Introduction
Gastric cancer is the fourth most common cancer type
and the second leading cause of cancer-associated mortality, with a
mortality rate of >700,000 individuals reported annually
(1). Currently, the highest
incidence rates are reported in China, Japan, Korea, Eastern Europe
and parts of Central and South America (2). Metastasis and post-surgery recurrence
rates in gastric cancer are >40%, and therefore how to
effectively deal with metastasis is a major challenge in gastric
cancer therapy (3).
DNA methylation is an essential feature of the
epigenetic regulation pathway, which is frequently perturbed in
human gastric cancer, and acts as a key epigenetic signature
implicated in transcriptional regulation, genomic imprinting and
the silencing of repetitive DNA elements, which occurs
predominantly within CpG sites (4–6). CpG
sites are underrepresented in the mammalian genome and tend to be
clustered within CpG islands (CGIs) located in the vicinity of the
transcription start sites (TSSs) of the majority of the human
protein-coding genes (7).
Inhibiting DNA methylation with cytidine analogues, including
decitabine (DAC), reactivated the expression of genes, which were
aberrantly silenced by hypermethylation. DAC is integrated into the
replicating DNA and forms irreversible covalent bonds with the
active site of DNA methyltransferase (8).
DAC was demonstrated to be active in several
hematological disorders, including myelodysplastic syndrome (MDS),
acute myelogenous leukemia (AML), chronic myelogenous leukemia
(CML) and sickle cell anemia (9–13).
Numerous previous studies in cancer cells demonstrated that DAC
inhibits cell proliferation and the motility of the cells, and it
is clinically administered as an antitumor drug to promote the
expression of tumor suppressor genes (8,9,12–15).
However, since demethylation across the entire genome alters the
expression of a large number of genes, the effects of DAC in
different tumor cell types are difficult to accurately predict
(16,17).
Neural precursor cell-expressed, developmentally
downregulated (NEDD)4 is a prominent member of the E3-ubiquitin
ligase family. NEDD4-1, a member of the NEDD4 subfamily, has a
catalytic HECT domain at the C-terminus, and C2 and WW domains at
the N-terminus, which are responsible for substrate recognition
(18,19). NEDD4-1 was reported to regulate a
number of cellular functions, including the development of the
neuromuscular junction (20) and
the central nervous system, and axon guidance (21,22),
in addition to exerting a role in brain diseases (23,24).
From the perspective of studying cancer, NEDD4-1 is highly
expressed in a wide range of tumor types, including colorectal
cancer, bladder cancer and gastric carcinoma, and it was
demonstrated to activate the phosphoinositide 3-kinase/Akt pathway
through the degradation of phosphatase and tensin homolog (PTEN)
(25–27). NEDD4-1 also promoted the migration
and invasion of glioma cells via the ubiquitination and subsequent
degradation of cyclic nucleotide-Ras guanine nucleotide exchange
factors (CNrasGEFs) (28). In a
major type of gastric cancer, gastric cardia adenocarcinoma,
NEDD4-1 was revealed to be an exceptional prognostic biomarker
(29).
In the present study, the effects of DAC in
promoting the invasion and migration of MGC803 gastric cancer
cells, or in inhibiting their proliferation, were investigated.
Furthermore, the present study aimed to investigate the mechanistic
role of NEDD4-1 in the invasion-promoting activity of DAC in the
MGC803 cells, and whether the underlying mechanism involved CGI
methylation, either in the NEDD4 promoter or in the first intron.
The effects of DAC on different gastric cancer cell lines were
subsequently discussed, including the therapeutic potential of DAC
in clinical applications.
Materials and methods
Materials
A primary antibody against NEDD4 [cat. no. 2740;
rabbit phosphoantibody (pAb); 1:500] was obtained from Cell
Signaling Technology, Inc. (Danvers, MA, USA), and an antibody
against β-actin [cat. no. sc-47778; horseradish peroxidase
(HRP)-conjugated; 1:500] was purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). HRP-conjugated
anti-rabbit/mouse secondary antibodies (A16072 and A16104) were
obtained from Thermo Fisher Scientific (Waltham, MA, USA). Antibody
against CNrasGEF (cat. no. WH0009693M1; mouse pAb; 1:400) and DAC
(5-Aza-2′-deoxycytidine; cat. no A3656) were obtained from
Sigma-Aldrich (St. Louis, MO, USA).
The pair of independent anti-NEDD4-1 stealth RNA
interference (RNAi) small interfering (si)RNAs used in the present
study, siN4-1 (5′-GAGTTCTTACAAGTGTGCAAACAAA-3′) and siN4-2
(5′-CCGATTGACAAGAGATGATTTCCTA-3′), Lipofectamine®
RNAiMAX reagent, Opti-MEM® reduced serum media, Gibco
fetal bovine serum (FBS) and SYBR® Select Master mix
were obtained from Thermo Fisher Scientific. HyClone™ RPMI-1640
medium was also obtained from Thermo Fisher Scientific. The
PrimeScript® reverse transcription (RT) reagent kit with
genomic (g)DNA Eraser was obtained from Takara Biotechnology Co.,
Ltd. (Dalian, China).
Cell culture and transfection
The human gastric cancer cell lines, MGC803, SGC7901
and NCI-N87, were purchased from the Cell Resource Center, IBMS,
CAMS/PUMC (Beijing, China). The cell lines were maintained in
either RPMI-1640 (SGC7901, NCI-N87) or in Dulbecco's modified
Eagle's medium (MGC803; Gibco; Thermo Fisher Scientific),
supplemented with 10% FBS and 100 U/ml penicillin/streptomycin
(HyClone™; Thermo Fisher Scientific) at 37°C in a humidified
atmosphere (5% CO2/95% air).
The siRNA transfections were performed according to
the manufacturer's protocols in 6-well plates, using
Lipofectamine® RNAiMAX transfection reagent.
Cell proliferation analysis
MGC803 or SGC7901 cells (2×105 cells/ml
in 96-well plates) were treated with phosphate-buffered saline
(PBS) or 1 µM DAC. The cell viability was measured using a
3-(4,5-dimethyl-2-thiazolyl)-2,5-di-phenyl-2-tetrazolium bromide
(MTT) cell viability detection kit (Beyotime Institute of
Biotechnology, Beijing, China), according to the manufacturer's
protocols.
The absorbance was measured at 490 nm using a
microplate spectrophotometer (Spectra Max M3; Molecular Devices,
LLC, Sunnyvale, CA, USA).
RNA extraction and RT-quantitative
polymerase chain reaction (qPCR)
For the RT-qPCR analyses, the total RNA was
extracted from cells using Invitrogen TRIzol® reagent
(Thermo Fisher Scientific), according to the manufacturer's
protocols. Clearance of the DNA contamination in RNA and cDNA
synthesis were performed using the PrimeScript® RT
reagent kit with gDNA Eraser, according to the manufacturer's
protocols (Takara Biotechnology Co., Ltd.). RT-qPCR was
subsequently performed using the ABI-7500 system (Applied
Biosystems; Thermo Fisher Scientific), using SYBR®
Select Master mix (Thermo Fisher Scientific) according to the
manufacturer's instructions. Primer sequences were as follows:
NEDD4-1, forward (F): 5'-GGTGGAGGTGTTCGGGCT-3′ and reverse (R):
5′-GCAAGGCCTATTCCGGCTA-3′; glyceraldehyde-3-phosphate dehydrogenase
(GAPDH), F: 5′-GAGTCAACGGATTTGGTCGT-3′ and R:
5′-GACAAGCTTCCCGTTCTCAG-3′.
Western blot analysis
The preparation of the whole cell lysates and
western blot analysis were performed, as previously described
(30). In brief, cells were
harvested and washed in ice-cold PBS, then the protein
concentration of the extracts was determined using bicinchoninic
acid reagent (Thermo Fisher Scientific). Equal quantities of
protein (30 g/lane) were loaded, separated using 12% SDS-PAGE and
transferred onto nitrocellulose membranes. Subsequent to being
blocked with 5% non-fat milk, membranes were incubated with the
primary antibodies at 4°C, overnight. Subsequent to incubation with
the respective secondary antibody, immune complexes were detected
using ECL western blotting reagents (Thermo Fisher Scientific). The
expression levels of β-actin were monitored as the internal
control, and band intensities were normalized to that of
β-actin.
Wound healing assay
The cells were seeded into 6-well plates
(5×105 cells) and were incubated until they had reached
90% confluence. At 24 h post-transfection, a vertical wound was
created using a 200 µl pipette tip. Subsequently, the cells
were washed three times with PBS and medium without serum was added
into the wells. Following a 48 h incubation with 1 µM DAC,
the wound was observed and random fields in each well were selected
for imaging. The captured images were analyzed using
ImageJ® software (National Institutes of Health,
Bethesda, MD, USA), and the distance of the wound closure was used
to estimate the migrational capacity of the cells.
Migration and invasion assay
The cell migrational and invasive abilities were
determined using 24-well Transwell plates. For the migration assay,
24 h following transfection, 5×104 cells/well were
seeded into the top chamber and maintained in serum-free medium.
Medium (600 µl) containing 10% FBS was added into the bottom
chamber. Following an incubation for 48 h at 37°C, the cells which
had migrated through the pore polycarbonate membrane were fixed
with methanol and stained with Giemsa (Sigma-Aldrich).
Subsequently, the cells that had migrated were observed and images
were captured using microscopy (IX70; Olympus Corporation, Tokyo,
Japan). For the invasion assay, prior to cell seeding, the Matrigel
was diluted in serum-free medium to a final concentration of 1
mg/ml. Diluted Matrigel (100 µl/well) was added into the top
chamber and incubated for 4 h at 37°C, followed by the identical
procedures as described for the migration assay.
Bisulfite sequencing analysis
Bisulfite sequencing PCR (BS-PCR) was performed with
gDNA extracted from MGC803, SGC7901 and NCI-N87 cells using an
EpiTect® Fast LyseAll Bisulfite kit (Qiagen, Hilden,
Germany). PCR reactions were performed using EpiTaq HS (Takara
Biotechnology Co., Ltd.) with primers as follows: NEDD4 promoter,
F: 5′-TTGTAGTGTTTTTTAGTAATAAGTTT-3′ and R:
5′-TCTTATAAAAATAACACCCTTAAC-3′; NEDD4 first intron, F:
5′-TTTTTTTTAATATTTTTTGAAGGAAATTG-3′ and R:
5′-AAAACTCTACTATCAACCCCTCCT-3′.
The PCR products were subcloned using a pMD-19 T
vector (Takara Biotechnology Co., Ltd.), according to the
manufacturer's protocols, and individual clones were subsequently
sequenced (Sangon Biotech Co. Ltd., Shanghai, China). Clones were
only accepted if at least 90% cytosine conversion occurred and all
possible clonalities were excluded based on the criteria included
in the BiQ Analyzer software (Max Planck Society, Munich, Germany).
At least 10 replicates were performed for each of the selected
regions in each cell line.
Statistical analysis
Each experiment was repeated at least in triplicate.
Statistical analyses (Student's t-test or one-way analysis of
variance) were performed using Microsoft Excel and statistical
add-on software (Microsoft Corporation, Redmond, WA, USA).
P<0.05 was considered to indicate a statistically significant
difference and the results are expressed as the mean ± standard
deviation.
Results
Decitabine promotes the invasion and
migration of MGC803 gastric cancer cell lines
To examine the effect of DAC on gastric cancer cell
invasion, wound-healing (Fig. 1A)
and Transwell (Fig. 1B) assays
were performed. The results revealed that 1 µM DAC
significantly promoted the invasion and migration of the MGC803
cells, although it failed to elicit a response in the SGC7901
gastric cancer cell line. Consistently with previously published
studies (31,32), DAC inhibited the proliferation of
each cell line, without any marked difference in its effectiveness
(Fig. 1C).
 | Figure 1DNA methylation inhibitor, DAC,
promotes MGC803 cell invasion and migration. (A) A wound-healing
assay, and (B) a Transwell migration and invasion assay of the
MGC03 and SGC7901 cells was performed following treatment with 1
µM DAC for 48 h; magnification, ×40. (C) The viability of
the MGC03 and SGC7901 cells was assessed following treatment with 1
µM DAC or phosphate-buffered saline, as measured using a
3-(4,5-dimethyl-2-thiazolyl)-2,5-di-phenyl-2-tetrazolium bromide
assay. The data are expressed as the mean ± standard deviation
(n>3; *P<0.05, **P<0.01 and
***P<0.001, compared with Veh). DAC, decitabine; OD,
optical density; Veh, vehicle. |
DAC promotes the expression of NEDD4-1 in
MGC803 cells
Since NEDD4-1 has been demonstrated to be involved
in glioma cell invasion (28), it
was hypothesized that NEDD4-1 may also mediate the DAC-promoted
invasion of the MGC803 gastric cancer cell line. At 48 h
post-incubation with a dilute concentration of DAC, NEDD4-1 was
upregulated in the MGC803 cells, and in agreement with the
hypothesis, NEDD4-1 was not upregulated in the SGC7901 cells in the
presence of DAC (Fig. 2A).
Treatment with 1 µM DAC for 24 h failed to elicit any
expression of NEDD4-1 in the MGC803 cell line (Fig. 2B, left), perhaps as a consequence
of the action of DAC-inhibiting DNA methyltransferases.
Furthermore, treatment with 1 µM DAC for 24 or 48 h failed
to upregulate the expression of NEDD4-1 in either the SGC7901 or
the NCI-N87 cell line (Fig. 2B,
middle and right, respectively). Subsequent to the RT-qPCR
analysis, DAC failed to induce the protein expression of NEDD4-1 in
the SGC7901 cells (Fig. 2C).
However, following an incubation for 48 h with a dilute
concentration of DAC, the protein expression level of NEDD4-1 was
upregulated in the MGC803 cells, whereas CNrasGEF, which has been
demonstrated to be a substrate of NEDD4-1, was downregulated
(Fig. 2D) (28).
 | Figure 2DAC promotes the expression of
NEDD4-1 in MGC803 cells. (A) The mRNA expression of NEDD4-1
following an incubation with DAC for 48 h at the concentrations
indicated in the figure is shown (left, MGC803; right, SGC7901).
(B) The mRNA expression of NEDD4-1 following an incubation with 1
µM DAC for 24 or 48 h is shown (left, MGC803; middle,
SGC7901; right, NCI-N87). Representative western blots illustrating
the protein expression levels of NEDD4-1 and CNrasGEF in (C)
SGC7901 and (D) MGC803 cells, following an incubation with DAC at
the indicated concentrations for 48 h. The bar-chart in (D) shows
the quantification of the data illustrated in the western blot for
the MGC803 cells. The data are expressed as the mean ± standard
deviation (n>3). (A) Left bar-chart, P<0.05; right bar-chart,
P>0.05 (one-way analysis of variance), compared with Veh; (B and
D) *P<0.05; **P<0.01;
***P<0.001 (Student's t-test), compared with time 0 h
or Veh. DAC, decitabine; Veh, vehicle; CNrasGEF, cyclic
nucleotide-Ras guanine nucleotide exchange factor; NEDD4-1, neural
precursor cell-expressed, developmentally downregulated 4-1; N.S.,
not significant. |
DAC promotes the cell-invasive behavior
in an NEDD4-1-dependent manner
To determine whether the expression of NEDD4-1 was
associated with the DAC-promoted cellular invasive properties of
the MGC803 cells, the siRNA silencing of NEDD4-1 was investigated.
The results from the RT-qPCR and western blotting experiments
suggested that the siRNAs (siN4-1 and siN4-2) almost completely
inhibited the mRNA and the protein expression levels of NEDD4-1
without DAC incubation (Fig. 3A).
Following transfection with the NEDD4-1 siRNAs, the migratory
ability of the cells on exposure to DAC was inhibited completely,
although in the vehicle-treated group, knocking down the NEDD4-1
protein failed to disturb the cell migration rate in the wound
healing assay (Fig. 3B).
Furthermore, the Transwell migration and invasion assays exhibited
a similar trend (Fig. 3C).
 | Figure 3DAC promotes cell-invasive behavior
in an NEDD4-1-dependent manner. (A) The mRNA and protein expression
levels of the MGC03 cells following transfection with 50 nM
anti-NEDD4-1 siRNAs, and subsequent treatment with 1 µM
DAC/Veh for 48 h. (B) The wound-healing assay of the MGC03 cells
following transfection with 50 nM anti-NEDD4-1 siRNAs, and
subsequent treatment with 1 µM DAC/Veh for 48 h;
magnification, ×40. (C) The Transwell migration and invasion assays
of the MGC03 cells following transfection with 50 nM anti-NEDD4-1
siRNAs, and subsequent treatment with 1 µM DAC/Veh for 48 h;
magnification, ×40. The data are expressed as the mean ± standard
deviation (n>3; *P<0.05, **P<0.01
and ***P<0.001, compared with siNC). CNrasGEF, cyclic
nucleotide-Ras guanine nucleotide exchange factor; Ctl, control;
DAC, decitabine; NEDD4-1, neural precursor cell-expressed,
developmentally downregulated 4-1; siNC, small interfering RNA
negative control; Veh, vehicle. |
DAC-mediated upregulation of NEDD4-1 is
not directly associated with the inhibition of methylation in CGIs
in the first intron of the NEDD4-1 promoter
To further examine whether DAC promoted the
expression of NEDD4-1 through the complete inhibition of DNA
methylation, CGIs in the NEDD4-1 promoter and its first intron were
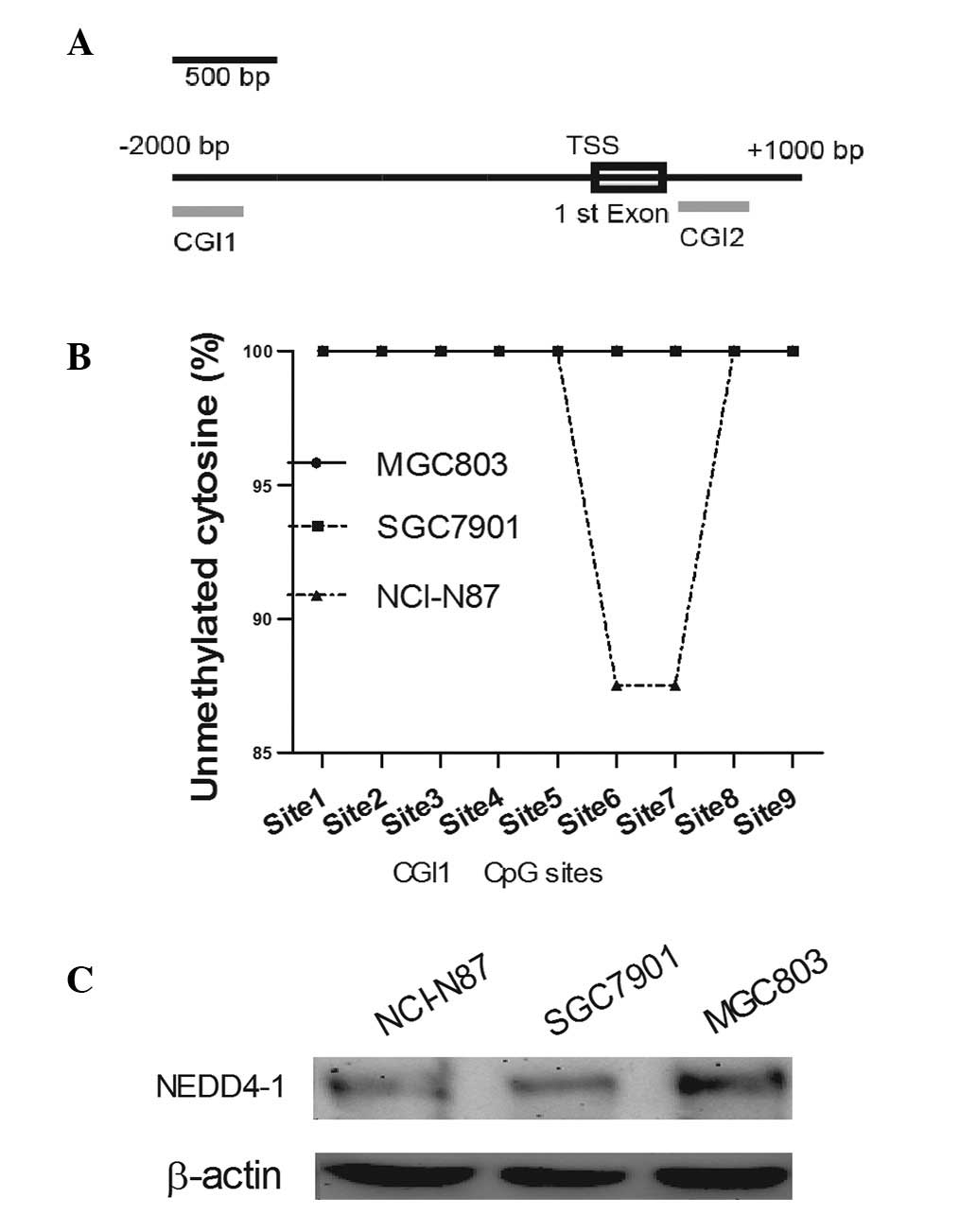
initially investigated using MethPrimer (Fig. 4A) (33). There are two CGIs, CGI1 and 2,
which are located in the promoter and in the first intron,
respectively. Subsequently, BS-PCR was performed to measure the
methylated cytosine content.
The results revealed that no clear DNA methylation
existed in the CGIs of the NEDD4-1 promoter (Fig. 4B) or the first intron (100%
unmethylated cytosine identified in 23 CpG sites). These data
indicated that the DAC-promoted NEDD4-1 expression was not mediated
by any direct alteration of the NEDD4-1 promoter or of intron
methylation. Among the NCI-N87, SGC7901 and MGC803 cell lines, the
expression of NEDD4-1 revealed no correlation with its methylation
level (Fig. 4C).
Discussion
DNA methylation is a crucial epigenetic signature
that is implicated in transcriptional regulation, which occurs
predominantly within CpG sites (3,4). CpG
sites are underrepresented in the mammalian genome and tend to be
clustered within CGIs located in the vicinity of the TSSs of the
majority of the human protein-coding genes (5). The inhibition of DNA methylation
using cytidine analogues, including DAC, reactivates the expression
of genes, which were aberrantly silenced by hypermethylation. DAC
integrates itself into replicating DNA, thereby forming
irreversible covalent bonds with the active sites of DNA
methyltransferase (6).
DAC was demonstrated to have activity in
hematological disorders, including MDS, AML, CML and sickle cell
anemia (9–13). Since it is administered in the
clinic as an antitumor agent, DAC functions by completely
inhibiting the proliferation and the motility of the cells
(8,9,12–15).
However, since demethylation in the genome may alter the gene
expression of a large number of genes, the effect of DAC in
different tumor cell types remained controversial (16,17).
In the present study, it was demonstrated for the
first time, to the best of our knowledge, that DAC may promote
cell-invasive behavior in a well-defined gastric cancer cell line,
MGC803, whereas it revealed no affect on the invasive properties of
another cell line, SGC7901. (Figs. 1A
and B), However, in both cell lines, DAC inhibited cell
proliferation (Fig. 1C). During
the incubation with DAC, an already proven cancer-associated gene,
NEDD4-1, was demonstrated to be upregulated in the MGC803 cells.
NEDD4-1 exerts its activity as an E3-ubiquitin ligase, belonging to
the NEDD4 family (24). Previous
reports indicated that NEDD4-1 promotes glioma cell migration and
invasion via the ubiquitination and degradation of CNrasGEFs
(28). The results of the present
study also suggested that DAC promotes the invasion of MGC803 cells
via the upregulation of the expression of NEDD4-1 and by
restricting the expression of CNrasGEFs. The MGC803, SGC7901 and
NCI-N87 cell lines exhibited a marked difference in their
aggressive behavior (M>S>N) (29), and the expression levels of NEDD4-1
correlated with this trend (Fig.
4C). Knocking down the NEDD4-1 protein completely inhibited the
invasion-promoting effect of DAC, however, notably, the cell
behavior upon treatment with vehicle caused no change (Figs. 3B and C). These results
collectively suggested that the constitutive expression of NEDD4-1
may not be a key factor in the aggressive behavior exhibited by
MGC803 cells. Furthermore, no clear discernible differences in the
mRNA (data not shown) and protein (Fig. 4C) expression levels were observed
in these three cell lines, and similarly, no marked differences
were observed in the DNA methylation status of the CGIs in the
promoter and the first intron. It is therefore considered that DAC
affects the expression level of NEDD4-1 in a
DNA-methylation-independent manner, and the expression of NEDD4-1
in gastric cell lines may not be predominantly controlled by
methylation.
Although PTEN was confirmed to be a NEDD4-1
substrate and it inhibits tumor cell proliferation, the
upregulation of NEDD4-1 was incapable of promoting cell
proliferation (Fig. 1C). This
result may be accounted for by one of two hypotheses: (i) With the
exception of NEDD4-1, DAC may alter the expression of other genes,
including proliferation-associated genes; (ii) Other pathways are
responsible for regulating PTEN, independently of NEDD4-1. In the
present study, three commercial antibodies of PTEN were examined,
none of which proved capable of successfully detecting the
expression of PTEN in the MGC803, SGC7901 or the NCI-N87 cell
lines.
Taken together, the present study suggested that DAC
promoted cell-invasive behavior in the MGC803 gastric cancer cell
line by upregulating the level of NEDD4-1 and restricting the level
of CNrasGEFs. However, cell proliferation was inhibited, a result
which was consistent with previous studies performed on other
cancer cell lines (34,35). Furthermore, CPI methylation in
neither the NEDD4 promoter nor the first intron was observed. This
study has revealed that DAC exerts different effects in different
gastric cancer cell lines, which may provide a novel aspect for the
consideration of clinical applications of DAC in the future.
Acknowledgments
The authors would like to thank other members of the
Yu laboratory for a critical assessment of the manuscript. The
present study was supported by grants from the health department of
Heilongjiang Province (no. 2012–669), the education department of
Heilongjiang Province (no. 12531270), and a grant from the National
Natural Science Foundation of China (no. 81172417).
References
|
1
|
Melton SD, Genta RM and Souza RF:
Biomarkers and molecular diagnosis of gastrointestinal and
pancreatic neoplasms. Nat Rev Gastroenterol Hepatol. 7:620–628.
2010.PubMed/NCBI
|
|
2
|
Correa P and Houghton J: Carcinogenesis of
Helicobacter pylori. Gastroenterology. 133:659–672. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Macdonald JS, Smalley SR, Benedetti J,
Hundahl SA, Estes NC, Stemmermann GN, Haller DG, Ajani JA,
Gunderson LL, Jessup JM and Martenson JA: Chemoradiotherapy after
surgery compared with surgery alone for adenocarcinoma of the
stomach or gastroesophageal junction. N Engl J Med. 345:725–730.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
5
|
Robertson KD: DNA methylation,
methyltransferases and cancer. Oncogene. 20:3139–3155. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo M and Yan W: Epigenetics of gastric
cancer. Methods Mol Biol. 1238:783–799. 2015. View Article : Google Scholar
|
|
7
|
Illingworth RS, Gruenewald-Schneider U,
Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews
R and Bird AP: Orphan CpG islands identify numerous conserved
promoters in the mammalian genome. PLoS Genet. 6:e10011342010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nie J, Liu L, Li X and Han W: Decitabine,
a new star in epigenetic therapy: The clinical application and
biological mechanism in solid tumors. Cancer Lett. 354:12–20. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kantarjian H, Issa JP, Rosenfeld CS,
Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C,
Ravandi F, et al: Decitabine improves patient outcomes in
myelodysplastic syndromes: Results of a phase III randomized study.
Cancer. 106:1794–1803. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Issa JP, Garcia-Manero G, Giles FJ,
Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS,
Cortes J and Kantarjian HM: Phase 1 study of low-dose prolonged
exposure schedules of the hypomethylating agent
5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies.
Blood. 103:1635–1640. 2004. View Article : Google Scholar
|
|
11
|
Sacchi S, Kantarjian HM, O'Brien S, Cortes
J, Rios MB, Giles FJ, Beran M, Koller CA, Keating MJ and Talpaz M:
Chronic myelogenous leukemia in nonlymphoid blastic phase: Analysis
of the results of first salvage therapy with three different
treatment approaches for 162 patients. Cancer. 86:2632–2641. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kantarjian HM, O'Brien S, Cortes J, Giles
FJ, Faderl S, Issa JP, Garcia-Manero G, Rios MB, Shan J, Andreeff
M, et al: Results of decitabine (5-aza-2′deoxycytidine) therapy in
130 patients with chronic myelogenous leukemia. Cancer. 98:522–528.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lubbert M, Suciu S, Baila L, Rüter BH,
Platzbecker U, Giagounidis A, Selleslag D, Labar B, Germing U,
Salih HR, et al: Low-dose decitabine versus best supportive care in
elderly patients with intermediate- or high-risk myelodysplastic
syndrome (MDS) ineligible for intensive chemotherapy: Final results
of the randomized phaseIII study of the European organisation for
research and treatment of cancer Leukemia group and the German MDS
study group. J Clin Oncol. 29:1987–1996. 2011. View Article : Google Scholar
|
|
14
|
Shin DY, Kim GY, Kim CG, Kim WJ, Kang HS
and Choi YH: Anti-invasive effects of decitabine, a DNA
methyltransferase inhibitor, through tightening of tight junctions
and inhibition of matrix metalloproteinase activities in AGS human
gastric carcinoma cells. Oncol Rep. 28:1043–1050. 2012.PubMed/NCBI
|
|
15
|
Liu WH, Sang MX, Hou SY, Zhang C and Shan
BE: Low-dose decitabine induces MAGE-A expression and inhibits
invasion via suppression of NF-κB2 and MMP2 in Eca109 cells. Biomed
Pharmacother. 68:745–750. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiao F, Bai SY, Ma Y, Yan ZH, Yue Z, Yu Y,
Wang X and Wang J: DNA methylation of heparanase promoter
influences its expression and associated with the progression of
human breast cancer. PLoS One. 9:e921902014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Borges S, Döppler HR and Storz P: A
combination treatment with DNA methyltransferase inhibitors and
suramin decreases invasiveness of breast cancer cells. Breast
Cancer Res Treat. 144:79–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rougier JS, Albesa M, Abriel H and Viard
P: Neuronal precursor cell-expressed developmentally downregulated
4-1 (NEDD4-1) controls the sorting of newly synthesized Ca (V) 1.2
calcium channels. J Biol Chem. 286:8829–8838. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kwak YD, Wang B, Pan W, Xu H, Jiang X and
Liao FF: Functional interaction of phosphatase and tensin homologue
(PTEN) with the E3 ligase NEDD4-1 during neuronal response to zinc.
J Biol Chem. 285:9847–9857. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Oppenheim RW, Sugiura Y and Lin W:
Abnormal development of the neuromuscular junction in
Nedd4-deficient mice. Dev Biol. 330:153–166. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kawabe H, Neeb A, Dimova K, Young SM Jr,
Takeda M, Katsurabayashi S, Mitkovski M, Malakhova OA, Zhang DE,
Umikawa M, et al: Regulation of Rap2A by the ubiquitin ligase
Nedd4-1 controls neurite development. Neuron. 65:358–372. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kawabe H and Brose N: The ubiquitin E3
ligase Nedd4-1 controls neurite development. Cell Cycle.
9:2477–2478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lackovic J, Howitt J, Callaway JK, Silke
J, Bartlett P and Tan SS: Differential regulation of Nedd4
ubiquitin ligases and their adaptor protein Ndfip1 in a rat model
of ischemic stroke. Exp Neurol. 235:326–335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kwak YD, Wang B, Li JJ, Wang R, Deng Q,
Diao S, Chen Y, Xu R, Masliah E, Xu H, et al: Upregulation of the
E3 ligase NEDD4-1 by oxidative stress degrades IGF-1 receptor
protein in neurodegeneration. J Neurosci. 32:10971–10981. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Trotman LC, Koppie T, Alimonti A,
Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo
C, et al: NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN.
Cell. 128:129–139. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Amodio N, Scrima M, Palaia L, Salman AN,
Quintiero A, Franco R, Botti G, Pirozzi P, Rocco G, De Rosa N and
Viglietto G: Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a
PTEN negative regulator, in non-small-cell lung carcinomas. Am J
Pathol. 177:2622–2634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim SS, Yoo NJ, Jeong EG, Kim MS and Lee
SH: Expression of NEDD4-1, a PTEN regulator, in gastric and
colorectal carcinomas. APMIS. 116:779–784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang H, Nie W, Zhang X, Zhang G, Li Z, Wu
H, Shi Q, Chen Y, Ding Z, Zhou X and Yu R: NEDD4-1 regulates
migration and invasion of glioma cells through CNrasGEF
ubiquitination in vitro. PLoS One. 8:e827892013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun A, Yu G, Dou X, Yan X, Yang W and Lin
Q: Nedd4-1 is an exceptional prognostic biomarker for gastric
cardia adenocarcinoma and functionally associated with metastasis.
Mol Cancer. 13:2482014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin F, Lin P, Zhao D, Chen Y, Xiao L, Qin
W, Li D, Chen H, Zhao B, Zou H, et al: Sox2 targets cyclinE, p27
and survivin to regulate androgen-independent human prostate cancer
cell proliferation and apoptosis. Cell Prolif. 45:207–216. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Wang H, Jiang N, Lu W, Zhang XF
and Fang JY: Effect of inhibition of MEK pathway on
5-aza-deoxycytidine-suppressed pancreatic cancer cell
proliferation. Genet Mol Res. 12:5560–5573. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang B, Li H, Yang R, Zhou S and Zou S:
Decitabine inhibits the cell growth of cholangiocarcinoma in
cultured cell lines and mouse xenografts. Oncol Lett. 8:1919–1924.
2014.PubMed/NCBI
|
|
33
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Everson RG, Antonios JP, Lisiero DN, Soto
H, Scharnweber R, Garrett MC, Yong WH, Li N, Li G and Kruse CA: et
al Efficacy of systemic adoptive transfer immunotherapy targeting
NY-ESO-1 for glioblastoma. Neuro Oncol. Sep 1–2015.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yan W, Herman JG and Guo M:
Epigenome-based personalized medicine in human cancer. Epigenomics.
Sep 7–2015.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|