Introduction
Liver fibrosis, characterized by excess production
and deposition of extracellular matrix (ECM) along with loss of
liver function and disruption of liver structure, is a
wound-healing response to chronic liver injury (1,2). It
is well known that hepatic stellate cells (HSCs) have an important
role in liver fibrosis. The activation and proliferation of
resident hepatic stellate cells (HSCs) has been considered as a
central event in the progression of liver fibrosis. During fibrosis
progression, quiescent (Q)-HSCs become activated and
transdifferentiate into myofibroblastic HSCs (MF-HSCs) that express
smooth-muscle α-actin (α-SMA) as a marker (3). Activation of HSC, the major cell type
promoting synthesis and deposition of ECM proteins, results in an
imbalance between ECM protein generation and their degradation in
the liver (4). Therefore, a
possible therapeutic strategy for treating liver fibrosis is to
inhibit the activation of HSCs.
Epithelial-to-mesenchymal transition (EMT) is the
process by which epithelial cells gradually lose their epithelial
signatures while acquiring the characteristics of mesenchymal cells
(5). A growing body of evidence
implied that EMT has an important role in liver fibrosis (6,7). In
particular, it has been confirmed that myofibroblasts can be
supplemented from hepatocytes by EMT during hepatic fibrosis
(8). EMT has emerged as a
promising therapeutic target for the attenuation of liver fibrosis.
Hedgehog (Hh) signaling has been identified to be involved in EMT.
Choi et al (9) demonstrated
that EMT is regulated by Hh signaling during myofibroblastic
transformation of rat hepatic cells in culture and cirrhosis.
Omenetti et al (7) found
that EMT responses to bile duct ligation were enhanced in
patched-deficient mice, which display excessive activation of the
Hh pathway. A previous study showed that leptin, a pro-EMT factor,
activates Hh signaling to alter the expression of genes which
control cell fate and have important implications in liver fibrosis
(10). These findings suggested
that Hh signaling contributes to EMT.
MicroRNAs (miRNAs) are endogenous small (18–22 nt)
non-coding RNAs that regulate the expression of proteins involved
in diverse cellular and developmental processes, including
differentiation, apoptosis and oncogenesis (11). miRNAs specifically bind to the
3′-untranslated region (3′-UTR) of their respective target mRNAs to
decrease their stability and translation to regulate protein
expression (12). During liver
fibrosis, miRNAs contribute to the activation status of HSCs and
function as HSC regulators in liver fibrosis. Sun et al
(13) reported that miR-200a
overexpression suppressed HSC activation and proliferation by
targeting TGF-β2 and β-catenin. Given the complexity of the EMT, it
is likely that miRNAs regulate most genes involved in the EMT.
Previously, it has been reported that the miR-200 family regulates
transforming growth factor (TGF)-β1-induced renal tubular EMT
through the Smad pathway by targeting the expression of zinc finger
E-box binding homeobox 1 (ZEB1) and ZEB2 (14). However, it has remained elusive
whether miR-200a regulates EMT in HSCs.
The present study assessed the effects of miR-200a
on the proliferation of activated HSCs; furthermore, western blot
analysis, reverse-transcription quantitative polymerase chain
reaction (RT-qPCR) and immunofluorescence were used to assess the
expression of epithelial marker E-cadherin and myofibroblastic
markers α-SMA, type I collagen and desmin in order to investigate
the effects of miR-200a on the EMT of cultured HSCs. In addition,
the effects of miR-200a on the expression of Hh pathway-associated
genes, including Hh-interacting protein (Hhip), Sonic Hh (Shh) and
GLI family zinc finger 1 (Gli1) and Gli2, as well as genes
associated with the EMT, including bone morphogenetic protein-7
(BMP-7), inhibitor of DNA binding 2 (Id2), Snail family zinc finger
1 (Snai1) and S100 calcium-binding protein A4 (S100A4) were
assessed. A luciferase reporter assay was then employed to
investigate whether Gli2 was a direct target of miR-200a. The
present study suggested that miR-200a suppressed EMT in rat HSCs,
at least in part, via Gli2.
Materials and methods
Materials
CCl4 was obtained from Sigma-Aldrich (St.
Louis, MO, USA) and antibodies, including rabbit polyclonal
anti-type I collagen (cat. no. ab34710), anti-desmin (cat. no.
ab15200) and anti-S100A4 (cat. no. ab27957), and mouse monoclonal
anti-E-cadherin (cat. no. ab76055), anti-α-SMA (cat. no. ab7817),
and anti-β-actin (cat. no. ab6276) were obtained from Abcam
(Cambridge, MA, USA). Antibodies including goat polyclonal
anti-BMP-7 (cat. no. sc-9305), anti-Id2 (cat. no. sc-26328), and
anti-Hhip (cat. no. sc-9406), and rabbit polyclonal anti-Snai1
(cat. no. sc-28199), anti-Shh (cat. no. sc-9024), anti-Gli1 (cat.
no. sc-20687) and anti-Gli2 (cat. no. sc-28674) were purchased from
Santa Cruz Biotechnology (Dallas, TX, USA).
Isolation and culture of rat HSCs
Adult male Sprague-Dawley rats (body weight, 400–500
g) were used for HSC isolation as described previously (15). The liver tissues were digested with
collagenase IV (0.5 g/l) and deoxyribonuclease I (0.03 g/l) prior
to fractionation on a discontinuous gradient of iodixanol. HSCs
were harvested from the 11.5% medium interface, washed and seeded
in tissue culture plates. Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific,
Waltham, MA, USA) with 10% fetal bovine serum (FBS; Sigma-Aldrich),
100 U/ml penicillin and 100 µg/ml streptomycin (Gibco;
Thermo Fisher Scientific, Inc.). The harvested primary HSCs were
used for subsequent experiments at day 3 after isolation. The
purity of cultures (>98%) was confirmed by immunocytochemical
staining for α-SMA. The present study was approved by the
University Animal Care and Use Committee of the Wenzhou Medical
University (Wenzhou, China).
miRNA transfection
Cells were seeded in a six-well plate at a density
of 1×106 cells per well. The following day, medium was
replaced with Opti-Minimum Essential Medium (Invitrogen; Thermo
Fisher Scientific) and cells were transiently transfected with
miR-200a mimics (60 nM) or the miR-negative control (NC; 60 nM;
GenePharma, Shanghai, China) using Lipofectamine 2000 (Invitrogen)
for 48 h in all experiments.
Rat model of CCl4-induced
liver injury
Male Sprague-Dawley rats (n=20; weighy, 180–220 g;
age, 7 weeks) were obtained from the Experimental Animal Center of
the Wenzhou Medical University, and housed in an environmentally
controlled room (23±2°C; 55±10% humidity) with a 12 h light/dark
cycle, and were provided ad libitum access to food and
water. Liver fibrosis was generated by a 6 week treatment with
CCl4 [CCl4/olive oil, 1:1 (v/v) per kg body
weight by intraperitoneal injection twice weekly] for six weeks as
previously described (16). Twenty
rats were randomly divided into two groups. Rats in group 1 (n=10)
received injections with olive oil (vehicle control) and rats in
group 2 (n=10) received injections of CCl4 twice weekly.
The animal experimental protocol was approved by the University
Animal Care and Use Committee (Wenzhou Medical University, Wenzhou,
China). Rats were sacrificed under anesthesia by intraperitoneal
injection of 10% chloral hydrate (Sigma-Aldrich; 4 ml/kg body
weight) after the last CCl4 injection and liver tissues
were harvested for further analysis.
Immunofluorescence microscopy
Primary HSCs on day 3 following inoculation were
plated on 18-mm cover glasses in DMEM at a density of
1×105 cells/well and incubated for 24 h. The cells were
then transfected with miR-200a mimics or miR-NC for 48 h. Following
washing with phosphate-buffered saline (PBS; Wenzhou Changfeng
Biotechnology Co., Ltd., Wenzhou, China) and fixing in acetic
acid/ethanol (30% ethanol and 10% acetic acid; Wenzhou Changfeng
Biotechnology Co., Ltd.) for 5 min at −20°C, non-specific binding
was blocked with 5% goat serum (Sigma-Aldrich) in PBS for 1 h at
room temperature and the cells were then incubated overnight at 4°C
with primary antibodies against α-SMA (1:200), type I collagen
(1:100), E-cadherin (1:100) or desmin (1:100) in a humidified
chamber. After washing twice in PBS, the cells were incubated for 1
h at room temperature with fluorescein-labeled secondary antibody
(1:50 dilution; Dianova, Hamburg, Germany) in antibody dilution
solution (Beyotime Institute of Biotechnology, Haimen, China) for 1
h at room temperature in the dark. The nuclei were stained with
4,6-diamidino-2-phenylindole (DAPI; Abcam) in the dark for 30 min
at room temperature. The slides were washed twice with PBS, covered
with DABCO (Sigma-Aldrich), and examined using confocal laser
scanning microscopy (FV-1000; Olympus, Tokyo, Japan) at 488 and 568
nm.
RT-qPCR
The cells were collected and total RNA was extracted
from cells using the miRNeasy Mini kit (Qiagen, Hilden, Germany)
and cDNA was synthesized according to the manufacturer's
instructions (Toyobo, Osaka, Japan). Gene expression was measured
by real-time PCR using cDNA, SYBR Green real-time PCR Master Mix
(Toyobo), and a set of gene-specific oligonucleotide primers
(Invitrogen; Thermo Fisher Scientific, Inc.): Id2 forward, 5′-CCT
CCT ACG AGC AGC ATGAA-3′ and reverse, 5′-GGC ACC AGT TCC TTGA
GCTT-3′; desmin forward, 5′-ATG TCAC AC CCA GTC GCTTT-3′ and
reverse, 5′-GAT GGC AGG GAA AAG GGT CA-3′; Gli1 forward, 5′-TTG CAG
CCA GGA GTT CGATT-3′ and reverse, 5′-GGA CTT CCG ACA GCC TTCAA-3′.
The sequences of primers for Col1A1, α-SMA, GAPDH, U6, E-cadherin,
BMP-7, S100A4, Snai1, Hhip, Shh and Gli2 identical to those used in
previous studies (9,17). To detect miR-200a expression, the
RT reaction was performed using the TaqMan MicroRNA assay (Applied
Biosystems, Thermo Fisher Scientific) according to the
manufacturer's instructions. RT-qPCR was performed in an ABI 7500
Real Time PCR system (Applied Biosystems), according to the
manufacturer's protocol, with the following cycles: 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 60 sec.
GAPDH and U6 small nuclear RNA levels were measured and used to
normalize the relative abundance of mRNAs and miRNA, respectively.
The 2−ΔΔCt method was used to determine the miRNA and
mRNA levels as described previously (18). In addition, the specificity of
RT-qPCR products was confirmed by melting curve analysis.
Protein extraction and western blot
analysis
The protein concentration of samples was determined
using a bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Total protein (20–50 µg) was subjected to
10–12% SDS-PAGE (Beyotime Institute of Biotechnology) and then
transferred onto Immobilon P membranes (Beyotime Institute of
Biotechnology). The membranes were incubated in blocking buffer
[(Beyotime Institute of Biotechnology) 5% non-fat milk powder in
Tris-buffered saline with Tween 20 (TBST; 100 mM Tris-HCl pH 7.5,
0.9% NaCl, 0.1% Tween 20)] for 3 h at room temperature, followed by
incubation overnight at 4°C with gentle agitation with specific
primary antibodies against type I collagen (1:1,000), desmin
(1:1,000), S100A4 (1:1,000), E-cadherin (1:500), α-SMA (1:500),
β-actin (1:2,000), BMP-7 (1:500), Id2 (1:1,000), Hhip (1:1,000),
Snai1 (1:500), Shh (1:500), Gli1 (1:500), and Gli2 (1:500).
Following washing with TBST, the membranes were incubated with
peroxidase-conjugated secondary antibodies (Fuzhou Maixin
Biological Technology Co., Ltd., Fujian, China) for 1 h at room
temperature. Unbound antibody was washed and removed with TBST, the
antigen-antibody complex was developed by enhanced
chemiluminescence using BeyoECL Plus (cat. no. P0018; Beyotime
Institute of Biotechnology), and images were captured using a Gel
Imager (WD-9413B; Beijing Liuyi Biotechnology Co., Ltd., Beijing,
China) in a dark room and analyzed for integral absorbance (IA) of
the protein bands using Quantity One 4.4 software (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Proliferation assay
Cell proliferation was determined using the MTT
assay (Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Briefly, the cells were seeded at a
density of 5×103 cells per well in 96-well culture
plates and transfected with miR-200a mimics or miR-NC. The cells
were incubated with 0.5% MTT for 4 h. Following removal of the
supernatant, 150 µl dimethyl sulfoxide (Sigma-Aldrich) was
added and plates were agitated for 5 min until the formazan
crystals had dissolved. The optical density was determined using a
microplate reader (Bio-Rad 550; Bio-Rad Laboratories, Inc.) at 490
nm wavelength. All experiments were performed in triplicate and
repeated at least three times.
Luciferase reporter assay
According to a target analysis with Targetscan
(http://www.targetscan.org/), sequences
containing rat Gli2 3′-UTR target sequence (located at 651–657 bp),
were amplified and cloned into the pMIR-Report™ Luciferase plasmid
(Applied Biosystems) using mouse cDNA as template to generate
pMIR-Gli2-200a vector. The primers for Gli2-3′-UTR (forward,
5′-TGCATCCATGAAGTTCGCCA-3′ and reverse, 5′-GAGAGGTCAG
GGGACCAGAA-3′) were obtained from Invitrogen (Thermo Fisher
Scientific, Inc.). The amplification conditions for Gli2-3′-UTR
were the same as those demonstrated in the RT-qPCR section.
pMIR-Gli2-200a-Mut was generated using a Site-Directed Mutagenesis
kit (Agilent Technologies, Inc., Santa Clara, CA, USA) according to
the manufacturer's instructions, using the primers containing the
desired mutation (19), provided
by Dr Zhou (Yuying Children's Hospital of Wenzhou Medical
University, Wenzhou, China). In addition, empty vector pMIR without
the inserts was used as a negative control. pMIR-Report β-gal
control plasmid was used for transfection normalization. Cells were
cultured in 24-well plates and transfected with 800 ng pMIR-200a or
pMIR together with 100 ng pMIR-β-gal and 20 pmol miR-200a precursor
or miRNA negative control (miR-NC) (GenePharma). Lipofectamine 2000
(Invitrogen) was used for transfection. Forty-eight hours after
transfection, luciferase and β-gal activity were measured using the
Dual-Light System (Applied Biosystems).
Statistical analysis
Values are expressed as the mean ± standard
deviation from at least three independent experiments. Statistical
analysis was performed using Student's t-test and P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed with SPSS software (version 13;
SPSS, Inc., Chicago, IL, USA).
Results
MiR-200a is decreased in fibrotic livers
and its upregulation suppresses the proliferation of HSCs
The present study used standard techniques to
isolate and culture HSC from the livers of healthy rats. The
expression of miR-200a was detected in primary HSCs using RT-qPCR,
which revealed that miR-200a expression gradually decreased with
increasing time in culture (Fig.
1A). In addition, a reduction of miR-200a expression was
detected in liver tissues from CCl4-treated rats
compared with that in the control animals (Fig. 1A). An MTT assay revealed that
overexpression of miR-200a significantly suppressed HSC
proliferation (P<0.05) (Fig.
1B).
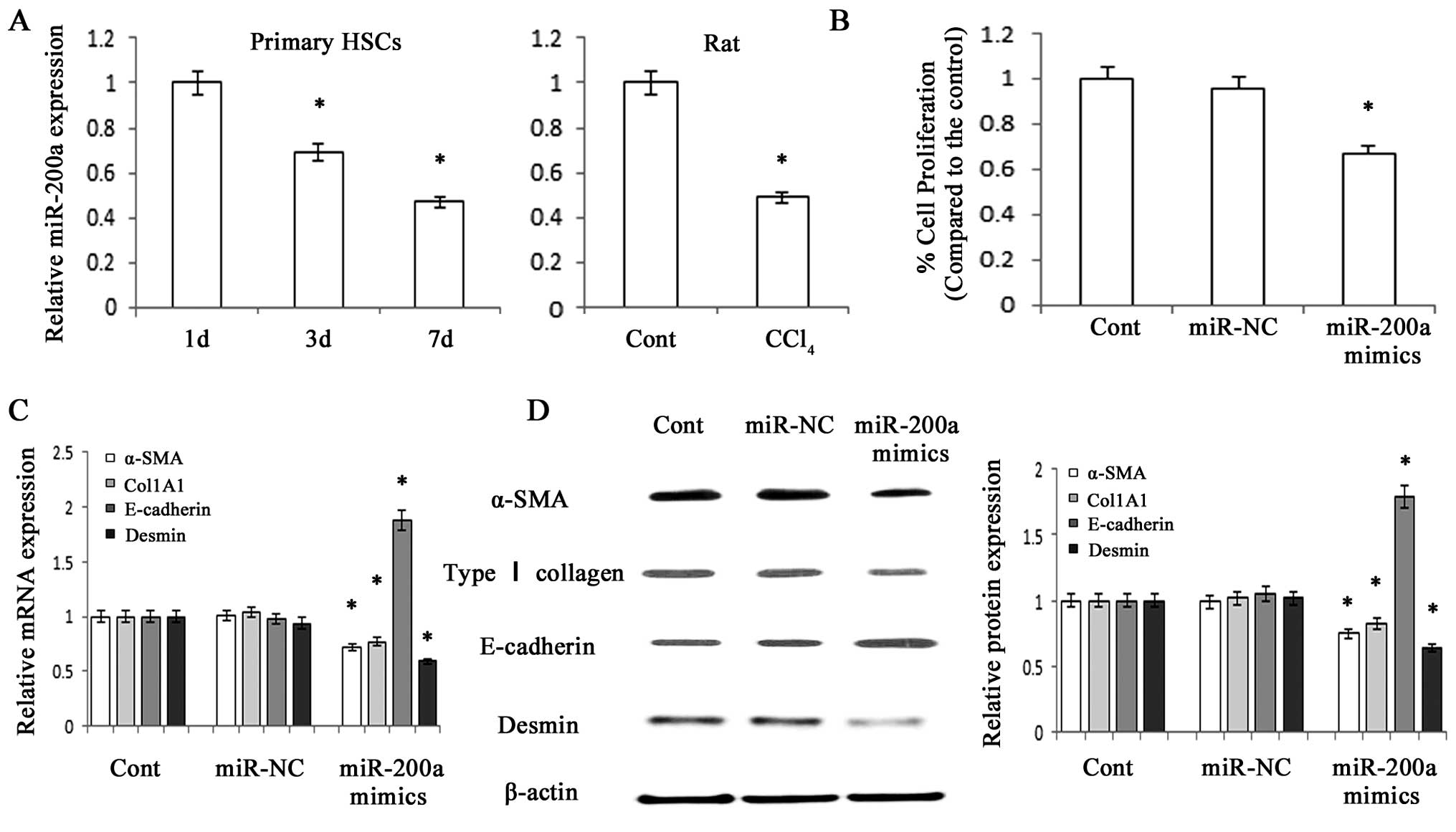 | Figure 1miR-200a is decreased during HSC
activation and contributes to the suppression of activated HSCs.
Primary HSCs at three days after isolation were transfected with
miR-200a mimics or miR-NC for 48 h. (A) miR-200a expression in
primary HSCs and CCl4-treated hepatic fibrotic tissues
was analyzed by RT-qPCR. (B) The growth rate, which was analyzed
using an MTT assay, was decreased by miR-200a mimics. (C) The mRNA
expression of α-SMA, Col1A1, E-cadherin and desmin in primary HSCs
was analyzed by RT-qPCR. (D) The protein expression of α-SMA, type
I collagen, E-cadherin and desmin was analyzed by western blot
analysis in primary HSCs. β-actin was used as an internal control.
Values are expressed as the mean ± standard deviation of three
experiments. *P<0.05 vs. control. miR, microRNA; HSC,
hepatic stellate cell; α-SMA, smooth-muscle α-actin; RT-qPCR,
reverse-transcription quantitative polymerase chain reaction; Cont,
control; NC, negative control. |
miR-200a suppresses EMT in activated
HSCs
Type I collagen, α-SMA and desmin are considered to
be myofibroblast-associated markers, while E-cadherin is an
epithelial cell marker. The present study therefore assessed the
protein and mRNA expression of these genes in order to elucidate
the effects of miR-200a on the EMT. RT-qPCR and western blot
analysis indicated that overexpression of miR-200a led to a
reduction of the mRNA and protein expression of the myofibroblastic
markers type I collagen and α-SMA and desmin, while the expression
of epithelial cell marker E-cadherin was upregulated (P<0.05)
(Fig. 1C and D). In analogy with
these results, immunofluorescence staining further confirmed that
the levels of type I collagen and α-SMA were reduced by miR-200a
overexpression, while E-cadherin expression was enhanced (Fig. 2). These results suggested that
miR-200a overexpression inhibited the EMT of HSCs, indicating that
miR-200a may attenuate liver fibrosis partly through inhibiting
EMT.
miR-200a prevents EMT by interfering with
Hh pathway activity
A previous study demonstrated that EMT is be
promoted by the activation of the Hh pathway in primary rat HSCs
(7). As the Hh pathway is involved
in the regulation of the EMT, the present study detected the
effects of miR-200a on the expression of Hhip (a competitive
antagonist of Hh ligands), Shh (Hh ligand), Gli1 and Gli2 (Hh
target genes) in primary HSCs. The results revealed that the
expression of Hhip, Shh and Gli1 was not affected by miR-200a
mimics (Fig. 3A and B). However,
the expression of Gli2, a downstream signaling molecule of the Hh
pathway, was markedly inhibited by miR-200a at the mRNA and protein
level (P<0.05) (Fig. 3A and B).
These results indicated that the Hh pathway was inhibited by
miR-200a. In general, upregulation of the Hh pathway is associated
with altered expression of several factors that regulate EMT,
including decreases in BMP-7 and Id2 (genes that inhibit EMT) and
increases in Snai1 and S100A4 (genes that promote EMT) (9). The present study further investigated
the effects of miR-200a on genes involved in the EMT. The mRNA and
protein expression of BMP-7 and Id2 was significantly upregulated
following miR-200a overexpression, while the mRNA expression of
Snai1 and S100A4 was downregulated (P<0.05) (Fig. 3C and D). These results suggested
that miR-200a suppressed EMT via the Hh pathway.
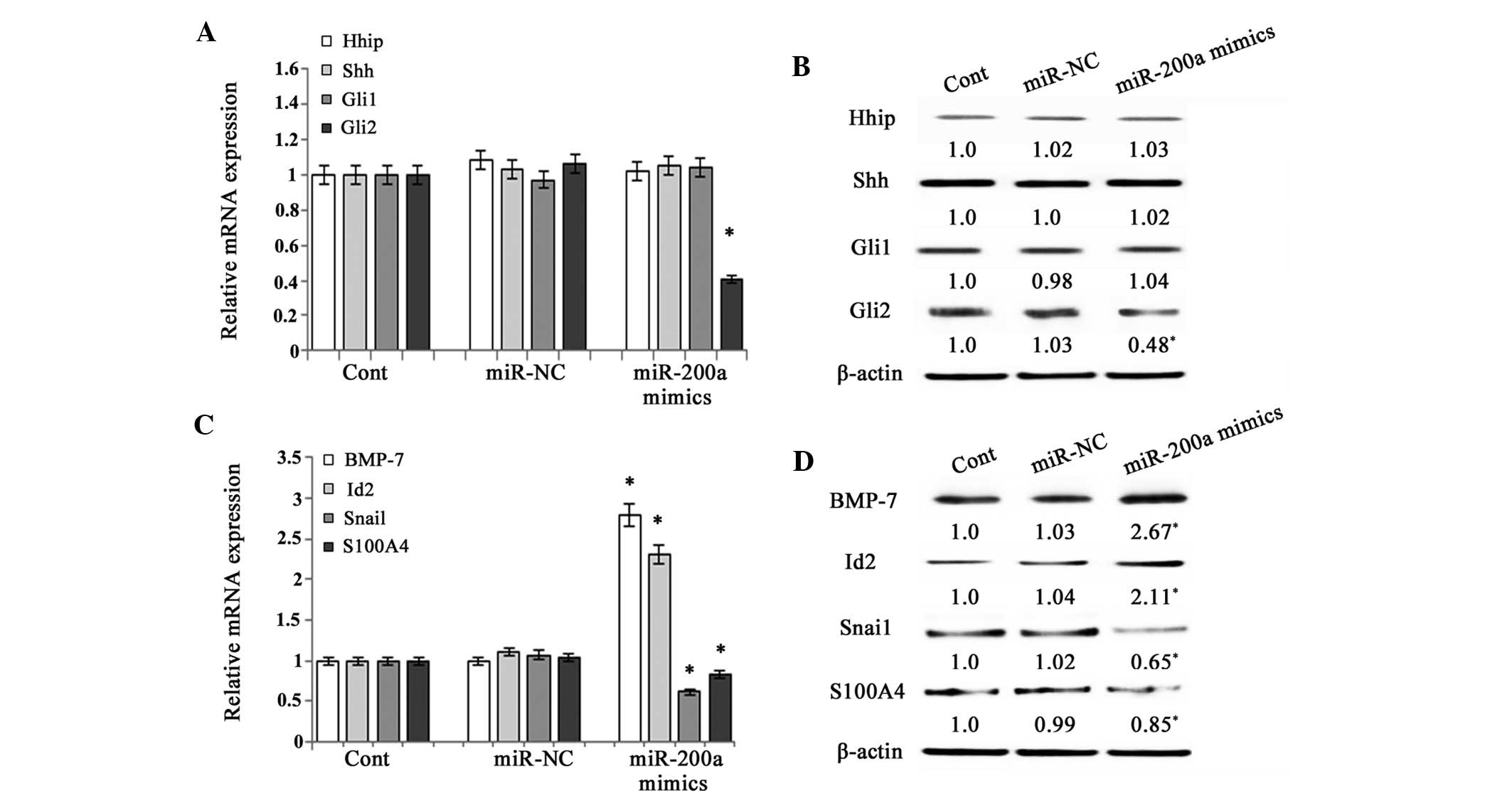 | Figure 3Effects of miR-200a on Hedgehog
pathway activity. Primary HSCs on day 3 after isolation and were
transfected with miR-200a mimics or miR-NC for 48 h. (A) The mRNA
expression of Hhip, Shh, Gli1 and Gli2 in primary HSCs was analyzed
by RT-qPCR. (B) The protein expression of Hhip, Shh, Gli1 and Gli2
in primary HSCs was assessed by western blot analysis. β-actin was
used as an internal control. (C) The mRNA expression of BMP-7,
Id-2, Snai1 and S100A4 in primary HSCs was analyzed by RT-qPCR. (D)
The protein expression of BMP-7, Id2, Snai1 and S100A4 was assessed
by western blot analysis in primary HSCs. β-actin was used as an
internal control. Values are expressed as the mean ± standard
deviation of three experiments. *P<0.05 compared with
the control. HSC, hepatic stellate cell; RT-qPCR,
reverse-transcription quantitative polymerase chain reaction; Cont,
control; NC, negative control; BMP, bone morphogenetic protein;
Id2, inhibitor of DNA binding 2; S100A4, S100 calcium-binding
protein A4; Shh, Sonic hedgehog; Hhip, Hedgehog-interacting
protein; GL1, GLI family zinc finger 1. |
Gli2 is a direct target of miR-200a in
rat HSC
To confirm the underlying mechanism of miR-200a
suppressing the Hh pathway, a bioinformatics analysis using
Targetscan was performed to search for targets of miR-200a. Gli2, a
downstream component of the Hh pathway, was predicted as a putative
target of miR-200a. Next, the present study used a luciferase
reporter assay to investigate whether Gli2 was a direct target of
miR-200a. The miR-200a-specific target region of the 3′UTR of Gli2
mRNA was cloned into the pMIR-Report™ Luciferase plasmid, which was
co-transfected into primary HSCs along with miR-200a precursor or
miR-NC (Fig. 4A and B). β-gal
reporter control plasmid was co-transfected to monitor transfection
efficiency. The results showed that the luciferase activity in the
reporter plasmid containing the wild-type 3′UTR of Gli2 was
significantly reduced by miR-200a precursor, while the luciferase
activities of the plasmid containing the mutated-type Gli2 3′UTR
and empty vector were not affected (Fig. 4C). In addition, Gli2 expression was
reduced by miR-200a mimics in primary HSCs (Fig. 3A and B). These results suggested
that Gli2 was a direct target of miR-200a.
Discussion
Emerging studies have indicated that the EMT is
involved in the transformation of Q-HSCs into MF-HSCs (9,20).
During the transformation of Q-HSC into MF-HSC in the present
study, HSCs acquired a more mesenchymal phenotype as characterized
by robust expression of several myofibroblastic markers, including
α-SMA and Col1A1. Simultaneously, the epithelial characteristics,
including E-cadherin expression, were lost in the EMT process. This
observation was in accordance with those of a previous study, which
reported that freshly isolated primary Q-HSC expressed epithelial
markers and subsequently acquired a myofibroblastic phenotype via
EMT under standard culturing conditions (9). For this reason, the present study
used freshly isolated primary HSCs (3 days following isolation) in
all experiments.
A previous study showed that miR-200a exhibits
activity against liver fibrosis (13); however, the underlying mechanism
and the possible involvement of the ECM had remained elusive.
Therefore, the present study assessed the mechanism of action of
miR-200a in HSCs and identified its direct target. Initially, the
present study revealed that miR-200a was reduced during activation
of primary HSCs and in HSCs isolated from a rat model of liver
fibrosis; furthermore, miR-200a upregulation and contributed to the
suppression of activated HSCs, leading to a reduction in cell
proliferation, ECM production and α-SMA expression. The results of
the present study were consistent with those of a previous study
(13). miRNAs are known to
function as key factors to regulate cell proliferation,
differentiation and apoptosis (21). miRNAs are also involved in
processes associated with the EMT. For example, a recent study
reported that the EMT of colon cancer cells was reversed by
resolving the inhibition of miR-200 cluster expression (22). However, few studies regarding the
regulation of the EMT by miRNAs in liver fibrosis have been
performed (23). The present study
assessed the effects of miR-200a on the EMT in primary HSCs. Of
note, myofibroblastic markers α-SMA, Col1A1 and desmin were reduced
by miR-200a in primary HSCs at the protein and mRNA level, while
the expression of epithelial marker E-cadherin was increased.
Immunofluorescence staining further confirmed these findings. These
results suggested that miR-200a inhibited the activation of HSCs
via the inhibition of the EMT.
Next, the present study investigated the molecular
mechanism by which miR-200a inhibited the EMT in primary HSCs.
Numerous studies have demonstrated that the Hh pathway is involved
in the EMT of activated HSCs, and that it modulates MF-HSC
accumulation and liver fibrosis (9,10,24,25).
For instance, increased expression of Shh ligand during liver
injury was shown to drive EMT by promoting MF-HSC proliferation and
viability (26). The Hh pathway is
important in the transition of Q-HSC to MF-HSC, as Hh ligands were
shown to be required for the transformation of Q-HSC into MF-HSC
and to be essential for MF-HSC to retain their fibroblastic
phenotype (24,26). These studies suggested that
activation of the Hh pathway may promote liver fibrosis via the
EMT. Therefore, when Hh ligands interact with their receptors, the
Hh pathway is activated, resulting in the activation and nuclear
localization of GLI family transcription factors (27). For this reason, the present study
assessed the effects of miR-200a on the activation of the Hh
pathway in primary HSCs. The results showed that miR-200a inhibited
the expression of the Hh downstream signaling molecule Gli2, while
Hhip, Shh and Gli1 were not affected. A previous study showed that
the suppression of the Hh pathway led to changes in the expression
of genes associated with the EMT, including an increase of EMT
inhibitors BMP-7 and Id2 and a reduction of EMT promoters Snai1 and
S100A4 (28). Consistent with the
results of this previous study, the present study found that
miR-200a mimics induced an increase of BMP-7 and Id2 while reducing
the expression of Snai1 and S100A4. These results confirmed the
inhibition of downstream signaling processes of the Hh pathway by
miR-200a. To identify the target of miR-200a within the Hh pathway,
a bioinformatics analysis with Targetscan was used. The downstream
signaling molecule Gli2 was indicated to be a direct target of
miR-200a, which was experimentally verified using a dual luciferase
reporter assay. A previous study indicated the role of miR-200
family in regulating TGF-β1-induced EMT in renal tubular cells
through the Smad pathway by targeting ZEB1 and ZEB2 expression
(14); however, the mechanism of
action of miR-200a in liver fibrosis as well as the effects of
other miR-200 family members on the EMT have remained elusive. The
present study suggested that miR-200a inhibited EMT via reducing Hh
signaling, at least in part, via its direct target Gli2. Targetscan
analysis indicated that other members of the Hh signaling pathway,
including Hhip, Gli1 and Gli2, were not targeted by any additional
miR-200 family members (i.e., miR-200b, miR-429, miR-200c and
miR-141) (data not shown). These results indicated that miR-200a
had a unique and vital role in the Hh pathway.
In conclusion, the results of the present study
provided novel insight into the role of miRs in liver fibrosis;
miR-200a inhibited the fibrosis-associated EMT process in HSCs, at
least in part, via blocking Gli2, a downstream signaling molecule
of the Hh pathway.
Acknowledgments
The authors of the present study are grateful to Dr
Zhou for the donation of specific primers containing the desired
mutation. The study was supported by the National Natural Science
Foundation of China (grant nos. 81000176/H0317, 81100292/H0317, and
81500458/H0317), the Zhejiang Provincial Natural Science Foundation
of China (grant nos. Y2090326, Y2110634 and LY16H030012), the Wang
Bao-En Liver Fibrosis Foundation (grant nos. 20100002 and 20120127)
and the Wenzhou Municipal Science and Technology Bureau (grant nos.
Y20110033 and Y20120127).
References
|
1
|
Yue HY, Yin C, Hou JL, Zeng X, Chen YX,
Zhong W, Hu PF, Deng X, Tan YX, Zhang JP, et al: Hepatocyte nuclear
factor 4alpha attenuates hepatic fibrosis in rats. Gut. 59:236–246.
2010. View Article : Google Scholar
|
|
2
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ogawa T, Iizuka M, Sekiya Y, Yoshizato K,
Ikeda K and Kawada N: Suppression of type I collagen production by
microRNA-29b in cultured human stellate cells. Biochem Biophys Res
Commun. 391:316–321. 2010. View Article : Google Scholar
|
|
4
|
Kawada N: The hepatic perisinusoidal
stellate cell. Histol Histopathol. 12:1069–1080. 1997.PubMed/NCBI
|
|
5
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Ann Rev Pathol. 6:425–456. 2011.
View Article : Google Scholar
|
|
7
|
Omenetti A, Porrello A, Jung Y, Yang L,
Popov Y, Choi SS, Witek RP, Alpini G, Venter J, Vandongen HM, et
al: Hedgehog signaling regulates epithelial-mesenchymal transition
during biliary fibrosis in rodents and humans. J Clin Invest.
118:3331–3342. 2008.PubMed/NCBI
|
|
8
|
Zeisberg M, Yang C, Martino M, Duncan MB,
Rieder F, Tanjore H and Kalluri R: Fibroblasts derive from
hepatocytes in liver fibrosis via epithelial to mesenchymal
transition. J Biol Chem. 282:23337–23347. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Choi SS, Omenetti A, Witek RP, Moylan CA,
Syn WK, Jung Y, Yang L, Sudan DL, Sicklick JK, Michelotti GA, et
al: Hedgehog pathway activation and epithelial-to-mesenchymal
transitions during myofibroblastic transformation of rat hepatic
cells in culture and cirrhosis. Am J Physiol Gastrointest Liver
Physiol. 297:G1093–G1106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choi SS, Syn WK, Karaca GF, Omenetti A,
Moylan CA, Witek RP, Agboola KM, Jung Y, Michelotti GA and Diehl
AM: Leptin promotes the myofibroblastic phenotype in hepatic
stellate cells by activating the hedgehog pathway. J Biol Chem.
285:36551–36560. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun X, He Y, Ma TT, Huang C, Zhang L and
Li J: Participation of miR-200a in TGF-β1-mediated hepatic stellate
cell activation. Mol Cell Biochem. 388:11–23. 2014. View Article : Google Scholar
|
|
14
|
Xiong M, Jiang L, Zhou Y, Qiu W, Fang L,
Tan R, Wen P and Yang J: The miR-200 family regulates
TGF-β1-induced renal tubular epithelial to mesenchymal transition
through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J
Physiol Renal Physiol. 302:F369–F379. 2012. View Article : Google Scholar
|
|
15
|
Weiskirchen R and Gressner AM: Isolation
and culture of hepatic stellate cells. Methods Mol Med. 117:99–113.
2005.PubMed/NCBI
|
|
16
|
Yao QY, Xu BL, Wang JY, Liu HC, Zhang SC
and Tu CT: Inhibition by curcumin of multiple sites of the
transforming growth factor-beta1 signalling pathway ameliorates the
progression of liver fibrosis induced by carbon tetrachloride in
rats. BMC Complement Altern Med. 12:1562012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng J, Wu C, Lin Z, Guo Y, Shi L, Dong
P, Lu Z, Gao S, Liao Y, Chen B and Yu F: Curcumin up-regulates
phosphatase and tensin homologue deleted on chromosome 10 through
microRNA-mediated control of DNA methylation-a novel mechanism
suppressing liver fibrosis. FEBS J. 281:88–103. 2014. View Article : Google Scholar
|
|
18
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY, Bin
Z, Wu JB, Tang LY and Gao SM: Long non-coding RNA HOTAIR modulates
c-KIT expression through sponging miR-193a in acute myeloid
leukemia. FEBS Lett. 589:1981–1987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi SS, Witek RP, Yang L, Omenetti A, Syn
WK, Moylan CA, Jung Y, Karaca GF, Teaberry VS, Pereira TA, et al:
Activation of Rac1 promotes hedgehog-mediated acquisition of the
myofibroblastic phenotype in rat and human hepatic stellate cells.
Hepatology. 52:278–290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: Key players in the immune system, differentiation,
tumorigenesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian Y, Pan Q, Shang Y, Zhu R, Ye J, Liu
Y, Zhong X, Li S, He Y, Chen L, et al: MicroRNA-200 (miR-200)
cluster regulation by achaete scute-like 2 (Ascl2): Impact on the
epithelial-mesenchymal transition in colon cancer cells. J Biol
Chem. 289:36101–36115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Csak T, Bala S, Lippai D, Kodys K,
Catalano D, Iracheta-Vellve A and Szabo G: MicroRNA-155 deficiency
attenuates liver steatosis and fibrosis without reducing
inflammation in a mouse model of steatohepatitis. PLoS One.
10:e01292512015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang L, Wang Y, Mao H, Fleig S, Omenetti
A, Brown KD, Sicklick JK, Li YX and Diehl AM: Sonic hedgehog is an
autocrine viability factor for myofibroblastic hepatic stellate
cells. J Hepatol. 48:98–106. 2008. View Article : Google Scholar :
|
|
25
|
Jung Y, Brown KD, Witek RP, Omenetti A,
Yang L, Vandongen M, Milton RJ, Hines IN, Rippe RA, Spahr L, et al:
Accumulation of hedgehog-responsive progenitors parallels alcoholic
liver disease severity in mice and humans. Gastroenterology.
134:1532–1543. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jung Y, Witek RP, Syn WK, Choi SS,
Omenetti A, Premont R, Guy CD and Diehl AM: Signals from dying
hepatocytes trigger growth of liver progenitors. Gut. 59:655–665.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kalderon D: Transducing the hedgehog
signal. Cell. 103:371–374. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View
Article : Google Scholar : PubMed/NCBI
|


















