit (3 µl
specific stem-loop RT primers, 100 mM dNTPs, 10X reverse
transcription buffer, 20 U/µl RNase inhibitor, 50 U/ml
MultiScribe™ Reverse Transcriptase and 4.16 µl nuclease-free
water to a final volume of 15 µl).
For qPCR, PCR products were amplified from cDNA
using the TaqMan® MicroRNA Assay/TaqMan®
Universal PCR Master mix (Applied Biosystems Life Technologies).
Briefly, a 1.33 µl aliquot of cDNA was combined with 1
µl TaqMan miRNA (consisting of the specific PCR primers, and
a 20X TaqMan® MGB probe, TaqMan® Universal
PCR Master mix II (2X), no UNG (10 µl) and nuclease-free
water (7.67 µl) to a final volume of 20 µl. The PCR
primers had the following sequences: Mature hsa-miR-15a,
UAGCAGCACAUAAUGGUUUGUG; reference gene RNU6B,
CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT. qPCR was performed
using a 7900HT Fast Real Time PCR system (Applied Biosystems Life
Technologies) with the following cycling conditions: 95°C for 10
min, followed by 95°C for 15 sec and 60°C for 60 sec for a total of
40 cycles.
The PCR of each sample was run in duplicate to
minimize experimental errors. The results were analyzed using
Sequence Detection Software version 1.7 (Applied Biosystems Life
Technologies). The expression levels of miR-15a were normalized to
those of RNU6B, a small nuclear RNA, the expression levels of which
were demon strated to be relatively abundant and constant across a
wide range of human tissues and cell line types. It is regarded as
one of the control genes with the least variability for miRNAs
assays (27) and has been widely
used in different fields including in diabetes research (28,29).
The fold change of miR-15a expression was determined
using the 2−ΔΔCt method. The change in threshold cycle
(ΔCt) was calcu lated by subtracting the Ct values of the reference
RNU6b from the Ct values of the target miR-15a. ΔΔCt was then
calculated by subtracting the average ΔCt values of the controls
from the average ΔCt values of the cases (diabetes or
pre-diabetes).
Statistical analysis
Comparisons between cases and controls were
conducted by χ2 or Student's unpaired t-test, and
presented as the mean ± standard deviation (SD). Multivariate
logistic regression analysis was used to assess the association
between miR-15a expression levels and T2D and pre-diabetes. The
area under the curve (AUC) using receiver operating characteristic
(ROC) analysis was calculated for miRNA-15a to assess the
predictive values. A two-tailed P<0.05 was considered to
indicated a statistically significant difference. The data were
analyzed using SPSS 19 (IBM, Armonk, NY, USA).
Results
Characteristics of the study
subjects
Patient characteristics are presented in Table I. No significant differences in
gender distribution and age were found among T2D patients,
pre-diabetes individuals (IFG/IGT), and healthy control subjects
(P>0.05). The body mass index (BMI) was significant higher in
patients with T2D, compared with control subjects (P>0.05), but
was not significantly different in individuals with pre-diabetes
compared with controls (P>0.05). The percentage of subjects with
hypertension differed significantly between T2D and pre-diabetes
compared with the controls (P<0.05). In the groups of patients
with T2D and pre-diabetes, significant differences were found for
levels of fasting glucose, OGTT glucose and HbA1c levels compared
with those of the control subjects (P<0.005). In addition,
patients with T2D had statistically different results for levels of
triglyceride (TG), high density lipoprotein cholesterol (HDL-C),
low density lipoprotein cholesterol (LDL-C) and total cholesterol
(TC) compared with controls (P<0.005). Conversely, pre-diabetes
individuals had statistically different results for levels of TG
(P<0.05), but not for HDL-C, LDL-C or TC (P>0.05), compared
with the controls.
 | Table ICharacteristics of the subjects. |
Table I
Characteristics of the subjects.
| Characteristic | T2D | Pre-diabetes | Controls |
|---|
| Number of
subjects | 24 | 22 | 24 |
| Age (years) | 52±6.0 | 49±7.7 | 49±9.1 |
| Gender (M/F) | 10/14 | 10/12 | 13/11 |
| BMI
(kg/m2) | 25.3±1.8a | 24.5±2.2 | 24.2±1.0 |
| Hypertension [n
(%)] | 14 (58.3)b | 9 (40.9)a | 1 (4.1) |
| FG (mmol/l) | 9.4±7.6b | 6.4±5.8b | 4.4±4 |
| 2 h OGTT
(mmol/l) | 14.6±2b | 8.8±0.8 | 6.1±0.6 |
| HbA1c (%) | 7.5±0.8b | 6.5±0.7b | 4.8±0.6 |
| TG (mmol/l) | 2.38
(1.0–3.5)b | 2.27
(1.0–3.3)b | 1.49 (0.9–1.6) |
| LDL-C (mmol/l) | 3.4±0.5b | 3.17±0.7 | 2.99±0.2 |
| HDL-C (mmol/l) | 1.0±0.1b | 1.29±0.2 | 1.31±0.2 |
| TC (mmol/l) | 5.26
(4.0–5.8)b | 4.97 (3.3–5.5) | 4.83 (3.8–5.0) |
Expression of miR-15a
Using RT-qPCR, the expression levels of miR-15a were
determined relative to the endogenous control RNU6B in the
peripheral whole blood of patients with T2D, pre-diabetes
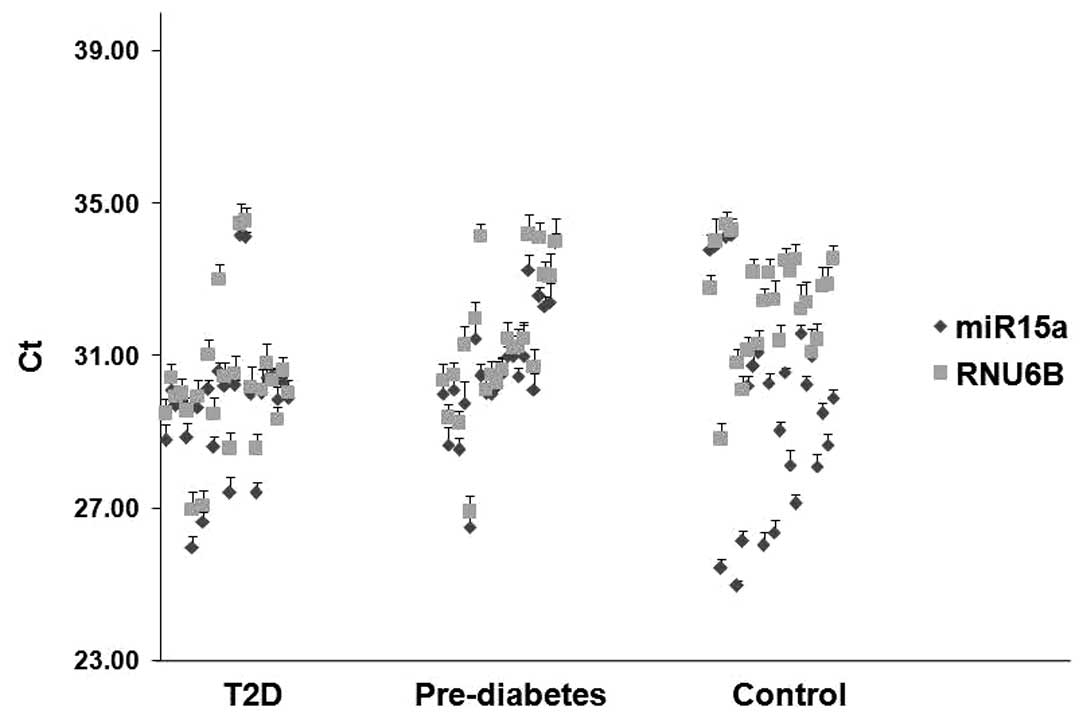
individuals, and healthy control subjects. miR-15a and RNU6B
exhibited reliable Ct values in all samples from T2D, pre-diabetes
and controls. No replicates with a Ct value >35 were detected
(Fig. 1).
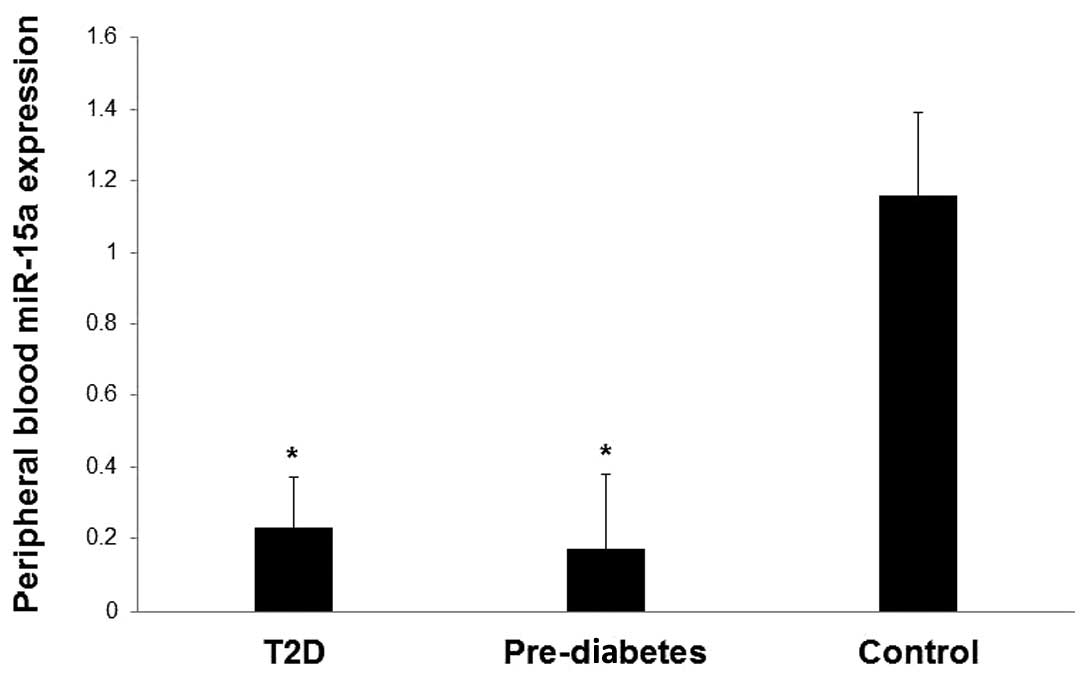
The expression levels of miR-15a in the blood were
significantly lower in patients with T2D and pre-diabetic
individuals compared with healthy controls (P<0.05; Fig. 2). The fold change of miR-15a
(calculated as the mean ± SD) was 0.23±0.14 for T2D, 0.17±0.21 for
pre-diabetes and 1.16±0.04 for healthy controls.
Association between miR-15a and T2D and
pre-diabetes
Multivariate logistic regression analysis (Table II) demonstrated strong association
between lower miR-15a expression levels and T2D [odds ratio (OR)
0.51; 95% confidence interval (CI) 0.16–0.73; P<0.05].
Similarly, lower miR-15a expression levels were strongly associated
with pre-diabetes (OR 0.56; 95% CI 0.23–0.79; P<0.05). This
association remained statistically significant for T2D and
pre-diabetes following adjustment for age and gender, and further
for BMI and hypertension, as well as levels of TG, LDL-C, HDL-C and
TC (Table II).
 | Table IIAssociation between lower miRNA-15a
expression levels and T2D and pre-diabetes. |
Table II
Association between lower miRNA-15a
expression levels and T2D and pre-diabetes.
| miR-15a
expression | T2D
| Pre-diabetes
|
|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Crude | 0.51
(0.16–0.73) | 0.02 | 0.56
(0.23–0.79) | 0.01 |
| Adjusted, model
1a | 0.55
(0.31–0.82) | 0.04 | 0.57
(0.28–0.83) | 0.05 |
| Adjusted, model
2a | 0.53
(0.19–0.79) | 0.03 | 0.58
(0.37–0.87) | 0.03 |
| Adjusted, model
3a | 0.57
(0.37–0.89) | 0.02 | 0.57
(0.35–0.80) | 0.05 |
Predictive potential of miR-15a
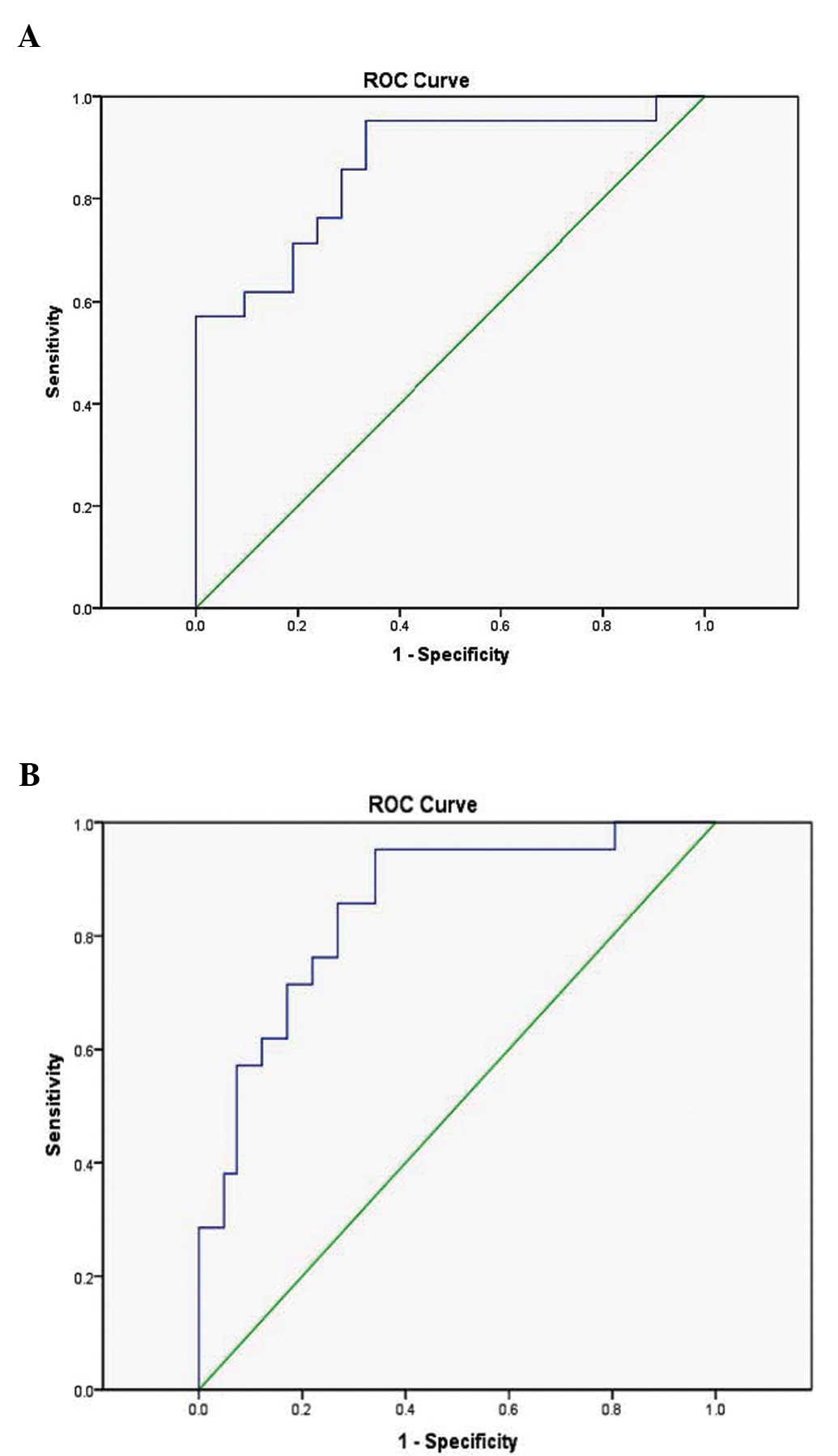
To evaluate the predictive potential of blood
miR-15a expression levels, ROC analysis was used. The AUC indicated
that blood miR-15a expression levels were able to distinguish
between patients with T2D and healthy control individuals (AUC
0.864; 95% CI 0.751–0.977; P<0.005) (Fig. 3A). Furthermore, blood miR-15a
expression levels distinguished individuals with pre-diabetes from
healthy controls (AUC 0.852; 95% CI 0.752–0.953; P<0.005)
(Fig. 3B).
Discussion
Previous studies have reported that serum and plasma
miRNAs may serve as novel biomarkers for various diseases,
including T2D (14,17–21).
Furthermore, miRNAs in peripheral whole blood have also shown
promise as a novel class of biomarkers for the detection of several
types of cancer, such as brain (22) and breast (23) cancer, as well as acute myocardial
infarction (24), and other human
diseases (25).
A study by Karolina et al (30) reported an miRNA marker in the
peripheral whole blood of patients with T2D patients and IFG. Their
study demonstrated that these blood miRNAs (miR-144, miR-146a,
miR-150, miR-182, miR-192, miR-29a, miR-30d and miR-320a) may serve
as unique biomarkers that are reflective and predictive of T2D
(30).
The present study examined the expression levels of
T2D-associated miR-15a (21) in
the peripheral whole blood of patients with T2D, pre-diabetes
individuals (IFG/IGT), and healthy control subjects, and
investigated the potential use of blood miR-15a as a biomarker for
predicting T2D and pre-diabetes.
The results demonstrated that blood miR-15a
expression levels were significantly lower in patients with T2D and
IFG/IGT individuals, compared with healthy controls. Furthermore, a
strong association was observed between lower miR-15a expression
levels and T2D, as well as pre-diabetes. Notably, this association
was not altered following adjustment for known risk factors and
certain other biochemical indicators.
In a previous population-based cohort study,
Zampetaki et al (21)
demonstrated a similar pattern of expression, with circulating
plasma miR-15a expression levels being lower in patients with T2D.
The results from the present study extended the association of
lower miR-15a expression levels to pre-diabetes individuals.
A previous study demonstrated that miR-15a is
implicated in controlling cell cycle and apoptosis in cancer cells
(31). The expression levels of
miR-15a were shown to inversely correlate with the expression
levels of cyclin D1, and negatively regulate those of B-cell
lymphoma 2, a key anti-apoptotic protein (32). Recently, miR-15a has been
demonstrated to positively regulate and promote insulin
biosynthesis by targeting and inhibiting endogenous uncoupling
protein-2 gene expression (33).
The overexpression of miR-15a in transfected MIN6 (mouse
insulinoma) cells increases insulin secretion, and its suppression
decreases insulin levels (33).
Therefore, miR-15a was suggested as an important mediator of β cell
function and insulin synthesis, and a possible therapeutic agent
for diabetes (33).
The progression from pre-diabetes (IFG and/or IGT)
to early T2D is marked by a decrease in β cell function, and thus a
decline in insulin secretion, followed by a failure of β cells to
compensate for insulin resistance with hyperinsulinemia that marks
the beginning of T2D (3,4). Individuals with IFG have a 20–30%
chance of developing T2D over the following 5–10 years, and the
risk is even higher if individuals with combined IFG and IGT
(34,35).
The observation of the present study that blood
miR-15a expression levels were decreased in patients with T2D and
IFG/IGT individuals may suggest that miR-15a expression correlates
with disease development.
In the present study, the ROC curves revealed that
blood miR-15a levels have a promising ability to distinguish
patients with T2D and IFG/IGT individuals from healthy controls,
which suggested a potential use of peripheral blood miR-15 as a
biomarker for T2D and pre-diabetes. To the best of our knowledge,
the present study demonstrated for the first time that peripheral
blood miR-15a may serve as a potential predictive biomarker in T2D
and pre-diabetes.
The high stability of miRNAs in human peripheral
blood renders them ideal biomarkers for disease detection (14). Given that lifestyle interventions
may be highly effective in delaying the onset and progression of
T2D (7), the evaluation of
peripheral blood biomarkers that are capable of distinguishing
between patients with T2D and healthy individuals may lead to a
novel preliminary screening method to prevent T2D, through the
early identification of individuals who are at a higher risk of
developing the disease, particularly those individuals with
pre-diabetes.
One of the limitations of the present study was the
small sample size, and therefore these results require further
investigation in a larger population sample. Additionally, the
present study focused exclusively on the association between blood
miR-15a expression levels and T2D/pre-diabetes, and did not
investigate other miRNAs. Nevertheless, further studies in our
laboratory are underway to examine the significance of other whole
blood miRNAs as predictive tools for T2D and pre-diabetes. Finally,
although the results of the present study demonstrated that blood
miR-15a is differentially expressed between patients with T2D,
pre-diabetes individuals, and healthy controls, the clinical
application of miR-15a in predicting T2D and pre-diabetes requires
further investigation and optimization.
In conclusion, the results of the present study
demonstrated that the expression levels of miR-15a were
significantly lower in the peripheral whole blood of patients with
T2D and pre-diabetes individuals, compared with healthy controls,
and that miR-15a in peripheral whole blood may serve as a potential
biomarker for the prediction of T2D and pre-diabetes.
Acknowledgments
The authors are grateful to all the participants of
the present study, as well as the staff of the Clinical Laboratory
of the King Abdullah Medical Centre in the Kingdom of Bahrain. The
present study was supported by a research grant from the College of
Medicine and Medical Sciences of the Arabian Gulf University,
Kingdom of Bahrain (grant no. 81).
References
|
1
|
Wild S, Roglic G, Green A, Sicree R and
King H: Global prevalence of diabetes: Estimates for the year 2000
and projections for 2030. Diabetes Care. 27:1047–1053. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alhyas L, McKay A and Majeed A: Prevalence
of type 2 diabetes in the states of the Co-operation council for
the Arab states of the Gulf: A systematic review. PLoS One.
7:e409482012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abdul-Ghani MA and DeFronzo RA:
Pathophysiology of prediabetes. Curr Diab Rep. 9:193–199. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cavaghan MK, Ehrmann DA and Polonsky KS:
Interactions between insulin resistance and insulin secretion in
the development of glucose intolerance. J Clin Invest. 106:329–333.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harris MI, Klein R, Welborn TA and Knuiman
MW: Onset of NIDDM occurs at least 4–7 yr before clinical
diagnosis. Diabetes Care. 15:815–825. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thompson TJ, Engelgau MM, Hegazy M, Ali
MA, Sous ES, Badran A and Herman WH: The Onset of NIDDM and its
relationship to clinical diagnosis in Egyptian adults. Diabet Med.
13:337–340. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tuomilehto J, Lindström J, Eriksson JG,
Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S,
Laakso M, Louheranta A, Rastas M, et al: Prevention of type 2
diabetes mellitus by changes in lifestyle among subjects with
impaired glucose tolerance. N Engl J Med. 344:1343–1350. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lynn FC, Skewes-Cox P, Kosaka Y, McManus
MT, Harfe BD and German MS: MicroRNA expression is required for
pancreatic islet cell genesis in the mouse. Diabetes. 56:2938–2945.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewasky N, Rorsman P
and Soffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He A, Zhu L, Gupta N, Chang Y and Fang F:
Overexpression of micro ribonucleic acid 29, highly up-regulated in
diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes.
Mol Endocrinol. 21:2785–2794. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Plaisance V, Abderrahmani A, Perret-Menoud
V, Jacquemin P, Lemaigre F and Regazzi R: MicroRNA-9 controls the
expression of Granuphilin/Slp4 and the secretory response of
insulin-producing cells. J Biol Chem. 281:26932–26942. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang X, Tang G and Ozcan S: Role of
microRNAs in diabetes. Biochimic Biophysic Acta. 1779:697–701.
2008. View Article : Google Scholar
|
|
14
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: Secreted microRNAs: A new form of intercellular communication.
Trends Cell Biol. 22:125–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kosaka N, Iguchi H, Yoshioka Y, Takeshita
F, Matsuki Y and Ochiya T: Secretory mechanisms and intercellular
transfer of microRNAs in living cells. J Biol Chem.
285:17442–17452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang X, Zeng Z, Hou Y, Yuan T, Gao C, Jia
W, Yi X and Liu M: MicroRNA-92a as a potential biomarker in
diagnosis of colorectal cancer: A systematic review and
meta-analysis. PLoS One. 9:e887452014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He
J, Qin YW and Jing Q: Circulating microRNA: A novel potential
biomarker for early diagnosis of acute myocardial infarction in
humans. Eur Heart J. 31:659–666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fichtlscherer S, de Rosa S, Fox H,
Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T,
Müller-Ardogan M, et al: Circulating microRNAs in patients with
coronary artery disease. Circ Res. 107:677–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zampetaki A, Kiechl S, Drozdov I, Willeit
P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E,
et al: Plasma microRNA profiling reveals loss of endothelial
miR-126 and other microRNAs in type 2 diabetes. Circ Res.
107:810–817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roth P, Wischhusen J, Happold C, Chandran
PA, Hofer S, Eisele G, Weller M and Keller A: A specific miRNA
signature in the peripheral blood of glioblastoma patients. J
Neurochem. 118:449–457. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7:e297702012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meder B, Keller A, Vogel B, Haas J,
Sedaghat-Hamedani F, Kayvanpour E, Just S, Borries A, Rudloff J,
Leidinger P, et al: MicroRNA signatures in total peripheral blood
as novel biomarkers for acute myocardial infarction. Basic Res
Cardiol. 106:13–23. 2011. View Article : Google Scholar
|
|
25
|
Keller A, Leidinger P, Bauer A, Elsharawy
A, Haas J, Backes C, Wendschlag A, Giese N, Tjaden C, Ott K, et al:
Toward the blood-borne miRNome of human diseases. Nat Methods.
8:841–843. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alberti KG and Zimmet PZ: Definition,
diagnosis and classification of diabetes mellitus and its
complications. Part 1: Diagnosis and classification of diabetes
mellitus provisional report of a WHO consultation. Diabet Med.
15:539–553. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wong L, Lee K, Russell I and Chen C:
Endogenous controls for realtime quantitation of miRNA using
TaqMan® MicroRNA assays. Macmillan Publishers Ltd; New
York: 2007
|
|
28
|
Roggli E, Britan A, Gattesco S, Lin-Marq
N, Abderrahmani A, Meda P and Regazzi1 R: Involvement of microRNAs
in the cytotoxic effects exerted by proinflammatory cytokines on
pancreatic beta-cells. Diabetes. 59:978–986. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Poy MN, Hausser J, Trajkovski M, Braun M,
Collins S, Rorsman P, Zavolan M and Stoffel M: miR-375 maintains
normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci
USA. 106:5813–5818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karolina DS, Armugam A, Tavintharan S,
Wong MT, Lim SC, Sum CF and Jeyaseelan K: MicroRNA 144 impairs
insulin signaling by inhibiting the expression of insulin receptor
substrate 1 in type 2 diabetes mellitus. PLoS One. 6:e228392011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bandi N, Zbinden S, Gugger M, Arnold M,
Kocher V, Hasan L, Kappeler A, Brunnet T and Vassella E: miR-15a
and miR-16 are implicated in cell cycle regulation in a
Rb-dependent manner and are frequently deleted or down-regulated in
non-small cell lung cancer. Cancer Res. 69:5553–5559. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun LL, Jiang BG, Li WT, Zou JJ, Shi YQ
and Liu ZM: MicroRNA-15a positively regulates insulin synthesis by
inhibiting uncoupling protein-2 expression. Diabetes Res Clin
Pract. 91:94–100. 2011. View Article : Google Scholar
|
|
34
|
Meigs JB, Muller DC, Nathan DM, Blake DR
and Andres R: Baltimore Longitudinal Study of Aging: The natural
history of progression from normal glucose tolerance to type 2
diabetes in the Baltimore longitudinal study of aging. Diabetes.
52:1475–1484. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bock G, Dalla Man C, Campioni M,
Chittilapilly E, Basu R, Toffolo G, Cobelli C and Rizza R:
Pathogenesis of pre-diabetes: Mechanisms of fasting and
postprandial hyperglycemia in people with impaired fasting glucose
and/or impaired glucose tolerance. Diabetes. 55:3536–3549. 2006.
View Article : Google Scholar : PubMed/NCBI
|