Introduction
Osteosarcoma is the most frequent type of primary
bone malignant tumor in children (1). It is considered to arise from
malignant mesenchymal cells, which produce osteoid or immature bone
(2). Although advanced treatment
for osteosarcoma consists of aggressive adjuvant chemotherapy, the
five-year survival rate of patients with high-grade osteosarcoma
remains <50% (3).
Ubiquitin-like with plant homeodomain (PHD) and RING
finger domain 1 (UHRF1) is a multi-domain protein, which was
initially identified as a nuclear protein associated with cell
proliferation (4). As a human
inverted CCAAT box-binding protein, UHRF1 is involved in regulating
the expression of topoisomerase IIα in proliferating cells
(5). UHRF1 has been found to
inhibit the mRNA and protein expression levels of Rb1 (6,7).
Deletion of Rb1 was found to suppress the expression of E-cadherin
and promote epithelial-to-mesenchymal transition (EMT) (8). Furthermore, UHRF1 is involved in the
methylation of newly synthesized CpG sequences during DNA
replication (9,10). UHRF1 binds preferentially to
dimethylated and trimethylated histone 3 lysine 9 peptides, and
this binding is required for the maintenance of DNA methylation
(11). Through interaction with
other nuclear proteins, including Tip60, histone deacetylase and
G9A, UHRF1 may function as a bridge between DNA methylation and the
histone methylation (12).
Furthermore, significant overexpression of UHRF1 has been observed
in several types of human tumor, including breast (13,14),
bladder (15), prostate (16) and lung cancer (17). The overexpression of UHRF1 has also
been reported to reduce the radiosensitivity of human breast cancer
cells and HeLa cells to γ-irradiation (18). Previous studies have demonstrated
that UHRF1 is an oncogene in hepatocellular carcinoma (19,20).
Furthermore, the overexpression of UHRF1 destabilizes and
delocalizes DNA (cytosine-5-)-methyltransferase 1 (DNMT1) and
causes DNA hypomethylation in cancer cells (19). Despite these findings, little is
known regarding the function of UHRF1 in human osteosarcoma cells.
In the present study, the effects of UHRF1 on the invasion of
osteosarcoma cells were examined, and the associated underlying
mechanisms were investigated.
Materials and methods
Cell culture
MG-63, Saos-2, U2OS and HOS human osteosarcoma cells
lines were obtained from American Type Culture Collection
(Manassas, VA, USA). HEK293T cells were obtained from the Institute
of Cell and Biochemistry Research of the Chinese Academy of Science
(Shanghai, China). The cell lines were cultured with Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) in a
humidified atmosphere containing 5% CO2 at 37°C.
Reagents
The mouse anti-human monoclonal antibodies targeting
UHRF1 (cat no. ab57083; 1:2,000 dilution) and GAPDH (cat no.
ab9484; 1:5,000 dilution) were purchased from Abcam (Cambridge, MA,
USA). Human Rb1-specific rabbit polyclonal antibody (cat. no. 9313;
1:2,000 dilution) was purchased from Cell Signaling Technology,
Inc. (Danvers, MA, USA). The mouse anti-human E-cadherin monoclonal
antibody (cat no. sc-21791; 1:2,000 dilution) was obtained from
Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). The pLenti-UHRF1
lentiviral expression vector and the control vector were purchased
from OriGene Technologies, Inc. (Beijing, China). PLKO.1 lentiviral
vectors containing short hairpin (sh)RNA inserts against Rb1 were
purchased from Sigma-Aldrich (St. Louis, MO, USA). The target
sequences were as follows: shRb-A, 5′-GCCTTTGATTCGTTCCTTCTT-3′ and
shRb-B, 5′-TGTGAAATACTGGCCCGAGAA-3′.
Transfection and lentiviral
transduction
Transfection of the cells was performed using FuGENE
transfection reagent (Roche Diagnostics, Indianapolis, IN, USA),
according to the manufacturer's protocol (3.5×106 cells
in a 10 cm dish with 20 µl FuGENE). The lentiviral
expression vector containing the pLenti-UHRF1/PLKO.1 or the
control/empty vector were transfected into the HEK293T cells. The
recombinant lentivirus was subsequently harvested, filtered through
Millipore Millex-HV 0.45 µM polyvinylidene difluoride
filters (Millipore, Billerica, MA, USA) and transduced into the
target cells (MG-63, Saos-2 and U2OS cells at 60% confluence) with
8 µg/ml polybrene (Sigma-Aldrich). After 48 h of incubation,
the cells were selected with fresh puromycin-containing media (2.0
µg/ml; Sigma-Aldrich). Following puromycin selection for 48
h, the expression levels of UHRF1 and Rb1 were quantified using
western blot analysis.
Cell Counting kit-8 (CCK-8) assay
Cell proliferation was determined using a CCK-8
assay (Dojindo, Kumamoto, Japan). Briefly, the cells were plated in
96-well plates at 2,500 cells/well and cultured in DMEM. Every 24
h, 10 µl CCK-8 was added to each well containing 100
µl DMEM once. The plates were then incubated for a further 2
h at 37°C. The cell growth was monitored every 24 h for 7 days
using the CCK-8 assay. Absorbance was measured at 450 nm using a
microplate reader (Infinite Pro 2000; Tecan GmbH, Grödig,
Austria).
Cell invasion assay
To assess the role of UHRF1 in cell invasion, a
total of 1×105 cells were suspended in 100 µl
DMEM supplemented with 10% FBS, and were seeded into the upper
compartment of Matrigel-coated (BD Biosciences, Franklin Lakes, NJ,
USA) Transwell chambers (24-well; 8 µm; Merck Millipore,
Darmstadt, Germany). The cells were incubated for 24 h at 37°C in a
5% CO2 chamber. The cells, which did not invade through
the pores were removed using a cotton swab. The cells on the lower
surface of the membrane were stained with a hematoxylin and eosin
staining kit (Baixu of Biotechnology, Shanghai, China) and counted
with a microscope (Leica DM 5000 B; Leica Microsystems, Wetzlar,
Germany). Results are presented as the average number of cells in
five randomly selected fields.
Western blot analysis
The cells were suspended in lysis buffer (Beyotime
Institute of Biotechnology, Nanjing, China) containing a mixture of
protease and phosphatase inhibitors (both from Roche Diagnostics
GmbH, Mannheim, Germany). The cell lysates were then centrifuged at
11,500 × g for 10 min at 4°C. Protein concentrations were estimated
using the Quick Start™ Bradford Protein Assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The samples were separated
by SDS-PAGE (4–29% gradient; Bio-Rad Laboratories, Inc.) and then
blotted onto a polyvinylidene difluoride membrane (Merck
Millipore). The membranes were blocked with 5% non-fat dried milk
in Tris-buffered saline with 1% Tween 20 (TBST; Sigma-Aldrich) for
1 h at room temperature prior to incubation with specific primary
antibodies overnight at 4°C. Following three washes with TBST, the
membrane was incubated with peroxidase-conjugated secondary
antibodies [anti-mouse immunoglobulin (Ig)G (cat no. 7076; 1:5,000
dilution) and anti-rabbit IgG (cat no. 7074) (both from Cell
Signaling Technology, Inc.)] for 30 min at 37°C and then washed
three times with TBST. The bound antibodies were detected using
chemiluminescent horseradish peroxidase substrate (Merck Millipore)
and images were captured using an LAS-4000 digital imaging system
(GE Healthcare Life Sciences, Little Chalfont, UK).
Statistical analysis
Values are expressed as the mean ± standard
deviation. GraphPad 6.01 Prism software (GraphPad, Inc., La Jolla,
CA, USA) was used for statistical analyses. Each experiment was
repeated three times. P<0.05 was considered to indicate a
statistically significant difference.
Results
UHRF1 promotes the proliferation of human
osteosarcoma cells
UHRF1 has been reported to be overexpressed in
various types of cancer (13). The
present study examined the expression levels of UHRF1 in four human
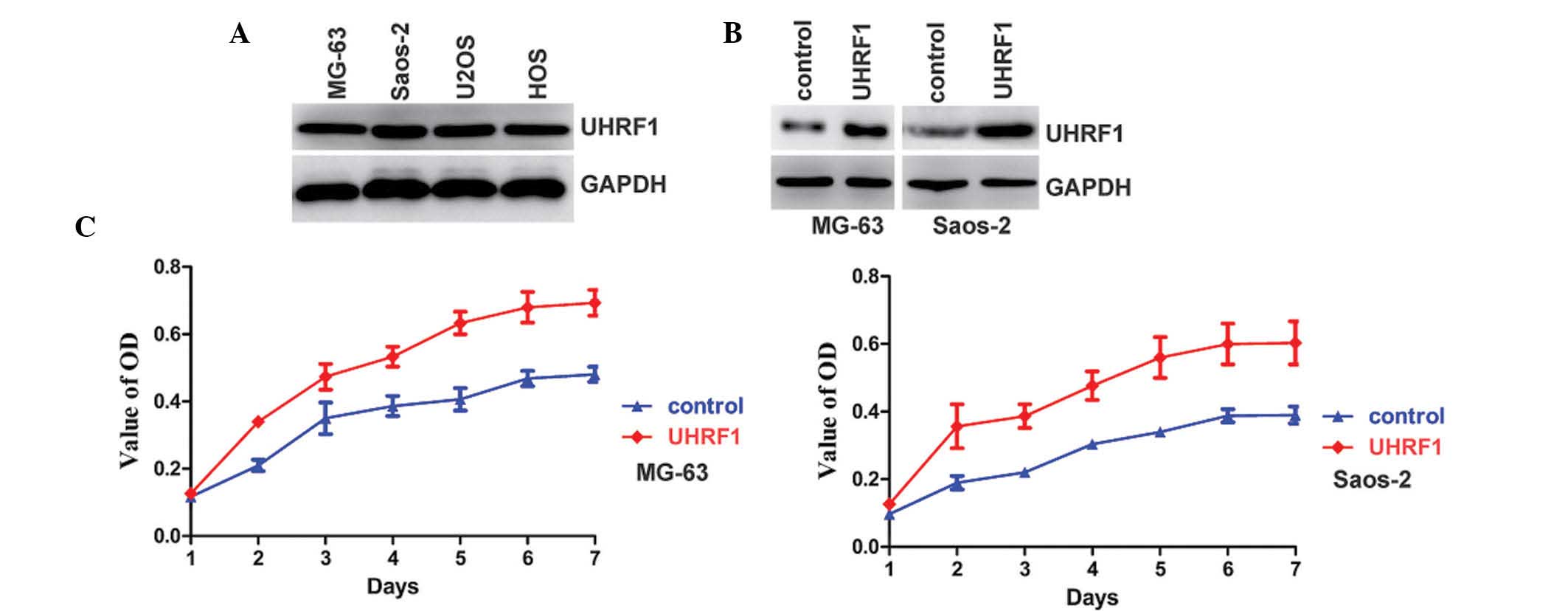
osteosarcoma cell lines. As shown in Fig. 1A, the expression of UHRF1 was
detected in all of the cell lines. Overexpression of UHRF1 can
increase cell proliferation (14,21),
therefore, the effects of UHRF1 on the proliferation of human
osteosarcoma cells were also investigated. UHRF1 was stably
overexpressed in the MG-63 and Saos-2 osteosarcoma cell lines
(Fig. 1B). As expected,
overexpression of UHRF1 increased the proliferation rate of the
MG-63 and Saos-2 osteosarcoma cell lines (Fig. 1C).
UHRF1 promotes the invasion of MG-63 and
U2OS human osteosarcoma cells, but not Saos-2 cells
In order to investigate the function of UHRF1 on the
invasion of osteosarcoma cells, Transwell invasion assays were used
with three human osteosarcoma cell lines in vitro. The
overexpression of UHRF1 significantly increased the invasion of
MG-63 and U2OS human osteosarcoma cells (Fig. 2). By contrast, the overexpression
of UHRF1 had no significant effect on the invasion of the Saos-2
cells (Fig. 2). Homozygous
deletion of the Rb1 gene has been identified in Saos-2 cells
(22,23), whereas MG-63 and U2OS cells exhibit
normal expression levels of Rb1 (23,24).
Therefore, the present study hypothesized that Rb1 may be involved
in regulating the invasion of osteosarcoma cells by UHRF1.
UHRF1 promotes the invasion of human
osteosarcoma cells in an Rb1-dependent manner
To test the hypothesis that Rb1 may be involved in
the regulation of the invasion of osteosarcoma cells by UHRF, the
expression levels of Rb1 were quantified in the osteosarcoma cell
lines. As expected, the MG-63 and U2OS cells exhibited normal
expression levels of Rb1. The expression of Rb1 in the Saos-2 cells
containing the homozygous deletion of Rb1 was not detected
(Fig. 3A). To further investigate
the mechanism underlying the effect of UHRF1 on the regulation of
osteosarcoma cell invasion, the expression of Rb1 was stably
knocked down in the MG-63 cells (Fig.
3B). The stable known-down of Rb1 resulted in UHRF1 being
overexpressed in the MG-63 cells (Fig.
3C). Following the knockdown of Rb1 in the Rb1-positive MG-63
cells, the invasion of the cells increased. By contrast, the
indiced overexpression of UHRF1 had no effect on the invasion of
the MG-63 cells following stable Rb1 knockdown (Fig. 3D). These results indicated that Rb1
appeared to be responsible for the regulation of cell invasion by
UHRF1.
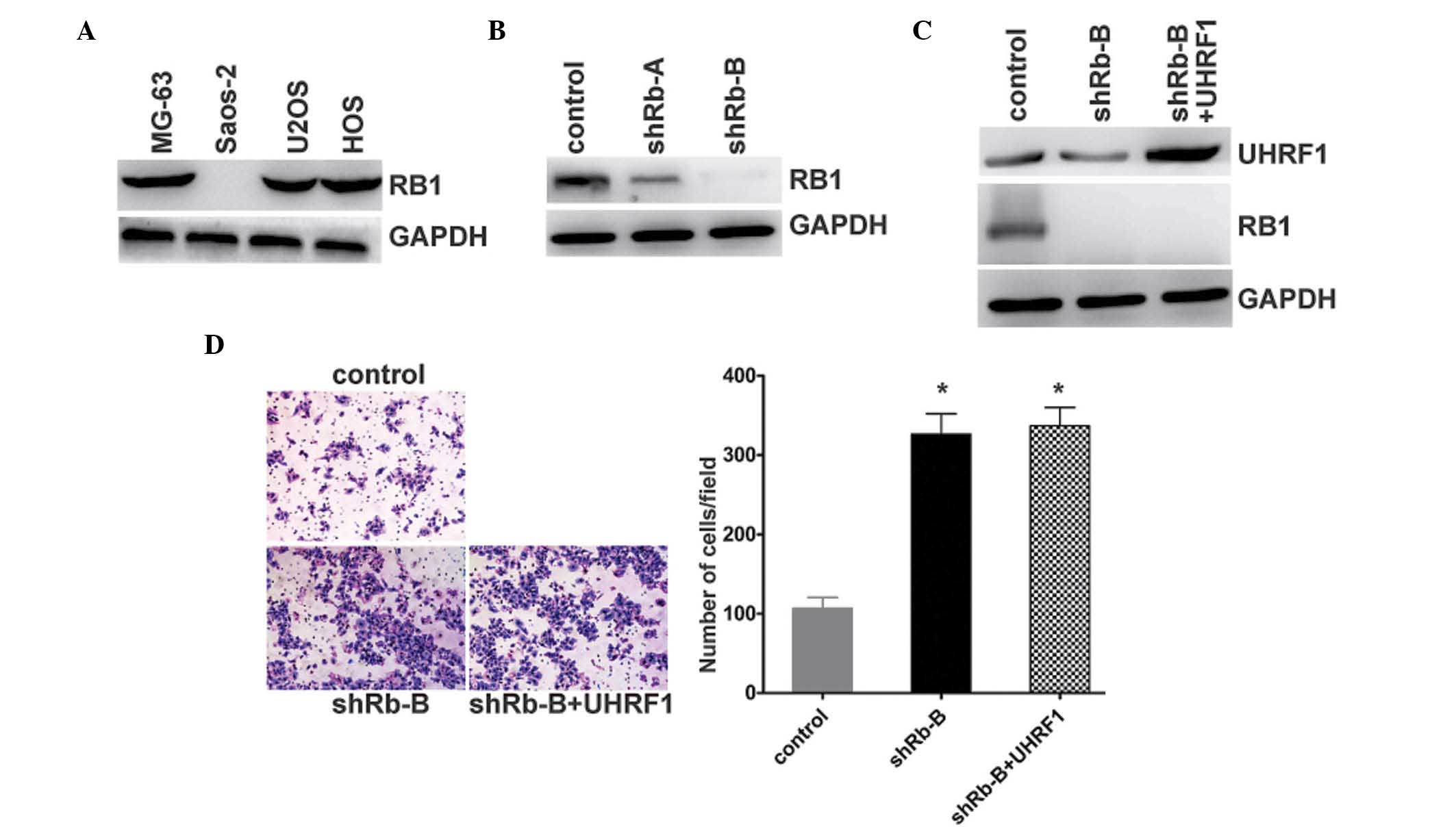 | Figure 3UHRF1 promotes the invasion of human
osteosarcoma cells in an Rb1-dependent manner. (A) Expression
levels of Rb1 and GAPDH were examined using western blot analysis
in the MG-63, Saos-2, U2OS and HOS human osteosarcoma cells lines.
(B) MG-63 cells were transfected with Rb1-shRNA (shRb-A and shRb-B)
or negative control, and the expression levels of Rb1 and GAPDH
were examined using western blot analysis. (C) UHRF1 was
overexpressed in the Rb1 stable knockdown (shRb-B) MG-63 cells. The
expression levels of UHRF1, Rb1 and GAPDH were examined using
western blot analysis. (D) MG-63 cells were transfected with
negative control (control) or Rb1-shRNA (shRb-B). Overexpression of
UHRF1 was observed in the Rb1-shRNA MG-63 cells (shRb-B + UHRF1).
The invasion potential of these cells was determined using a
Transwell assay. Representative images are shown on the left
(magnification, ×40), and quantification of five randomly-selected
fields is shown on the right. Data are presented as the mean ±
standard deviation. *P<0.0002, vs. control. UHRF1,
ubiquitin-like with plant homeodomain and RING-finger domain 1;
Rb1, retinoblastoma 1; shRNA, short hairpin RNA. |
UHRF1 suppresses the expression of
E-cadherin and promotes epithelial-mesenchymal transition (EMT)
through the inhibition of Rb1
UHRF1 can inhibit the expression levels of Rb1 at
the protein and mRNA levels (6).
Furthermore, the inhibition of Rb1 suppresses the expression of
E-cadherin and increases EMT (8).
The present study hypothesized that UHRF1 may inhibit the
expression of Rb1 and thereby suppress the expression of
E-cadherin. E-cadherin is known to suppress the invasion of cancer
cells (25). Therefore, the
UHRF1-mediated promotion of invasion may be the result of
E-cadherin inhibition. The results of the present study
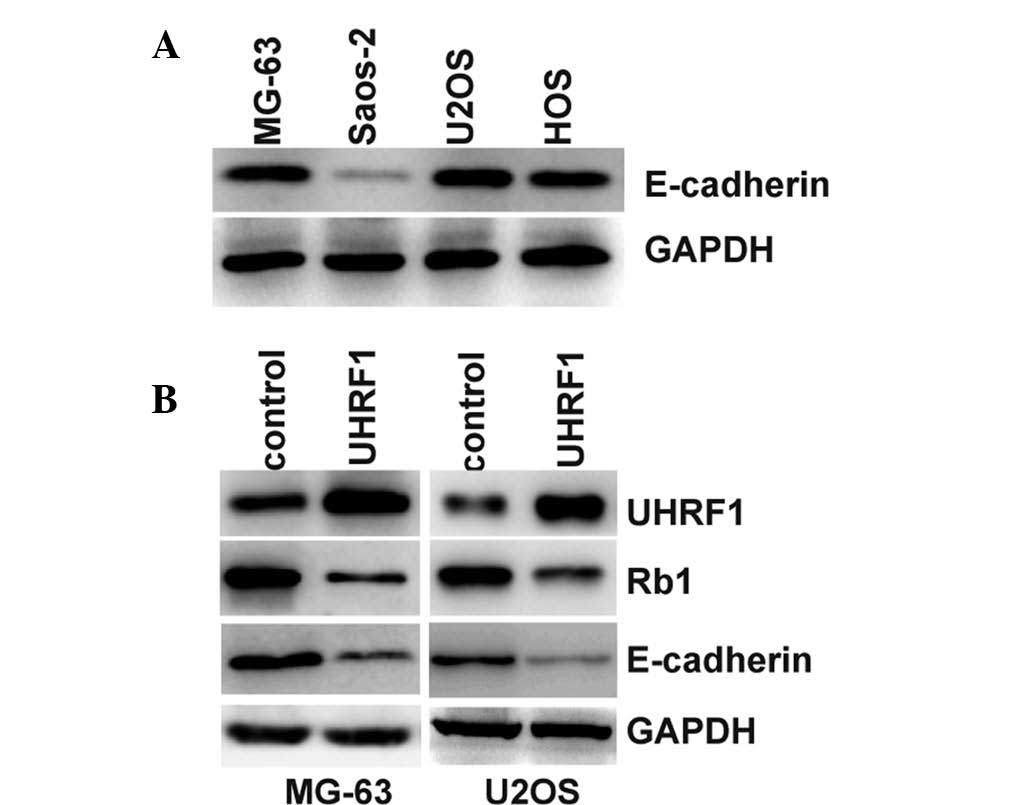
demonstrated that the expression levels of E-cadherin were
significantly lower in the Saos-2 cells, compared with the other
Rb1-positive cells (Fig. 4A).
Furthermore, overexpression of UHRF1 in the MG-63 and U2OS cells
inhibited the expression of Rb1 and E-cadherin (Fig. 4B). The loss of E-cadherin is the
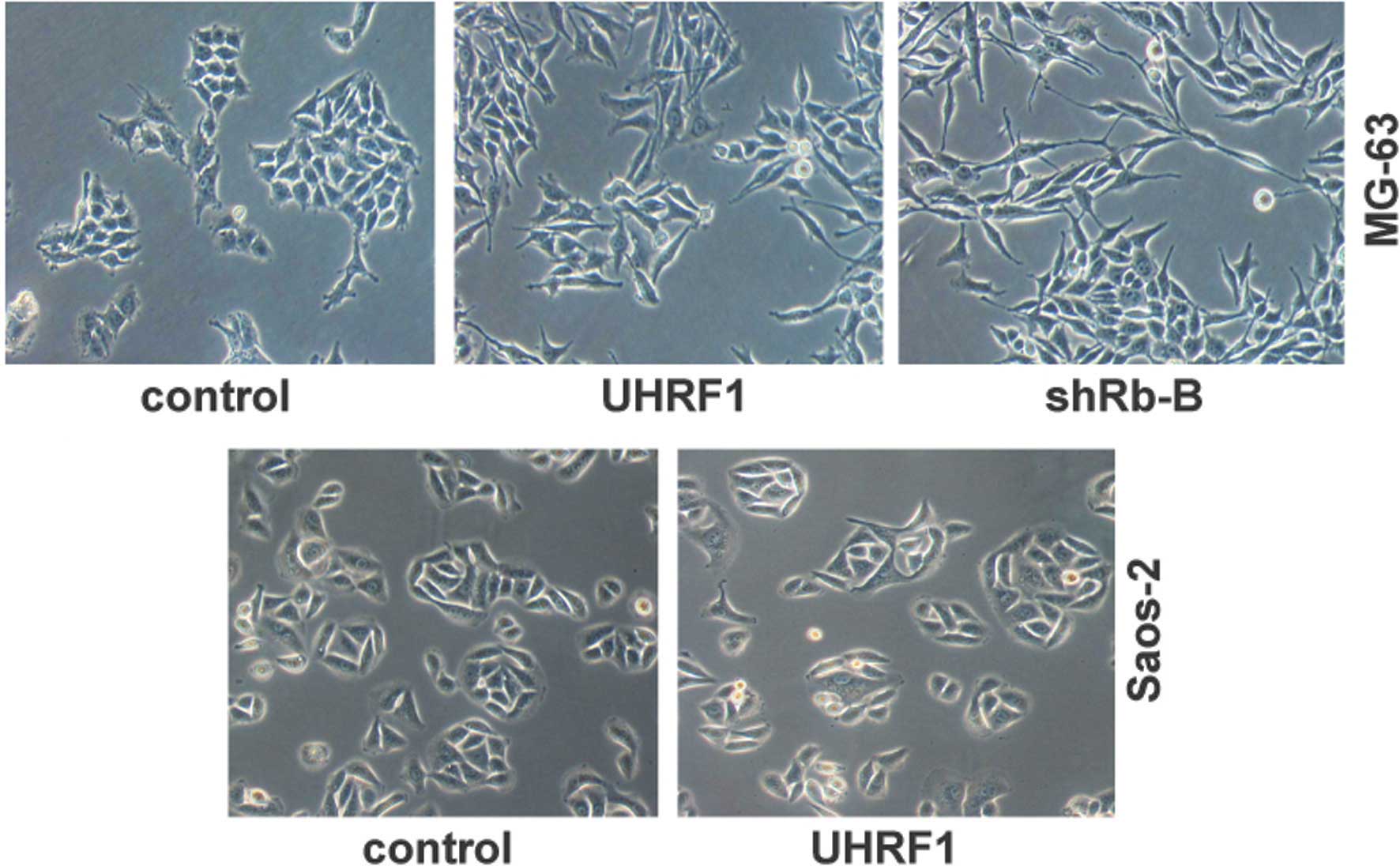
initial or primary cause for EMT (26). In the present study, the
UHRF1-overexpressing MG-63 cells exhibited marked changes in cell
morphology, with transformation of the cobblestone-like epithelial
cells to an elongated fibroblast-like morphology, and with
pronounced cellular scattering that indicated EMT (Fig. 5). Similarly, knockdown of Rb1 in
the MG-63 cells also showed EMT characteristic patterns (Fig. 5). However, UHRF1 had no effect on
the cell morphology of the Saos-2 cells with the Rb1 deletion
(Fig. 5). These results further
supported the hypothesis that UHRF1 inhibits the expression of
E-cadherin and promotes EMT through the suppression of Rb1.
Discussion
Osteosarcoma arises from malignant mesenchymal
cells, which produce osteoid or immature bone, and is the most
frequent type of primary bone malignant tumor (27). Despite advanced treatment for
osteosarcoma, which combines chemotherapy, surgery and occasionally
radiotherapy, the five-year survival rate for patients with
high-grade osteosarcoma remains <50% (3). Therefore, novel targeted molecular
therapeutic techniques for osteosarcoma are required.
UHRF1, also known as inverted CCAAT box-binding
protein of 90 kDa, contains different domains, including an
E3-ligase RING domain, a SET and RING-associated (SRA) domain, a
PHD finger domain and a tandem tudor domain (28). The SRA domain of UHRF1 specifically
binds to hemimethylated CpG following DNA replication, and recruits
DNMT1 to methylate the newly synthesized DNA strand (29). UHRF1 is overexpressed and
associated with tumor stages, and predicts poor prognosis in
various types of cancer (15,21).
UHRF1 has previously been identified as an oncogene in
hepatocellular carcinoma (19,20).
Furthermore, the overexpression of UHRF1 destabilizes and
delocalizes DNMT1, and causes DNA hypomethylation in cancer cells
(19). Although several studies
have demonstrated the function of UHRF1 in tumorigenesis and tumor
progression (14,17,20),
little is known regarding the function of UHRF1 in human
osteosarcoma cells. In the present study, the expression of UHRF1
was detected in all of the human osteosarcoma cell lines examined.
Furthermore, the overexpression of UHRF1 increased the
proliferation of rates of the osteosarcoma cell lines.
EMT, during which epithelial cells are
transdifferentiated to a mesenchymal state, is considered to be
important in the initiation of the invasion and metastasis of
cancer cells (30,31). Loss of E-cadherin is considered to
be the most fundamental event during EMT (32). Deregulation in the expression of
several genes or microRNAs has been reported to downregulate the
expression of E-cadherin (33,34).
As an oncogene, UHRF1 is able to bind to methylated DNA and recruit
transcriptional repressors to suppress the transcription of several
tumor suppressor genes (35,36).
Therefore, UHRF1 may regulate the transcription of E-cadherin. The
deletion of Rb1 is associated with downregulation of the expression
of E-cadherin and increased EMT (8). UHRF1 binds to the Rb1 gene promoter
and inhibits the expression of Rb1 (6). The results of the present study
demonstrated that UHRF1 promoted the invasion of osteosarcoma cells
of the MG-63 and U2OS cell lines, but not of the Saos-2 cell line.
Saos-2 cells undergo homozygous deletion of the Rb1 gene (22,23),
therefore the present study hypothesized that Rb1 may be involved
in the regulation of the invasion of osteosarcoma cells by UHRF1.
Further investigations demonstrated that knockdown of the
expression of Rb1 in the Rb1-positive cells eliminated the
regulation of invasion by UHRF1. Furthermore, the expression levels
of E-cadherin were consistent with the Rb1 status. The results of
the present study also demonstrated that the overexpression of
UHRF1 significantly downregulated the expression levels of Rb1 and
E-cadherin in the Rb1-positive cells. Similarly, overexpression of
UHRF1 in the MG-63 cells resulted in marked changes in cell
morphology, indicating EMT, although this was not observed in the
Saos-2 cells. The knockdown of Rb1 led to the observation of
similar changes in EMT in the UHRF1-overexpressing MG-63 cells.
In conclusion, the present study revealed that UHRF1
promoted human osteosarcoma cell invasion by downregulating the
expression of E-cadherin and increasing EMT, in an Rb1-dependent
manner.
Acknowledgments
The present study was supported by grants from the
Science and Technology Planning Project of Jinzhong City (grant.
no. N1312) and from the National Natural Science Foundations of
China (grant. no. 81100293).
References
|
1
|
Goguet-Surmenian E, Richard-Fiardo P,
Guillemot E, Benchetrit M, Gomez-Brouchet A, Buzzo P,
Karimdjee-Soilihi B, Alemanno P, Michiels JF, Schmid-Alliana A and
Schmid-Antomarchi H: CXCR7-mediated progression of osteo-sarcoma in
the lungs. Br J Cancer. 109:1579–1585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Y, Wu Y, Gu S, Sun Z, Rui Y, Wang J,
Lu Y, Li H, Xu K and Sheng P: Prognostic role of CD44 expression in
osteosarcoma: Evidence from six studies. Diagn Pathol. 9:1402014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chu J, Loughlin EA, Gaur NA, SenBanerjee
S, Jacob V, Monson C, Kent B, Oranu A, Ding Y, Ukomadu C and Sadler
KC: UHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is
required for zebrafish embryogenesis. Mol Biol Cell. 23:59–70.
2012. View Article : Google Scholar :
|
|
5
|
Hopfner R, Mousli M, Jeltsch JM, Voulgaris
A, Lutz Y, Marin C, Bellocq JP, Oudet P and Bronner C: ICBP90, a
novel human CCAAT binding protein, involved in the regulation of
topoisomerase IIalpha expression. Cancer Res. 60:121–128.
2000.PubMed/NCBI
|
|
6
|
Jeanblanc M, Mousli M, Hopfner R, Bathami
K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD and
Bronner C: The retinoblastoma gene and its product are targeted by
ICBP90: A key mechanism in the G1/S transition during the cell
cycle. Oncogene. 24:7337–7345. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benavente CA, Finkelstein D, Johnson DA,
Marine JC, Ashery-Padan R and Dyer MA: Chromatin remodelers HELLS
and UHRF1 mediate the epigenetic deregulation of genes that drive
retinoblastoma tumor progression. Oncotarget. 5:9594–9608. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arima Y, Inoue Y, Shibata T, Hayashi H,
Nagano O, Saya H and Taya Y: Rb depletion results in deregulation
of E-cadherin and induction of cellular phenotypic changes that are
characteristic of the epithelial-to-mesenchymal transition. Cancer
Res. 68:5104–5112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arita K, Ariyoshi M, Tochio H, Nakamura Y
and Shirakawa M: Recognition of hemimethylated DNA by the SRA
protein UHRF1 by a base-flipping mechanism. Nature. 455:818–821.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hashimoto H, Vertino PM and Cheng X:
Molecular coupling of DNA methylation and histone methylation.
Epigenomics. 2:657–669. 2010. View Article : Google Scholar
|
|
11
|
Rothbart SB, Krajewski K, Nady N, Tempel
W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT,
Fuchs SM, et al: Association of UHRF1 with methylated H3K9 directs
the maintenance of DNA methylation. Nat Struct Mol Biol.
19:1155–1160. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Unoki M: Current and potential anticancer
drugs targeting members of the UHRF1 complex including epigenetic
modifiers. Recent Pat Anticancer Drug Discov. 6:116–130. 2011.
View Article : Google Scholar
|
|
13
|
Alhosin M, Sharif T, Mousli M,
Etienne-Selloum N, Fuhrmann G, Schini-Kerth VB and Bronner C:
Down-regulation of UHRF1, associated with re-expression of tumor
suppressor genes, is a common feature of natural compounds
exhibiting anti-cancer properties. J Exp Clin Cancer Res.
30:412011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li XL, Xu JH, Nie JH and Fan SJ: Exogenous
expression of UHRF1 promotes proliferation and metastasis of breast
cancer cells. Oncol Rep. 28:375–383. 2012.PubMed/NCBI
|
|
15
|
Zhang Y, Huang Z, Zhu Z, et al:
Upregulated UHRF1 promotes bladder cancer cell invasion by
epigenetic silencing of KiSS1. PLoS One. 9:e1042522014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Babbio F, Pistore C, Curti L, et al: The
SRA protein UHRF1 promotes epigenetic crosstalks and is involved in
prostate cancer progression. Oncogene. 31:4878–4887. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Daskalos A, Oleksiewicz U, Filia A, et al:
UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell
lung cancer. Cancer. 117:1027–1037. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li XL, Meng QH and Fan SJ:
Adenovirus-mediated expression of UHRF1 reduces the
radiosensitivity of cervical cancer HeLa cells to
gamma-irradiation. Acta Pharmacol Sin. 30:458–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mudbhary R, Hoshida Y, Chernyavskaya Y,
Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson
RT, et al: UHRF1 overexpression drives DNA hypomethylation and
hepatocellular carcinoma. Cancer Cell. 25:196–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhuo H, Tang J, Lin Z, et al: The aberrant
expression of MEG3 regulated by UHRF1 predicts the prognosis of
hepatocellular carcinoma. Mol Carcinog. Jan 16–2015.Epub ahead of
print. View
Article : Google Scholar
|
|
21
|
Zhou L, Zhao X, Han Y, Lu Y, Shang Y, Liu
C, Li T, Jin Z, Fan D and Wu K: Regulation of UHRF1 by miR-146a/b
modulates gastric cancer invasion and metastasis. FASEB J.
27:4929–4939. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Manning AL, Yazinski SA, Nicolay B, Bryll
A, Zou L and Dyson NJ: Suppression of genome instability in
pRB-deficient cells by enhancement of chromosome cohesion. Mol
Cell. 53:993–1004. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rosemann M, Gonzalez-Vasconcellos I, Domke
T, Kuosaite V, Schneider R, Kremer M, Favor J, Nathrath M and
Atkinson MJ: A Rb1 promoter variant with reduced activity
contributes to osteosarcoma susceptibility in irradiated mice. Mol
Cancer. 13:1822014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ory B, Blanchard F, Battaglia S, Gouin F,
Rédini F and Heymann D: Zoledronic acid activates the DNA S-phase
checkpoint and induces osteosarcoma cell death characterized by
apoptosis-inducing factor and endonuclease-G translocation
independently of p53 and retinoblastoma status. Mol Pharmacol.
71:333–343. 2007. View Article : Google Scholar
|
|
25
|
Lau MT, Klausen C and Leung PC: E-cadherin
inhibits tumor cell growth by suppressing PI3K/Akt signaling via
β-catenin-Egr1-mediated PTEN expression. Oncogene. 30:2753–2766.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lombaerts M, van Wezel T, Philippo K,
Dierssen JW, Zimmerman RM, Oosting J, van Eijk R, Eilers PH, van de
Water B, Cornelisse CJ and Cleton-Jansen AM: E-cadherin
transcriptional downregulation by promoter methylation but not
mutation is related to epithelial-to-mesenchymal transition in
breast cancer cell lines. Br J Cancer. 94:661–671. 2006.PubMed/NCBI
|
|
27
|
Cao ZQ, Shen Z and Huang WY: MicroRNA-802
promotes osteosarcoma cell proliferation by targeting p27. Asian
Pac J Cancer Prev. 14:7081–7084. 2013. View Article : Google Scholar
|
|
28
|
Cheng J, Yang Y, Fang J, Xiao J, Zhu T,
Chen F, Wang P, Li Z, Yang H and Xu Y: Structural insight into
coordinated recognition of trimethylated histone H3 lysine 9
(H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain
(TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger
domains, 1) protein. J Biol Chem. 288:1329–1339. 2013. View Article : Google Scholar :
|
|
29
|
Sharif J, Muto M, Takebayashi S, Suetake
I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T,
Okamura K, et al: The SRA protein Np95 mediates epigenetic
inheritance by recruiting Dnmt1 to methylated DNA. Nature.
450:908–912. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Puisieux A, Brabletz T and Caramel J:
Oncogenic roles of EMT-inducing transcription factors. Nat Cell
Biol. 16:488–494. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiong H, Hong J, Du W, Lin YW, Ren LL,
Wang YC, Su WY, Wang JL, Cui Y, Wang ZH and Fang JY: Roles of STAT3
and ZEB1 proteins in E-cadherin down-regulation and human
colorectal cancer epithelial-mesenchymal transition. J Biol Chem.
287:5819–5832. 2012. View Article : Google Scholar :
|
|
33
|
Sun XJ, Liu H, Zhang P, Zhang XD, Jiang ZW
and Jiang CC: miR-10b promotes migration and invasion in
nasopharyngeal carcinoma cells. Asian Pac J Cancer Prev.
14:5533–5537. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang YP, Wang MZ, Luo YR, Shen Y and Wei
ZX: Lentivirus-mediated shRNA interference targeting SLUG inhibits
lung cancer growth and metastasis. Asian Pac J Cancer Prev.
13:4947–4951. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim JK, Estève PO, Jacobsen SE and Pradhan
S: UHRF1 binds G9a and participates in p21 transcriptional
regulation in mammalian cells. Nucleic Acids Res. 37:493–505. 2009.
View Article : Google Scholar :
|
|
36
|
Guan D, Factor D, Liu Y, Wang Z and Kao
HY: The epigenetic regulator UHRF1 promotes ubiquitination-mediated
degradation of the tumor-suppressor protein promyelocytic leukemia
protein. Oncogene. 32:3819–3828. 2013. View Article : Google Scholar :
|