Introduction
Hearing loss is one of the most common type of
sensory disorder, affecting ~120 million patients worldwide and can
be caused by gene mutations and external factors, including
aminoglycoside antibiotics (1–3). The
well-known A1555G mutation in the human mitochondrial 12S rRNA gene
has been associated with aminoglycoside-induced and non-syndromic
hearing loss (AINHL) in numerous pedigrees all over the world
(4–6). Aminoglycoside inevitably induces
hearing impairment in individuals carrying the 12S rRNA A1555G
mutation. In the absence of aminoglycosides, matrilineal relatives
in families carrying the A1555G mutation exhibited a wide range of
clinical phenotypes, age of onset as well as various degrees of
penetrance and severity of hearing loss (5). In addition, the mitochondrial
tRNASer(UCN) gene was shown to be a hotspot for pathogenetic
mutations associated with deafness of the sensorineural type, with
mutations including A7445G, 7472insC, T7510C and T7511C (7–9).
Furthermore, the G7444A mutation in the cytochrome c oxidase
sub-unit I (COI)/tRNASer(UCN) gene is highly conserved between
various species (10).
With the aim of elucidating the molecular basis of
hearing loss, an extensive genomic screening analysis for mutations
in the mitochondrial (mt)-tRNASer(UCN) and 12S rRNA genes was
performed in the Shaoxing area (China). The present study presented
the case of a patient from this study cohort who had a family
history of AINHL. Sequence analysis of the patient's mitochondrial
genome revealed the presence of COI/tRNASer(UCN) G7444A and 12S
rRNA A1555G mutations.
Patients and methods
The patient was a 19 year-old male from Zhejiang
Province (China) who was treated for hearing loss at Shaoxing
People's Hospital (Shaoxing, China). The medical history of the
patient and his family was assessed and a physical examination was
performed to identify any syndromic manifestations. The patient had
a history of treatment with aminoglycoside (3–5 mg/kg gentamycin
every 8 h) after hospitalization due to pneumonia with fever at the
age of 15 years and developed a bilateral hearing impairment two
months later. In addition, the patient's mother, who was also
impaired of hearing, had a history of using aminoglycosides
(kanamycin) during pregnancy. Informed consent to participate in
the present study was obtained from the patient, and the protocol
of the present study was approved by the Ethics Committee of
Shaoxing People's Hospital (Shaoxing, China). In addition, 268
healthy individuals residing in the Shaoxing area were recruited as
controls, whose the DNA was obtained at the Department of
Otolaryngology (Shaoxing People's Hospital, Shaoxing, China) with
written informed consent provided by all individuals.
Auditory examinations, including pure-tone
audiometry, auditory brainstem response, immittance testing and
determination of distortion product otoacoustic emissions were
performed. The degree of hearing loss was classified into five
levels: Normal, <26 dB; mild, 26–40 dB; moderate, 41–70 dB;
severe, 71–90 dB; and profound, >90 dB.
The genomic DNA was isolated from the blood of the
patient using the Puregene DNA isolation kit (Gentra Systems,
Minneapolis, MN, USA). DNA fragments containing the mt-tRNASer(UCN)
and 12S rRNA genes were amplified by polymerase chain reaction as
previously described (11). The
primers used were as follows: tRNASer(UCN) forward, 5′-ACG AGT ACA
CCG ACT ACG GC-3′ and reverse, 5′-TGG GTG GTT GGT GTA AAT GA-3′;
12S rRNA forward, 5′-CGA TCA ACC TCA CCA CCT CT-3′ and reverse,
5′-TGG ACA ACC AGC TAT CAC CA-3′. In addition, the coding regions
of connexin 26 (GJB2) gene mutations were amplified using
the following primers: Forward, 5′-TAT GAC ACT CCC CAG CAC AG-3′
and reverse, 5′-GGG CAA TGC TTA AAC TGG C-3′ (9). The PCR primers were supplied by BGI
(Shenzhen, China) and the PCR mixture included 200 µM dNTP,
10X buffer, Taq DNA polymerase and 15 mmol/l Mg2+
(Takara Biotechnology Co., Ltd., Dalian, China). Each amplified DNA
sample was purified and analyzed using the ABI 3700 automated DNA
sequencer and the Big Dye Terminator Cycle sequencing reaction kit
(Applied Biosystems; Thermo Fisher Scientific, Waltham, MA, USA).
For screening of the mutations in the mitochondrial genome, the
sequence data were compared with the reversed Cambridge sequence
(GenBank accession no. NC_012920) (12) and for screening the mutations in
the GJB2 gene, the results were compared with the wild-type
GJB2 sequence (GenBank accession no. M86849).
Results and Discussion
Clinical evaluation showed that the patient had a
profound hearing impairment. The patient had been treated with
gentamycin at the age of 15 years and developed a bilateral hearing
impairment two months after the drug administration. The patient's
father's hearing was normal; however, the patient's mother had a
hearing impairment and had been treated with kanamycin during
pregnancy. Due to these clinical characterizations, the
mitochondrial genome of the patient was screened for mutations. The
mitochondrial gene sequence data (Fig.
1) were compared with the Mitomap database (http://www.mitomap.org/MITOMAP), which revealed
the presence of the homoplasmic 12S rRNA A1555G and tRNASer(UCN)
G7444A mutations. To further elucidate the putative role of
GJB2 gene mutations in the phenotypic outcome of the A1555G
mutation, the patient was subjected to mutational screening of
GJB2; however, as no sequence variants in the GJB2
gene were identified, it is unlikely to be involved. Other nuclear
genes may contribute to the phenotypic outcome of the A1555G
mutation.
In 1993, Prezant et al investigated the
underlying molecular mechanisms of AINSHL in three Chinese families
and a large Arab-Israeli family with maternally inherited hearing
impairment by complete mitochondrial genome analysis to identify an
A-to-G replacement at the 1555 position in the 12S rRNA gene
(13). In fact, the homoplasmic
A1555G mutation was localized at the highly evolutionarily
conserved aminoacyl-tRNA acceptor site (A-site) of the small
ribosomal sub-unit (14). This
mutation was found in numerous families with maternally inherited,
non-syndromic hearing loss and also in patients with hearing loss
following use of amino-glycosides (15). Biochemical characterization of
cybrid cells containing the A1555G mutation revealed that they
exhibited reduced mitochondrial protein synthesis, oxygen
consumption and growth rate in galactose medium (16). However, individuals carrying the
A1555G mutation presented with a variety of clinical phenotypes,
including incomplete penetrance and varying degrees of hearing
loss, which indicated that other factors, including environmental
factors, nuclear genes and mitochondrial haplogroups, may
contribute to the clinical manifestation of hearing impairment
associated with the A1555G mutation (17). However, the present study did not
detect any common variants in the GJB2 gene, which suggested
that this nuclear gene may not be involved in the manifestation of
hearing loss due to the A1555G mutation.
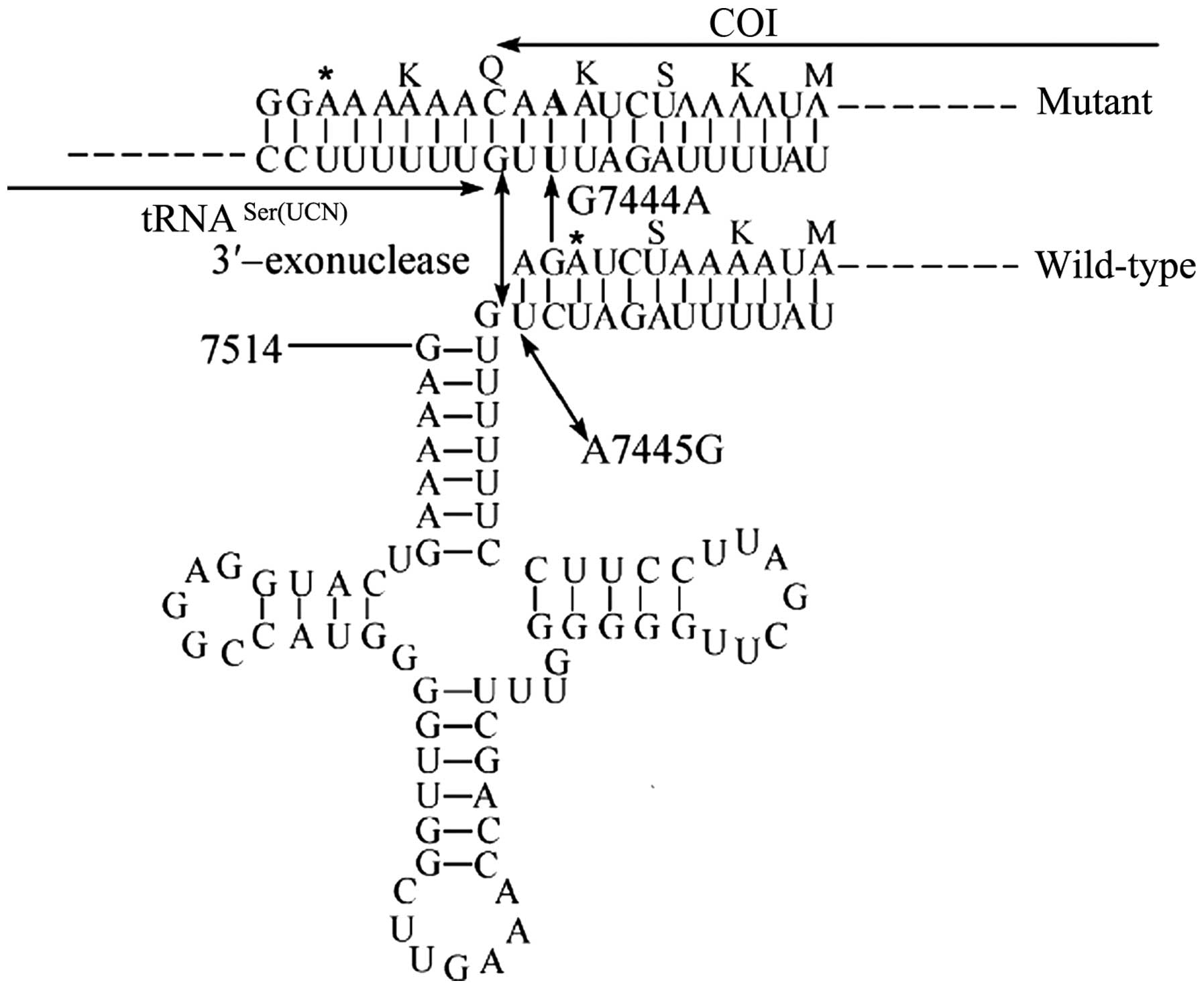
Sequence analysis of the mitochondrial tRNASer(UCN)
gene led to the identification of a homoplasmic G7444A mutation
(Fig. 1). The G7444A mutation was
located in the COI/precursor of tRNASer(UCN) genes; this mutation
was previously found to be associated with Leber's Hereditary Optic
Neuropathy and is considered to be a secondary aberration leading
to increases in the penetrance of the primary mutation, whereas the
G7444A mutation alone did not produce the clinical phenotype
(18). Furthermore, the
homoplasmic A7445G mutation was reported to reduce tRNASer(UCN)
levels by ~70% and to cause a 45% reduction in mitochondrial
protein synthesis in cybrid cells containing this mutation
(19). Structurally, the G7444A
mutation is similar to the A7445G mutation and causes a
read-through of the stop-codon AGA in the COI sequence, leading to
the addition of three amino acids (Lys-Gln-Lys) to the C-terminus
of the polypeptide (Fig. 2). This
leads to the hypothesis that the G7444A mutation may inhibit
mitochondrial protein synthesis, which in turn affects the
synthesis of adenosine triphosphate, increases the production of
reactive oxygen species and consequently leads to damage of hair
cells and cochlear neurons in the ear. Therefore, the combination
of the G7444A and A1555G mutations may be responsible for the
hearing impairment of the patient of the present study.
Acknowledgments
The present study was supported by a grant from the
Foundation of the Shaoxing Bureau of Science and Technology Project
(no. 2013B70066).
References
|
1
|
Brown SD, Hardisty-Hughes RE and Mburu P:
Quiet as a mouse: Dissecting the molecular and genetic basis of
hearing. Nat Rev Genet. 9:277–290. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fischel-Ghodsian N: Genetic factors in
aminoglycoside toxicity. Pharmacogenomics. 6:27–36. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guan MX: Prevalence of mitochondrial 12S
rRNA mutations associated with aminoglycoside ototoxicity. Volta
Rev. 105:211–237. 2005.
|
|
4
|
del Castillo FJ, Rodríguez-Ballesteros M,
Martín Y, Arellano B, Gallo-Terán J, Morales-Angulo C,
Ramírez-Camacho R, Cruz Tapia M, Solanellas J, Martínez-Conde A, et
al: Heteroplasmy for the 1555A>G mutation in the mitochondrial
12S rRNA gene in six Spanish families with non-syndromic hearing
loss. J Med Genet. 40:632–636. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Young WY, Zhao L, Qian Y, Wang Q, Li N,
Greinwald JH Jr and Guan MX: Extremely low penetrance of hearing
loss in four Chinese families with the mitochondrial 12S rRNA
A1555G mutation. Biochem Biophys Res Commun. 328:1244–1251. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao L, Wang Q, Qian Y, Li R, Cao J, Hart
LC, Zhai S, Han D, Young WY and Guan MX: Clinical evaluation and
mitochondrial DNA sequence analysis in two Chinese families with
aminoglycoside-induced and non-syndromic hearing loss. Biochem
Biophys Res Commun. 336:967–973. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tiranti V, Chariot P, Carella F, Toscano
A, Soliveri P, Girlanda P, Carrara F, Fratta GM, Reid FM, Mariotti
C and Zeviani M: Maternally inherited hearing loss, ataxia and
myoclonus associated with a novel point mutation in mitochondrial
tRNASer(UCN) gene. Hum Mol Genet. 4:1421–1427. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hutchin T, Haworth I, Higashi K,
Fischel-Ghodsian N, Stoneking M, Saha N, Arnos C and Cortopassi G:
A molecular basis for human hypersensitivity to aminoglycoside
antibiotics. Nucleic Acids Res. 21:4174–4179. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li R, Greinwald JH Jr, Yang L, Choo DI,
Wenstrup RJ and Guan MX: Molecular analysis of mitochondrial 12S
rRNA and tRNASer(UCN) genes in paediatric subjects with
nonsyndromic hearing loss. J Med Genet. 41:615–620. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rydzanicz M, Wróbel M, Cywińska K,
Froehlich D, Gawecki W, Szyfter W and Szyfter K: Screening of the
general Polish population for deafness-associated mutations in
mitochondrial 12S rRNA and tRNA Ser(UCN) genes. Genet Test Mol
Biomarkers. 13:167–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rieder MJ, Taylor SL, Tobe VO and
Nickerson DA: Automating the identification of DNA variations using
quality-based fluorescence re-sequencing: Analysis of the human
mitochondrial genome. Nucleic Acids Res. 26:967–973. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andrews RM, Kubacka I, Chinnery PF,
Lightowlers RN, Turnbull DM and Howell N: Reanalysis and revision
of the cambridge reference sequence for human mitochondrial DNA.
Nat Genet. 23:1471999. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prezant TR, Agapian JV, Bohlman MC, Bu X,
Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, et
al: Mitochondrial ribosomal RNA mutation associated with both
antibiotic-induced and non-syndromic deafness. Nat Genet.
4:289–294. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ruiz-Pesini E and Wallace DC: Evidence for
adaptive selection acting on the tRNA and rRNA genes of human
mitochondrial DNA. Hum Mutat. 27:1072–1081. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding Y, Leng J, Fan F, Xia B and Xu P: The
role of mitochondrial DNA mutations in hearing loss. Biochem Genet.
51:588–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guan MX, Fischel-Ghodsian N and Attardi G:
Biochemical evidence for nuclear gene involvement in phenotype of
non-syndromic deafness associated with mitochondrial 12S rRNA
mutation. Hum Mol Genet. 5:963–71. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu J, Qian Y, Li Z, et al: Mitochondrial
haplotypes may modulate the phenotypic manifestation of the
deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion.
10:69–81. 2010. View Article : Google Scholar
|
|
18
|
Brown MD, Torroni A, Reckord CL and
Wallace DC: Phylogenetic analysis of Leber's hereditary optic
neuropathy mitochondrial DNA's indicates multiple independent
occurrences of the common mutations. Hum Mutat. 6:311–325. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guan MX, Enriquez JA, Fischel-Ghodsian N,
Puranam RS, Lin CP, Maw MA and Attardi G: The deafness-associated
mitochondrial DNA mutation at position 7445, which affects
tRNASer(UCN) precursor processing, has long-range effects on NADH
dehydrogenase ND6 subunit gene expression. Mol Cell Biol.
18:5868–5879. 1998. View Article : Google Scholar : PubMed/NCBI
|