Introduction
Aplastic anemia (AA), a rare bone marrow disease,
which leads to pancytopenia, anemia, leukopenia and
thrombocytopenia mainly occurs in teenagers worldwide (1). Although the pathogenesis of AA has
been associated with chemicals, drugs (2), radiation (3), infection and immune diseases
(4), the precise cause remains
unknown in half of the cases of AA (5). Without effective treatment, AA is
associated with a high risk of mortality (6), therefore, studies regarding the
underlying pathogenesis are considered to be necessary and
relevant.
Bone marrow mesenchymal stem cells (BMSCs) are
multi-potent stromal cells, which differentiate into numerous types
of cell, such as osteoblasts, chondrocytes and adipocytes (7). The capability to support
hematopoiesis and immunomodulatory characteristics render BMSCs
vital in the bone marrow hematopoietic microenvironment (8). Since the abnormal alteration of BMSCs
was observed in patients with AA (9), various studies have investigated the
association between BMSCs and the pathogenesis of AA. Zhao et
al (10) showed that AA BMSCs
were prone to differentiate into adipocytes rather than
osteoblasts. However, treatment with arsenic trioxide partially
reversed the differentiation imbalance. Wang et al (11) treated BMSCs (obtained from patients
with AA) with rapamycin at varying concentrations and identified
that rapamycin was vital in the suppression of BMSC proliferation,
cell cycle progression and adipogenesis (8). However, the underlying mechanisms of
the influence of BMSCs on AA treatment by activating growth factor
remains unclear and require further investigation.
Recently, the investigation of AA-associated BMSC
differentiation at the gene level has become increasingly
prevalent. Jiang et al (12) demonstrated that basic fibroblastic
growth factor (FGF) was expressed at a low level in the BMSCs of
infants presenting with AA and subsequently inferred that low FGF
expression may be involved in the pathogenesis of AA. FGFs, are a
family of pluripotent growth factors that affect mitosis, cell
regulation and morphology, as well as the endocrine system. Thus
far, 22 members of the FGF family have been identified and verified
to be structurally associated with molecular signaling (13). Furthermore, FGF1, encoded by FGF1,
exerts potent activity on cell survival, embryonic development, as
well as tissue repair (14).
Stegmann (15) identified that
FGF-1 promoted neoangiogenesis in the hypoxic heart muscle of
humans and demonstrated the angiogenic effect of FGF-1. Cao et
al (16) reported that FGF1
and FGF2 exhibited more potent efficacy on angiogenesis compared
with vascular endothelial or platelet-derived growth factors, and
induced the formation of stable vascular networks. On the basis of
previous research, the aim of the present study was to investigate
the regulatory mechanism of FGF1 in BMSCs to provide a novel
insight into the management of AA.
Long non-coding (lnc) RNAs are non-protein coding
transcripts which contain >200 nucleotides (17). lncRNAs have been reported to be
significant in dosage compensation effects, the regulation of
epigenetics, the cell cycle and cell differentiation in mammals
(18). Due to their unknown, but
potentially efficacious applications, researchers worldwide have
focused on establishing databases of lncRNAs at a genome-wide level
(19). Thus far, the constructed
lncRNA databases are as follows: lncRNABase (20), ChIPBase (21), LNCipedia (22), lncRNAdb (23), NONCODE (24) and NRED (25).
In the present study, the potential association
between FGF1 and BMSCs in patients with AA was investigated, and
the regulatory mechanism of FGF1 by lncRNAs was evaluated to
provide a novel insight into the treatment of AA.
Materials and methods
Isolation and culture of BMSCs
Marrow was obtained from patients diagnosed with
aplastic anemia (AA), which had been preserved in the Cancer Tissue
Bank between 2007 and 2013 at Changzhou First People's Hospital
(Jiangsu, China). Among the 24 selected tumor samples, 12 were from
male patients and 12 were from female patients. The average age of
the patients was 36 years. Informed consent for the experimental
use of surgical samples was obtained from all patients. The study
was approved by the ethics committee of The First People's Hospital
of Changzhou, Changzhou, China. Following heparinization (3,000
units; 0.2 ml), 1 ml of marrow, was added to 5 ml Red Blood Cell
Lysis Buffer (Beyotime Institute of Biotechnology, Nanjing, China)
and the homogeneous mixture was centrifuged at 2,930 × g for 10
min. The supernatant was discarded and the precipitate was rinsed
twice with phosphate-buffered saline (pH 7.2). The BMSCs were
isolated by an additional centrifugation of the mixture and
isometric percoll lymphocyte separation medium (Ficoll-lsopaque,
Pharmacia, Piscataway, NJ, USA) (ρ=1.072 g/ml) was added. The
mixture was cultured in α-minimum essential medium (α-MEM)
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
(Sigma-Aldrich, St. Louis, MO, USA) and 100 U/ml streptomycin
(Sigma-Aldrich) at 37°C with 5% CO2 for 24 h. Finally,
the isolated BMSCs were subcultured every 3 days according to
whether the ratio of the original medium to the fresh medium was
1:2 (v:v).
Reconstruction of the adenovirus
vector
The pSileneerl.0-shFGF1 and pShuttle vectors (BD
Biosciences, Palo Alto, CA, USA) were cut using BamHI and
HindIII restriction enzymes (Promega Corporation, Madison,
WI, USA). Then the fragments of shFGF1 cDNA (0.3 kb) and pShuttle
(4.2 kb) were retrieved and ligated using T4 DNA ligase for 4 h at
22°C. DH5α™ competent cells were transformed and the plasmids were
extracted following screening for positive colonies in
Luria-Bertani (LB) medium supplemented with kanamycin. The
combination of the materials was termed pShuttle-shFGF1 To
construct the recombinant adenovirus vector, cells were transfected
with pAdxsi vector as well as pShuttle-shFGF1. Superstratum was
covered with 5% gelose and cultured at 37°C with 5% CO2
for 10 days. Following connection of the retrieved plasmids and
fragments with their target genes [FGF1 or small interfering
(si)RNA-testis development related gene 1 (TDRG1), designed and
synthesized by Invitrogen Life Technologies (San Diego, CA, USA)],
the DH5α was transformed and coated onto LB medium with ampicillin
(Marsan Pharmaceuticals, Cherry Hill, NJ, USA). The positive
colonies were selected by sending to Sigma-Aldrich for sequencing.
The cultured BMSCs (5×105/well) were seeded into a
6-well plate filled with Dulbecco's modified Eagle's medium (DMEM;
Life Technologies, Inc., Gaithersburg, MD, USA) and infected using
Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA) until the cells covered 80–90% of the plate. Two days later,
the virus was collected using the cytopathic effect and the titer
was determined with a hemolytic plaque assay using the following
model: Virus titer = No. of plaques/dilution factor × volume of
diluent. For comparison, a sham control (medium only) was included
and underwent the above-mentioned procedure. All culture processes
were conducted in an atmosphere of 5% CO2 at 37°C and
the experiments were performed in triplicate.
Cell proliferation assay
The suspension of BMSCs, FGF1-BMSCs, TDRG1
siRNA-BMSCs, FGF1-TDRG1 siRNA-BMSCs, scramble siRNA-BMSCs and
FGF1-scramble siRNA-BMSCs were plated in 96-well plates at a
concentration of 100 ml/well in α-MEM with 10% FBS. The cells were
then cultured by incubation at 37°C in a 5% CO2
atmosphere for 24, 36, 48, 60 and 72 h. Cell Counting kit-8 (CCK-8;
Dojindo Laboratory, Kumamoto, Japan) reagent (10 ml) was added to
the well and incubated for 24, 36, 48, 60 or 72 h. After a 2-h
incubation, the optical density (OD) values of corresponding cells
were measured using an ultraviolet spectrophotometer (Varian
Medical Systems, Inc., Palo Alto, CA, USA) at 450 nm. The results
were recorded for further comparison.
Immunofluorescence
After being fixed on a 48-well plate using 4%
paraformaldehyde, the infected cells were permeabilized using 0.2%
Triton X-100 and then sealed using 5% goat serum for 30 min.
Incubation with anti-FGF1 primary antibody (Abcam, Cambridge, MA,
USA; cat. no. ab9588; dilution, 1:200) and a fluorescein
isothiocyanate (FITC)-labeled secondary antibody (cat. no.
ABIN101988; Upstate Biotechnology, Lake Placid, NY, USA) was
conducted at 37°C. The cell nucleus was counterstained with
4′,6-diamino-2-phenylindole and the plate was sealed using
glycerinum. Microscopy (Olympus IX71, Tokyo, Japan) was performed
to observe and obtain images the cells.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To investigate the expression of target genes, total
RNA was extracted and isolated from BMSCs using TRIzol reagent
(Invitrogen Life Technologies). RNA was reverse transcribed using
M-MLV Reverse Transcriptase (Promega Corporation). RNA quality was
assessed with the ThermoScientific NanoDrop1000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). RT-qPCR was performed using
the QuantiTect Primer assay (Qiagen GmbH, Hilden, Germany) and
QuantiTect SYBR Green RT-PCR kit (Qiagen GmbH) on a LightCycler 480
Instrument (Roche Diagnostics, Mannheim, Germany). The detection
and quantification contained the following steps: Reverse
transcription was performed for 30 min at 55°C and initial
activation for 15 min at 95°C; followed by 40 cycles of
denaturation conducted at 94°C for 15 sec, annealing for 30 sec at
55°C and extension for 30 sec at 72°C. The target gene primers were
designed by Invitrogen Life Technologies and primer sequences were
as follows: Forward: 5′-CAGTACTTGGCCATGGACA-3′ and reverse:
5′-AGTGAGTCCGAGGACCGC-3′. The outcome of the RT-qPCR was assessed
using the 2−ΔΔCt method and GAPDH served as a reference
for normalizing the target gene expression.
Chromatin immunoprecipitation (ChIP)
BMSCs were cross-linked with 1% formaldehyde for 10
min at room temperature. The cross-linking was terminated by adding
125 mM glycine and the cells were washed twice with ice-cold PBS.
The cells were solubilized in a buffer containing 10 mM Tris-HCl
(pH 8.0), 1% Triton X-100, 1% sodium deoxycholate, 1 mM
phenylmethanesulfonyl fluoride and protease inhibitor cocktail for
10 min at 4°C. Sonication using a Bioruptor® Sonicator
(Diagenode s.a., Seraing, Belgium) was performed to shear chromatin
into 500-bp fragments. The supernatant was obtained by
centrifugation (16,000 × g for 10 min at 4°C) and equally divided
into six tubes (100 µl/tube). The appropriate antibody
[anti-histone deacetylase (HDAC) 3 (Abcam; cat. no. ab7030;
dilution: 1:500) or HDAC4 (Abcam; ab53331; dilution, 1:500] was
added into each tube and incubated for 3 h at 4°C.
Immunoprecipitation was performed using ChIP-grade agarose beads
with protein G (Cell Signaling Technology, Inc., Danvers, MA, USA),
and the cells were blocked with 1% bovine albumin and 1% salmon
sperm DNA. Finally, ChIP-grade agarose beads, protein G, cells,
bovine albumin and salmon sperm DNA, were collected and the DNA was
isolated by sedimentation velocity for qPCR.
Statistical analysis
Data were processed using SPSS 12.0 statistical
software (SPSS, Inc., Chicago, IL, USA) and recorded as the mean ±
standard error of the mean. P<0.05 was considered to indicate a
statistically significant difference. In addition, one-way analysis
of variance was adopted to assess the data. All of the experiments
were performed in triplicate for the purposes of comparison.
Results
FGF1 promotes the proliferation of
BMSCs
The expression of FGF1 was identified in the BMSCs
by immunofluorescence and the cells infected with Ad-FGF1 were
observed to divide vigorously (Fig. 1A
and B). According to the results of the CCK-8 assay in Fig. 1C, BMSCs of patients with AA
infected with FGF1 grew more markedly when compared with the
non-treated BMSCs (control group) with significantly higher OD
values from 24 h during culture. Therefore, it was inferred that
FGF1 may promote the proliferation of BMSCs in patients with
AA.
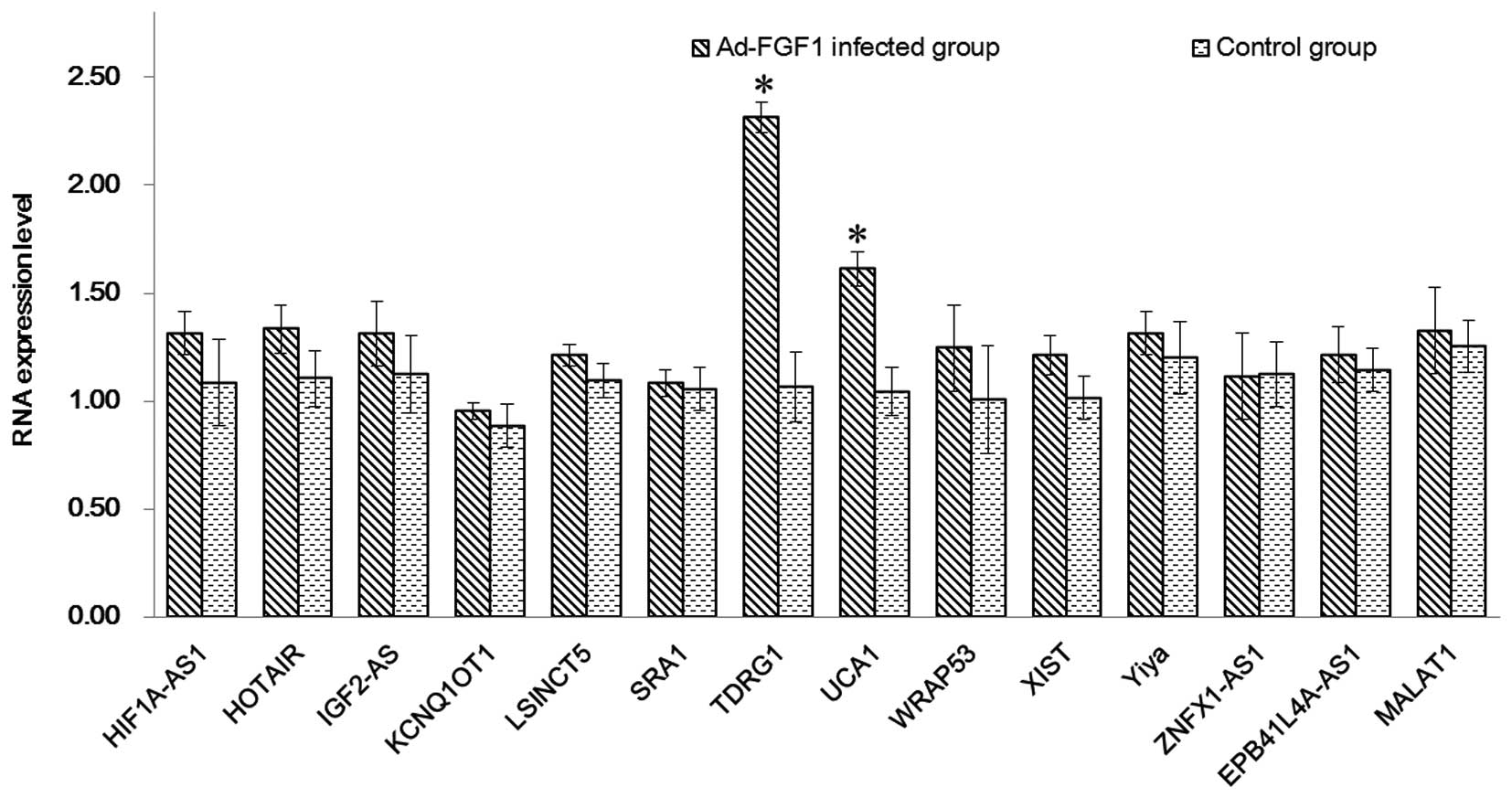
FGF1 influences the expression of lncRNA
TDRG1
The expression levels of lncRNAs in BMSCs infected
with/without Ad-FGF1 were detected by RT-qPCR (Fig. 2). The upregulated expression of all
of the selected genes was observed in the two groups. Compared with
the control group, the genes in Ad-FGF1-infected BMSCs were
observed to be upregulated and expressed at higher levels. The two
genes that were significantly upregulated compared with the control
group were TDRG1 and urothelial carcinoma-associated 1 (UCA1;
P<0.05), which indicated that the infection with Ad-FGF1 may
impact the proliferation of BMSCs by influencing lncRNA. In order
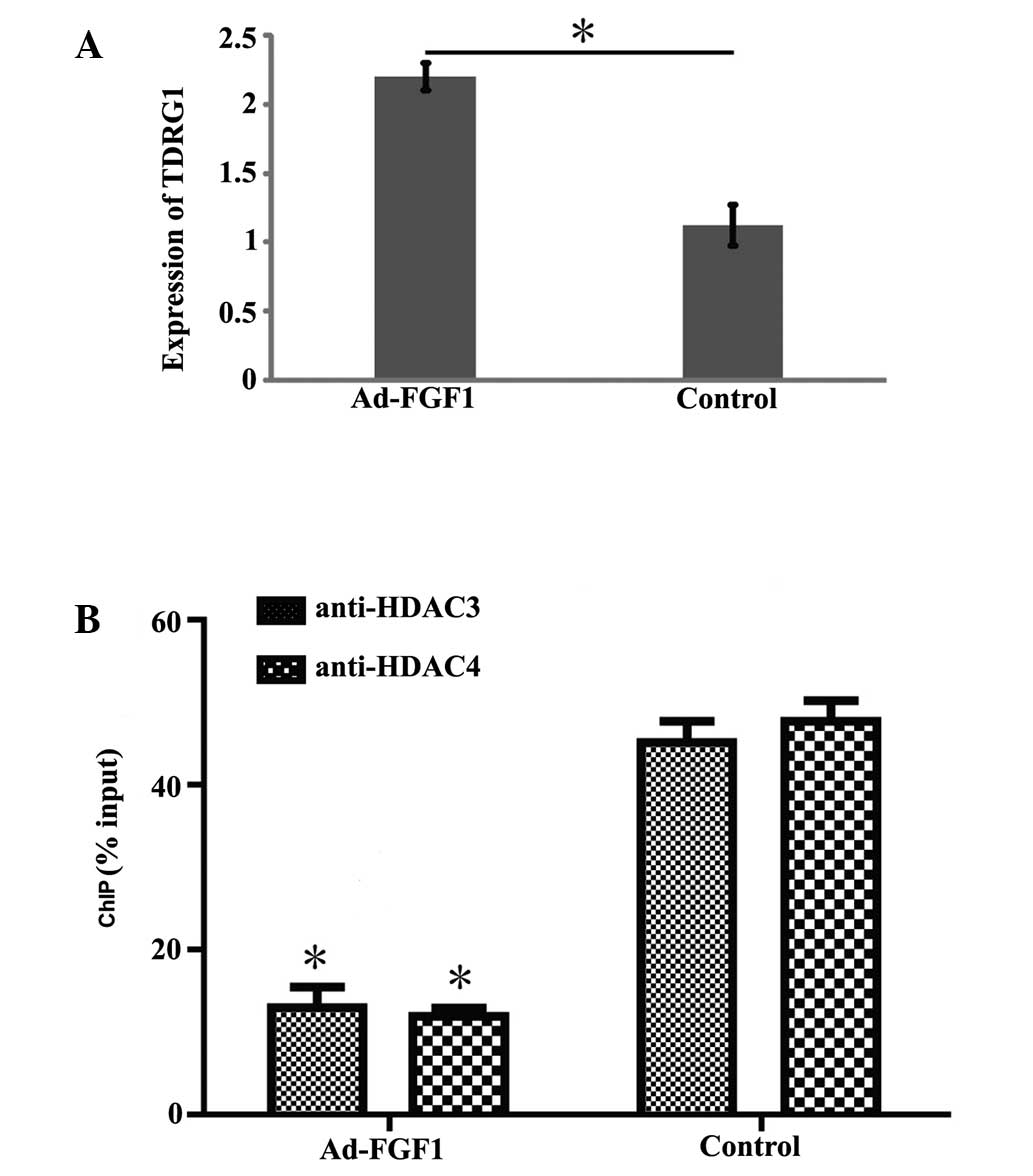
to confirm the result described above, the expression level of
TDRG1 was determined by RT-qPCR. The expression of TDRG1 was
observed to be significantly higher in BMSCs infected with Ad-FGF1
than that in the control group (Fig.
3A; P<0.05). This finding illustrates that FGF1 influences
the expression of lncRNA TDRG1.
FGF1 promotes acetylation in the TDRG1
gene promoter
HDAC3 and HDAC4, which are members of the histone
deacetylase/acetylase family was encoded by HDAC3 and HDAC4,
respectively. HDAC3 and HDAC4 possess deacetylase activity and
regulate the process of acetylation/deacetylation in order to alter
chromosome structure and impact the access of transcription factor
to DNA. Therefore, it was hypothesized that the acetylation level
on the promoter region of the TDRG1 gene may be regulated by the
overexpression of FGF1 in BMSCs. This hypothesis was tested via
ChIP experiments using anti-HDAC3 and anti-HDAC4 antibodies. As
shown in Fig. 3B, the
deacetylation level of the TDRG1 gene promoter was significantly
reduced in BMSCs overexpressing FGF1 when compared with the control
group, which indicates that decreased enrichment of HDAC3 and HDAC4
results in the overexpression of FGF1, leading to acetylation of
the TDRG1 gene promoter.
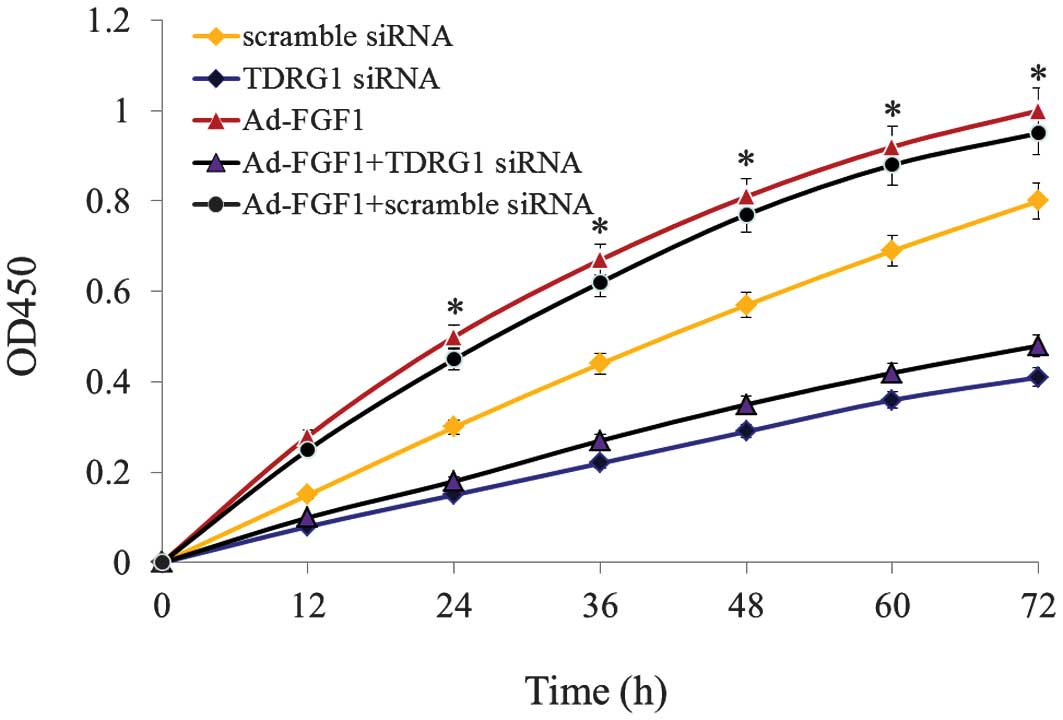
FGF1 enhances the proliferation of BMSCs
through lncRNA TDRG1
In order to analyze the function of TDRG1 lncRNA, an
RNA interference molecule was designed and TDRG1 lncRNA was
silenced in BMSCs infected with/without Ad-FGF1. The results are
presented in Fig. 4. Following
TDRG1 lncRNA silencing, cell proliferation was observed to be
significantly downregulated in BMSCs that were infected
with/without Ad-FGF1, when compared with the control groups
(P<0.05). All of these results indicate that FGF1 enhances the
proliferation of BMSCs via regulating the expression of TDRG1
lncRNA.
Discussion
AA is a rare disease in which the normal generation
of red blood cells and various white cell lines in the bone marrow
is disrupted (26,27). Various studies have focused on
investigating the pathogenesis of AA by observing the regulation of
gene expression levels. Zhang et al (28) demonstrated that the increased
expression of T cell immunoglobulin and immunoreceptor
tyrosine-based inhibition motif domain ameliorated immune-mediated
bone marrow failure of AA via the depressed function of
CD4+ T cells. Furthermore, Wang et al (29) confirmed the harmful and protective
function of human leukocyte antigen alleles in Chinese patients
exhibiting severe AA. In the current study, it was inferred that
FGF1 is a potential regulator of BMSC differentiation in patients
with AA on the basis of experiments in which low expression of FGF
was observed the BMSCs of patients with AA. As BMSCs are
significant in the hematopoietic microenvironment, an investigation
concerning the underlying mechanism of FGF1 in the differentiation
of BMSCs was considered to be essential and necessary.
The association between FGF1 and BMSCs has been
reported in various studies. Eom et al (30) demonstrated that FGF4 and FGF2
exerted an autocrine effect by regulating the proliferation of
BMSCs. Chen et al (31)
demonstrated that the mutation of Ser252Trp in FGF receptor 2 was
vital in FGF signaling and reduced the proliferation of BMSCs. In
the present study, research was initiated with the study of the
association between FGF1 and BMSC proliferation. The increased OD
value of the cultured FGF1-BMSCs (when compared with the control
group) revealed the increased proliferative capability of the cells
infected with Ad-FGF1 and illustrated that FGF1 was a potential
regulator of BMSC proliferation. Furthermore, this finding was
supported by results of immunofluorescence in which vigorous
proliferation and FGF1 expression were observed in FGF1-BMSCs.
In recent years, lncRNA has become increasingly
popular in the investigation of the pathogenesis of certain
malignant diseases. Metastasis associated lung adenocarcinoma
transcript 1 (MALAT1) and UCA1 were found to be abnormally
expressed in melanoma metastases (32). X-inactive specific transcript was
identified as a biomarker for membranous nephropathy (33) and WD repeat containing, antisense
to TP53 was considered as a candidate for a cancer susceptibility
gene (34). The bi-functional
lncRNA, steroid receptor activator RNA 1 was identified to be a
component of the steroid receptor coactivator-1 acetyltransferase
complex and indicated to be an epigenetic regulatory component
(35). Insulin-like growth factor
2 antisense RNA was observed to be a potent stimulator in cancer
cell lines of diabetic patients (36). Furthermore, HOX transcript
antisense RNA and MALAT1 were identified as the specific lncRNAs in
laryngeal squamous cell carcinoma (37). The overexpression of hypoxia
inducible factor 1-antisense RNA 1 (HIF1A-AS1) was associated with
non-papillary renal HIF1A carcinomas (38). The disease-related lncRNA of
erythrocyte membrane protein band 4.1 like 4A-AS1, zinc finger,
NFX1-type containing 1-AS1, Yiya and long stress-induced non-coding
transcript 5 were selected from lncRNABase together with the
above-mentioned lncRNAs, and determined in the present study using
RT-qPCR to observe the potential regulatory role of lncRNAs in
FGF1-BMSCs. According to the findings of the PCR, all the genes
were observed to be upregulated in the FGF1-BMSCs; however, two
genes (TDRG1 and UCA1) exhibited significant increases in
expression. It was indicted that FGF1 promoted BMSC proliferation
via regulation of the lncRNAs of TDRG1 and UCA1.
Although numerous studies have documented their
association with certain diseases, investigations involving the
role of TDRG1 and UCA1 in Ad-FGF1-infected BMSCs are considered to
be limited. According to the result of Fig. 3, the expression of TDRG1 at the
gene level in FGF1-BMSCs increased significantly compared with
control cells. When the cells were infected with TDRG1 siRNA,
proliferation was markedly inhibited, this further confirmed the
role of TDRG1 in BMSC differentiation in patients with AA. In
Fig. 4, the expression of HDAC3
and HDAC4 (indicators of acetylation levels in cells) was observed
to be significantly reduced in FGF1-BSMCs. Since HDAC3 and HDAC4
possess the potent activity of deacetylation, a decrease in their
expression levels would lead to enhanced acetylation, which
introduces the acetyl functional group into a chemical compound and
is considered to be an important modification process of proteins
in cell biology.
Therefore, it was concluded in the present study
that FGF1 promotes the proliferation of BMSCs to alleviate AA by
enhancing acetylation of the lncRNA of the TDRG1 gene promoter.
The findings of the current study provide a
promising insight into the therapeutic treatment of AA. However,
further validation via in vivo studies is required.
Acknowledgments
The present study was supported by Shanghai Jiaotong
University School of Medicine Science Fund Project (grant no.
2013236).
References
|
1
|
Sheng W, Liu C, Fu R, Wang H, Qu W, Ruan
E, Wang G, Liu H, Wu Y, Song J, et al: Abnormalities of quantities
and functions of linker for activations of T cells in severe
aplastic anemia. Eur J Haemato. 93:214–223. 2014.
|
|
2
|
Young NS, Scheinberg P and Calado RT:
Aplastic anemia. Current Opin Hematol. 15:162–168. 2008. View Article : Google Scholar
|
|
3
|
Cohen T and Creger WP: Acute myeloid
leukemia following seven years of aplastic anemia induced by
chloramphenicol. Am J Med. 43:762–770. 1967. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Binder D, van den Broek MF, Kägi D,
Bluethmann H, Fehr J, Hengartner H and Zinkernagel RM: Aplastic
anemia rescued by exhaustion of cytokine-secreting CD8+ T cells in
persistent infection with lymphocytic choriomeningitis virus. J Exp
Med. 187:1903–1920. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xing L, Liu C, Fu R, Wang H, Wang J, Liu
X, Feng L, Li L, Liu H, Wang H, et al: CD8+HLA-DR+ T cells are
increased in patients with severe aplastic anemia. Mol Med Rep.
10:1252–1258. 2014.PubMed/NCBI
|
|
6
|
Feng X, Scheinberg P, Biancotto A, Rios O,
Donaldson S, Wu C, Zheng H, Sato K, Townsley DM, McCoy JP and Young
NS: In vivo effects of horse and rabbit antithymocyte globulin in
patients with severe aplastic anemia. Haematologica. 99:1433–1440.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kato RB, Roy B, De Oliveira FS, Ferraz EP,
De Oliveira PT, Kemper AG, Hassan MQ, Rosa AL and Beloti MM:
Nanotopography directs mesenchymal stem cells to osteoblast lineage
through regulation of microRNA-SMAD-BMP-2 circuit. J cell Physiol.
229:1690–1696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding L, Zhu H, Yang Y, Wang ZD, Zheng XL,
Yan HM, Dong L, Zhang HH, Han DM, Xue M, et al: Functional
mesenchymal stem cells remain present in bone marrow
microenvironment of patients with leukemia post-allogeneic
hematopoietic stem cell transplant. Leuk lymphoma. 55:1635–1644.
2014. View Article : Google Scholar
|
|
9
|
Sun ZM, Liu HL, Geng LQ, Wang XB, Yao W,
Liu X, Ding KY, Han YS, Yang HZ, Tang BL, et al: HLA-matched
sibling transplantation with G-CSF mobilized PBSCs and BM decreases
GVHD in adult patients with severe aplastic anemia. J Hematol
Oncol. 3:512010. View Article : Google Scholar
|
|
10
|
Zhao J, Wang C, Song Y and Fang B: Arsenic
trioxide and microRNA-204 display contrary effects on regulating
adipogenic and osteogenic differentiation of mesenchymal stem cells
in aplastic anemia. Acta Biochim Biophys Sin (Shanghai).
46:885–893. 2014. View Article : Google Scholar
|
|
11
|
Wang X, Ma FX, Lu SH, Chi Y, Chen F, Li X,
Li JJ, Du WJ, Feng Y, Cui JJ, et al: Effects of rapamycin on
biological characteristics of bone marrow mesenchymal stem cells
from patients with aplastic anemia. Zhongguo Shi Yan Xue Ye Xue Za
Zhi. 22:762–766. 2014.In Chinese. PubMed/NCBI
|
|
12
|
Jiang SY, Xie XT, Jiang H, Zhou J, Li FX
and Cao P: Low expression of basic fibroblastic growth factor in
mesenchymal stem cells and bone marrow of children with aplastic
anemia. Pediatric Hematol Oncol. 31:11–19. 2014. View Article : Google Scholar
|
|
13
|
Tursi A, Elisei W, Inchingolo CD, Nenna R,
Picchio M, Ierardi E and Brandimarte G: Chronic diverticulitis and
Crohn's disease share the same expression of basic fibroblastic
growth factor, syndecan 1 and tumour necrosis factor-α. J Clin
Pathol. 67:844–846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marinucci L, Bodo M, Balloni S, Locci P
and Baroni T: Sub-toxic nicotine concentrations affect
extracellular matrix and growth factor signaling gene expressions
in human osteoblasts. J Cell Physiol. 229:2038–2048. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stegmann TJ: New approaches to coronary
heart disease: Induction of neovascularisation by growth factors.
Bio Drugs. 11:301–308. 1999.
|
|
16
|
Cao R, Bråkenhielm E, Pawliuk R, Wariaro
D, Post MJ, Wahlberg E, Leboulch P and Cao Y: Angiogenic synergism,
vascular stability and improvement of hind-limb ischemia by a
combination of PDGF-BB and FGF-2. Nat Med. 9:604–613. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kapranov P, Willingham AT and Gingeras TR:
Genome-wide transcription and the implications for genomic
organization. Nat Rev Genet. 8:413–423. 2007. View Article : Google Scholar
|
|
18
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Muers M: RNA: Genome-wide views of long
non-coding RNAs. Nat Rev Genet. 12:7422011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang JH, Li JH, Shao P, Zhou H, Chen YQ
and Qu LH: StarBase: A database for exploring microRNA-mRNA
interaction maps from Argonaute CLIP-Seq and Degradome-Seq data.
Nucleic Acids Res. 39:D202–D209. 2011. View Article : Google Scholar
|
|
21
|
Yang JH, Li JH, Jiang S, Zhou H and Qu LH:
ChIPBase: A database for decoding the transcriptional regulation of
long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic
Acids Res (Database Issue). 41:D177–D178. 2013. View Article : Google Scholar
|
|
22
|
Volders PJ, Helsens K, Wang X, Menten B,
Martens L, Gevaert K, Vandesompele J and Mestdagh P: LNCipedia: A
database for annotated human lncRNA transcript sequences and
structures. Nucleic Acids Res (Database Issue). 41:D246–D251. 2013.
View Article : Google Scholar
|
|
23
|
Amaral PP, Clark MB, Gascoigne DK, Dinger
ME and Mattick JS: lncRNAdb: A reference database for long
noncoding RNAs. Nucleic Acids Res (Database Issue). 39:D146–D151.
2011. View Article : Google Scholar
|
|
24
|
Bu D, Yu K, Sun S, Xie C, Skogerbø G, Miao
R, Xiao H, Liao Q, Luo H, Zhao G, et al: NONCODE v3.0: Integrative
annotation of long noncoding RNAs. Nucleic Acids Res (Database
Issue). 40:D210–D215. 2012. View Article : Google Scholar
|
|
25
|
Dinger ME, Pang KC, Mercer TR, Crowe ML,
Grimmond SM and Mattick JS: NRED: A database of long noncoding RNA
expression. Nucleic Acids Res (Database Issue). 37:D122–D126. 2009.
View Article : Google Scholar
|
|
26
|
Self SG, Longton G, Kopecky KJ and Liang
KY: On estimating HLA/disease association with application to a
study of aplastic anemia. Biometrics. 47:53–61. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baldwin JL, Storb R, Thomas ED and Mannik
M: Bone marrow transplantation in patients with gold-induced marrow
aplasia. Arthritis Rheum. 20:1043–1048. 1997. View Article : Google Scholar
|
|
28
|
Zhang T, Wang J, Zhou X, Liang R, Bai Q,
Yang L, Gu H, Gao G, Dong B, Zhu H and Chen X: Increased Expression
of TIGIT on CD4+ T cells ameliorates immune-mediated bone marrow
failure of aplastic anemia. J Cell Biochem. 115:1918–1927.
2014.PubMed/NCBI
|
|
29
|
Wang M, Nie N, Feng S, Shi J, Ge M, Li X,
Shao Y, Huang J and Zheng Y: The polymorphisms of human leukocyte
antigen loci may contribute to the susceptibility and severity of
severe aplastic anemia in Chinese patients. Hum Immunol.
75:867–872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eom YW, Oh JE, Lee JI, Baik SK, Rhee KJ,
Shin HC, Kim YM, Ahn CM, Kong JH, Kim HS and Shim KY: The role of
growth factors in maintenance of stemness in bone marrow-derived
mesenchymal stem cells. Biochem Biophys Res Commun. 445:16–22.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen P and Zhang L, Weng T, Zhang S, Sun
S, Chang M, Li Y, Zhang B and Zhang L: A Ser252Trp mutation in
fibroblast growth factor receptor 2 (FGFR2) mimicking human Apert
syndrome reveals an essential role for FGF signaling in the
regulation of endochondral bone formation. PloS One. 9:e873112014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
Malat-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang YS, Hsieh HY, Shih HM, Sytwu HK and
Wu CC: Urinary Xist is a potential biomarker for membranous
nephropathy. Biochem Biophys Res Commun. 452:415–421. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sedaie Bonab A, Pouladi N,
Hosseinpourfeizi MA, Ravanbakhsh Gavgani R, Dehghan R, Azarfam P,
Montazeri V and Fakhrjou A: Single-strand conformational
polymorphism analysis of a common single nucleotide variation in
WRAP53 gene, rs2287499 and evaluating its association in relation
to breast cancer risk and prognosis among Iranian-Azeri population.
Med Oncol. 31:1682014. View Article : Google Scholar
|
|
35
|
Bilinovich SM, Davis CM, Morris DL, Ray
LA, Prokop JW, Buchan GJ and Leeper TC: The C-terminal domain of
SRA1p has a fold more similar to PRP18 than to an RRM and does not
directly bind to the SRA1 RNA STR7 region. J Mol Biol.
426:1753–1765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sciacca L, Cassarino MF, Genua M, Vigneri
P, Giovanna Pennisi M, Malandrino P, Squatrito S, Pezzino V and
Vigneri R: Biological effects of insulin and its analogs on cancer
cells with different insulin family receptor expression. J Cell
Physiol. 229:1817–1821. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen H, Xin Y, Zhou L, Huang JM, Tao L,
Cheng L and Tian J: Cisplatin and paclitaxel target significant
long noncoding RNAs in laryngeal squamous cell carcinoma. Med
Oncol. 31:2462014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deb S, Johansson I, Byrne D, Nilsson C,
Investigators K, Constable L, Fjällskog ML, Dobrovic A, Hedenfalk I
and Fox SB: Nuclear HIF1A expression is strongly prognostic in
sporadic but not familial male breast cancer. Mod Pathol.
27:1223–1230. 2014. View Article : Google Scholar : PubMed/NCBI
|