Introduction
Rapid and efficient inactivation of genes in
microorganisms is an important process, which enables the analysis
of physiological and genetic characteristics of bacteria. In recent
years, chromosome gene deletion technology of Escherichia
coli has developed rapidly. The method of λ Red and FLP
recombination, as described by Datsenko and Wanner (1), is a popular method to delete genes in
E. coli. Baba et al (2) reported that the single gene mutation
of various strains of E. coli was constructed using this
method. The process of building multiple gene deletion strains, in
order to remove the resistance genes in the mutant strains,
required repeated transformation or removal of the auxiliary
plasmid, which complicates the experimental process and elongates
the experimental cycle. Dif sites are identified by the Xer
recombinant enzyme in E. coli, which simplifies the
procedure. It has been reported that one gene was successfully
deleted from an E. coli chromosome using the λ Red and Xer
recombination system (3). However,
whether it is suitable for multiple gene deletion in different
strains and the efficiency of such deletions have not, to the best
of our knowledge, been reported.
During bacterial growth specific signal molecules
are secreted allowing the bacteria to monitor the population
density in the surrounding environment and adjust the expression of
multiple genes. This is known as quorum sensing (QS) and uses
signal molecules, termed autologous inducers (AIs). In type II QS,
luxS and pfs genes are significant genes, involved in
AI-2 synthesis (4). The pathogenic
association between bacteria and host is complex and diverse;
numerous studies demonstrate that the QS system of pathogenic
bacteria aids in a variety of biological functions, including
biofilm formation, virulence factor production, drug resistance and
adhesion (5).
Biofilms are formed by bacteria adhering to surfaces
and extracellular matrices (6).
The biofilms of certain pathogenic bacteria are formed on the
surface of tissues and organs, as well as on the surface of medical
devices, which results in complications due to infection (7). According to statistical analyses, the
majority of bacterial infections are caused by biofilms, which
result in drug resistance, but also compromise the host immune
system, markedly impacting the prevention and treatment of
bacterial infection (8).
Enterotoxigenic E. coli (ETEC) is the main cause of piglet
diarrhea, a particularly serious piglet disease, and an important
cause of piglet mortality. The current study focuses on
enterohemorrhagic (EHEC) and enteropathogenic E. coli
(EPEC). In addition, there are few studies, to the best of our
knowledge, regarding ETEC, thus the present study investigates the
effect of luxS and pfs gene deletion on biofilm
formation, in order to further elucidate the effects of virulence
factor gene expression and the association with drug
resistance.
Materials and methods
DNA manipulation
Pfu DNA polymerase (Agilent Technologies, Inc.,
Santa Clara, CA, USA) was used to generate polymerase chain
reaction (PCR) products for cloning and gene insertion, and
ReddyMix (Thermo Fisher Scientific, Beijing, China) was used for
screening of colonies by PCR, according to standard PCR protocols.
Restriction enzymes (Takara Biotechnology Co., Ltd., Changchun,
China), T4 DNA ligase (Takara Biotechnology Co., Ltd.), and the PCR
Purification Clean-Up kit (Takara Biotechnology Co., Ltd.) were
used according to the manufacturer's instructions.
Bacterial strains and media
Four strains of ETEC served as targets in the gene
integration experiments. ETEC and E. coli DH5α (Takara
Biotechnology Co., Ltd.) were cultured in Luria-Bertani (LB) medium
(Junfeng Bioengineering Co., Ltd., Beijing, China) and antibiotic
medium (Junfeng Bioengineering Co., Ltd.), in liquid broth and 1.5%
agar plates containing 100 µg/ml ampicillin (Junfeng
Bioengineering Co., Ltd.). All bacterial cultures were incubated at
37°C, with agitation at 200 rpm for liquid cultures. Strains and
plasmids used in the present study are described in Table I.
 | Table IBacterial strains and plasmids used
in the current study. |
Table I
Bacterial strains and plasmids used
in the current study.
| Strain or
plasmid | Description | Serial number | Source |
|---|
| Strain | | | |
| E. coli
K88 | Type F4
adhesin | CVCC1525 | CVMMC |
| E. coli
K99 | Type F5
adhesin | CVCC232 | CVMMC |
| E. coli
987P | Type F6
adhesin | CVCC209 | CVMMC |
| E. coli
F41 | Type F41
adhesin | CVCC231 | CVMMC |
| V. harveyi
BB120 | AI-2 detection | BAA-1116 | ATCC |
| V. harveyi
BB170 | AI-2 detection | BAA-1117 | ATCC |
| E. coli
DH5α | Host of clone
plasmid | | Takara
Biotechnology, Co., Ltd. |
| Plasmid | | | |
| pMD18-T | Vector for cloning
polymerase chain reaction products | | Takara
Biotechnology, Co., Ltd. |
| pJN105 | Cloning gentamicin
resistance gene | | Invitrogen Life
Technologies |
| pKD46 | Promote E.
coli chromosomal gene integration | | Invitrogen Life
Technologies |
Preparation of the homology arms with the
gentamicin (GM) resistance gene
The luxS and pfs genes from four types
of ETEC were amplified using the primers, luxS forward (F), luxS
reverse (R), pfs F and pfs R. These PCR products were cloned into
pMD-18T to create plasmids, p18T-luxS or p18T-pfs. The GM
resistance gene was amplified from the pJN105 plasmid using
primers, difGM F and difGM R (Table
II.) This primer incorporated a 3′ region of a gene encoding GM
resistance and a 5′ tail that included a 28-bp E. coli dif
site (dif E. coli: GGT GCG CAT AAT GTA TAT TAT GTT AAAT).
The plasmids, p18T-luxS and p18T-pfs were digested using
EcoRV restriction enzyme, and the GM resistance gene with
dif sites was cloned into the plasmids forming
p18T-luxS-difGM or p18T-pfs-difGM plasmids.
 | Table IIPCR primers used in the current
study. |
Table II
PCR primers used in the current
study.
| Name | Size (nt) | Sequence (5′ to
3′) | Function of PCR
product |
|---|
| luxS | | | |
| F | 22 |
ATGCCGTTGTTAGATAGCTTCA | |
| R | 22 |
GATGTGCAGTTCCTGCAACTTC | luxS gene
amplification |
| Pfs | | | |
| F | 22 |
TGGTAAACTATGCCTTCAAATC | |
| R | 19 |
GTACGACAACAAACGGGAC | pfs gene
amplification |
| GM | | | |
| F | 28 |
GGTGCGCATAATGTATATTATGTTAAAT | Gentamicin
resistance and gene amplification |
| R | 27 |
ATTTAACATAATATACATTATGCGCACC |
| difGM | | | |
| F | 56 |
ACTTCCTAGAATATATATTATGTAAACT | Gentamicin
resistance gene and mplification with dif site |
|
GGTGCGCATAATGTATATTATGTTAAAT |
| R | 55 |
AGTTTACATAATATATATTCTAGGAAGT |
|
ATTTAACATAATATACATTATGCGCACC |
The p18T-luxS-difGM and p18T-pfs-difGM plasmids were
digested with both EcoRI and PstI restriction
enzymes, and the DNA fragments were recovered and amplified using
the luxS F, luxS, Pfs F and Pfs R primers to transform the E.
coli.
Preparation of competent cells and
plasmid transformation
The plasmid, pKD46 was transformed into ETEC, as
described by De Mey et al (9). The four strains of ETEC, with the
pKD46 plasmid, were cultured in LB medium at 30°C and oscillated at
180 rpm for 12 h. Bacteria (0.1 ml) was added to 1 ml LB medium
containing 1 µg/ml ampicillin and 2 mmol/l L-arabinose
(Spectrum Technology Co., Ltd., Shanghai, China), and cultured at
30°C, with oscillation at 180 rpm. Bacteria were grown until
reaching an optical density (OD600) value of ~0.6. The
culture was then placed in ice-cold water for 20 min, and the
bacteria were centrifuged at 6,000 × g for 10 min, washed once with
ice-cold water and washed twice with ice-cold 10% glycerol
(Shanghai Seagull Trading Co., Ltd., Shanghai, China) prior to
collection for electroporation.
Bacterial transformation and ETEC
chromosomal gene integration
The luxS or pfs gene DNA fragment (on
plasmids, including the GM resistance gene and dif sites)
were mixed with competent cells and placed in an electroporation
cuvette (Takara Biotechnology Co., Ltd.). An electrical pulse was
applied to the competent cells at 1,800 V for 5 µsec and
were then placed in LB broth containing 1 mmol/l L-arabinose. The
cells were cultured at 30°C, oscillated at 180 rpm for 1 h and
plated on GM-selective agar (Thermo Fisher Scientific, Beijing,
China). The bacteria was inoculated in LB medium with GM, cultured
at 30°C and oscillated at 180 rpm for 24 h. This procedure was
repeated twice following which the cells were appropriately diluted
with LB culture without GM and plated on a solid LB agar to exclude
the strains that were unable to grow on the agar with GM. These
bacteria were further identified by PCR.
Crystal violet (CV) method for
quantification of biofilm formation
Biofilm assays on 6-well plates were performed, as
described by Hossain and Tsuyumu (10) with certain modifications. For
quantitative analysis of biofilm production, an overnight culture
was grown in biofilm-inducing medium (Junfeng Bioengineering Co.,
Ltd.) to an OD600 of ~2.0 and diluted at 1:20. Culture
medium was added to each plate and incubated for 24 h at 30°C
without agitation. To remove the loosely associated bacteria, the
culture was removed from the wells and rinsed three times with
sterile distilled water. CV (2%; Beijing Noble Laser Technology
Co., Ltd., Beijing, China) solution was added to each well to stain
the bacteria and the culture was incubated at room temperature for
15 min. The wells were rinsed with Milli-Q water (Shanghai Seagull
Trading Co., Ltd.) and 95% ethanol was added to each CV-stained
well. Absorbance (600 nm) was measured using a QFLC-7001
spectrophotometer (HKY Technology Co.,Ltd, Beijing, China).
Six-well plates were used per experiment, and the entire experiment
was performed in triplicate.
AI-2 activity assay
The AI-2 activity in cell-free E. coli
culture fluids was measured using the Vibrio harveyi BB170
bioluminescence reporter assay, at 6,000 × g for 10 min (11,12).
Cell-free culture fluids were prepared by filtration of liquid
cultures (13,14) or by centrifugation, as described
above. AI-2 activity was evaluated as light production compared to
background light obtained with the appropriate E. coli
growth medium. Liquid supernatant of V. harveyi BB120 served
as the control group and E. coli DH5α served as the negative
control.
Statistical analysis
Results were expressed as means ± standard deviation
and analyzed using SPSS 16.0 software (SPSS, Inc., Chicago, IL,
USA). Group differences were compared using Student's t-test and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Gene integration and selectable marker
excision
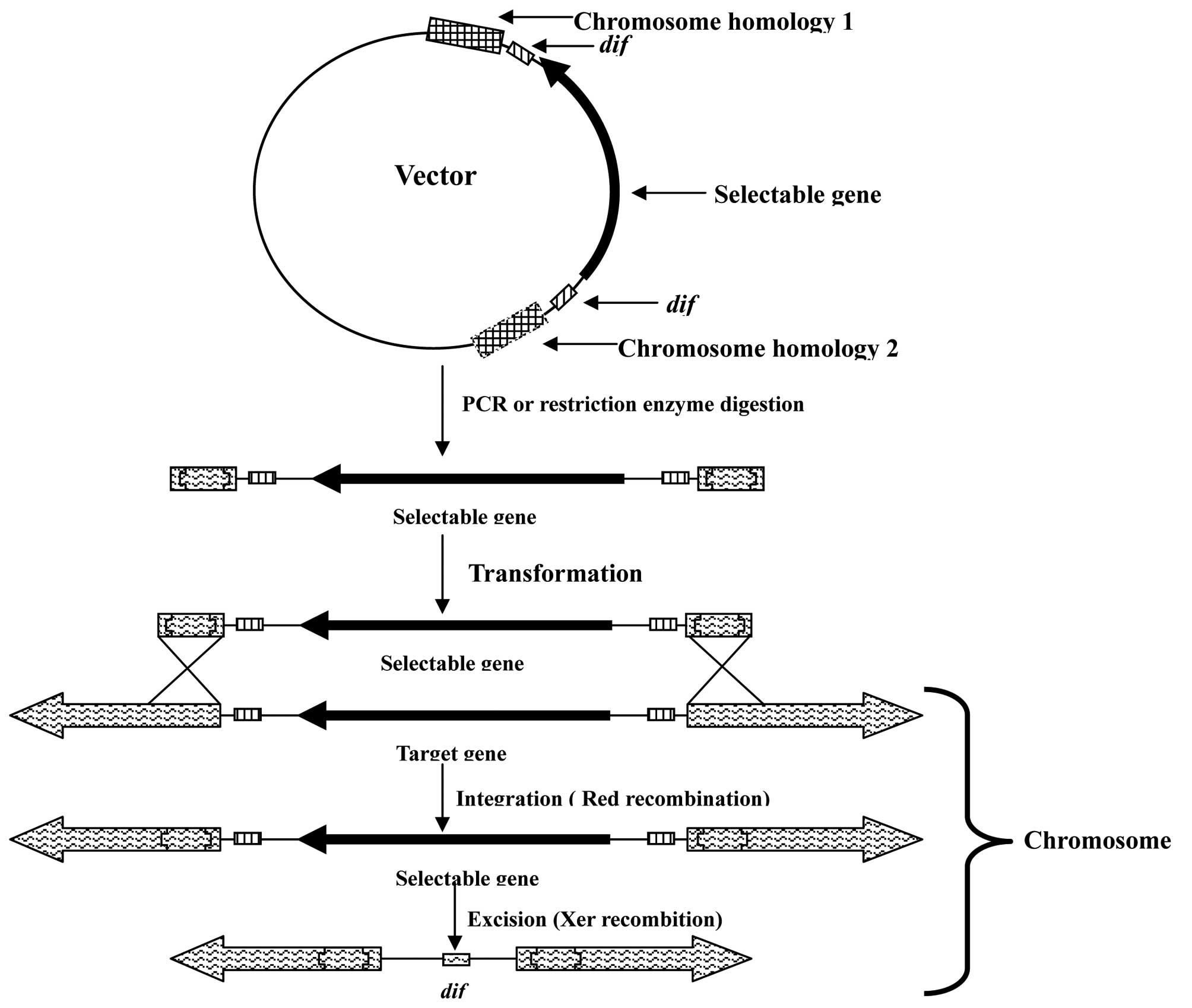
A schematic of the gene integration approach in
E. coli using λ Red and Xer recombination is presented in
Fig. 1. The target gene from the
ETEC bacteria was amplified and cloned into the pMD18-T vector.
Appropriate restriction sites in the target gene were selected;
however, when there was no suitable restriction site, reverse PCR
amplification was used to obtain two homologous arm vectors.
Fragments of the antibiotic resistance genes with dif sites
at the two terminals were inserted into the target gene (in the
present study, a GM resistance gene was used). The fragment,
including the selectable gene, dif sites and homologous
arms, was obtained from the vector by PCR amplification or
restriction enzyme digestion. The fragment and pKD46 vector were
subsequently transformed into ETEC. The target gene was excised as
the mutated ETEC recombines with the dif site to excise the
selectable gene using Xer recombination.
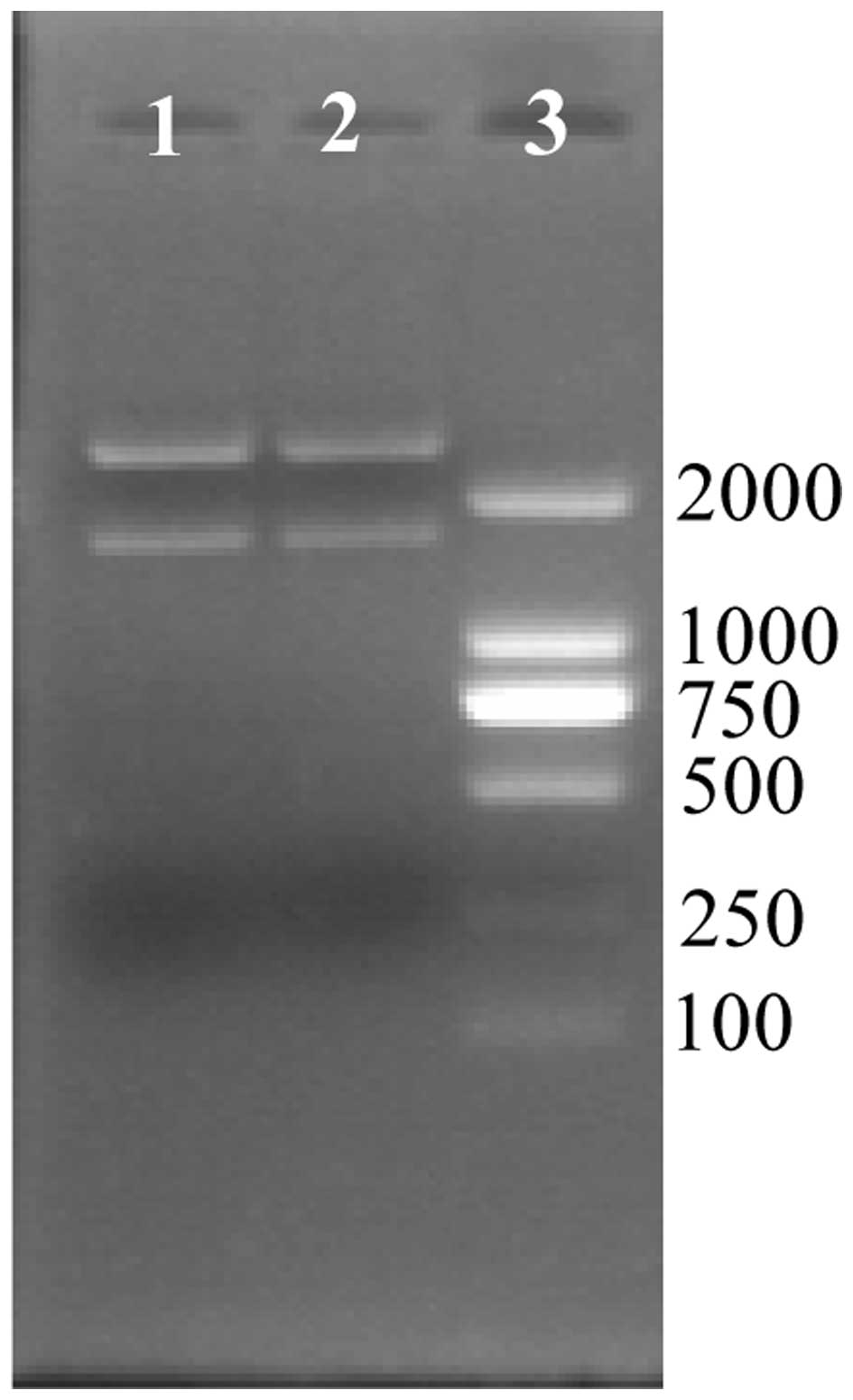
Construction of recombinant vector
The mutation method for the luxS and
pfs genes was the same, therefore, the method for
luxS gene inactivation is presented. The luxS gene
was amplified with primers luxS F and luxS R, and cloned into the
vector, pMD18-T. The vector, pMD-luxS, was then digested by the
restriction enzyme, EcoRV, and the fragment of GM resistance
gene with dif sites at the two terminals was cloned into the
vector. EcoRI and HindIII restriction enzymes were
used to digest the vector of pMD-luxS-difGM in order to obtain the
fragment from homologous recombination (Fig. 2). Two products are produced
following digestion; the vector, pMD18-T and the fragment of
homologous recombination.
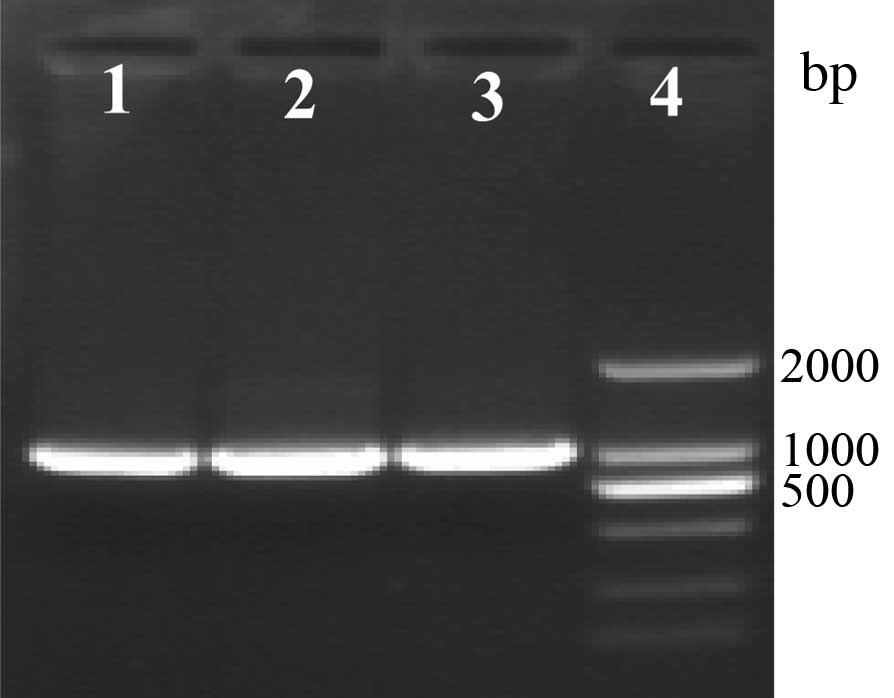
Recombination of four strains of
ETEC
Four strains of ETEC produced the same results,
thus, the data presented is of E. coli K88. Three randomly
selected bacteria were inoculated in LB medium with GM, amplified
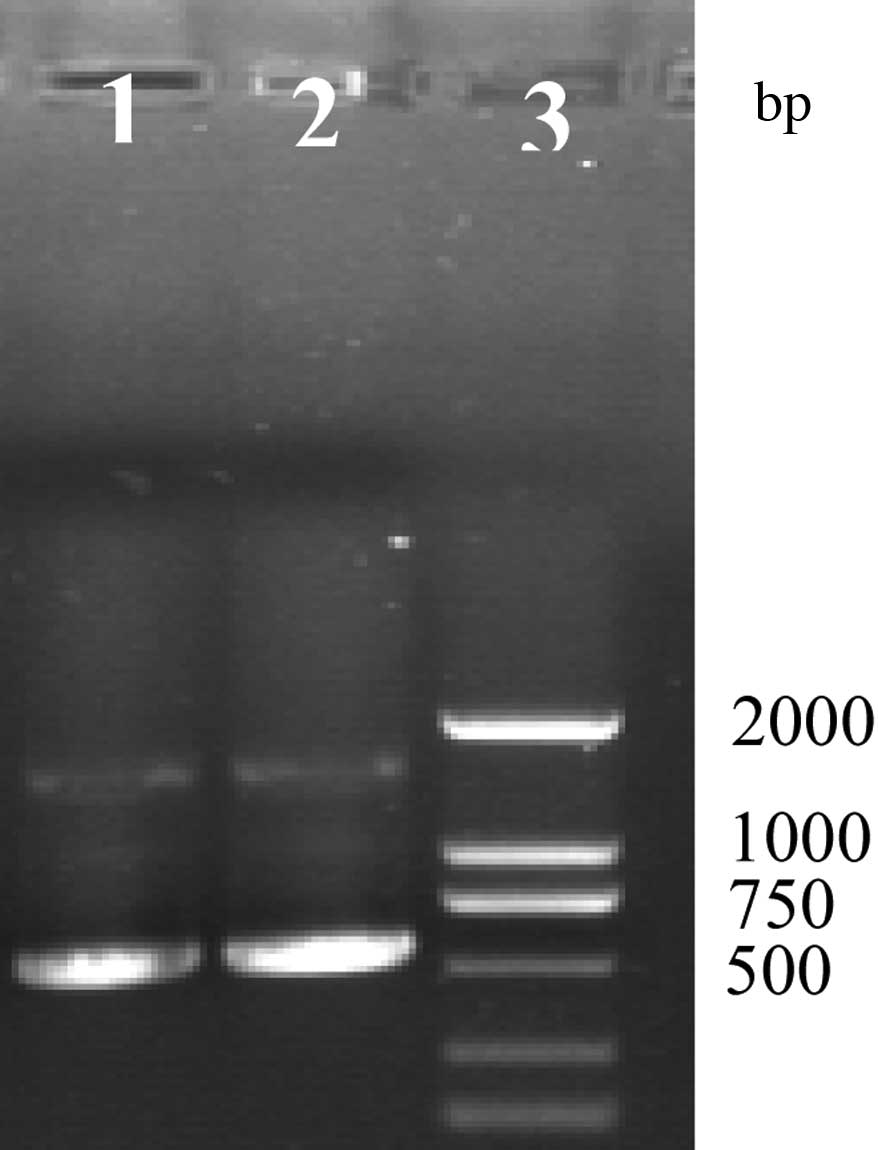
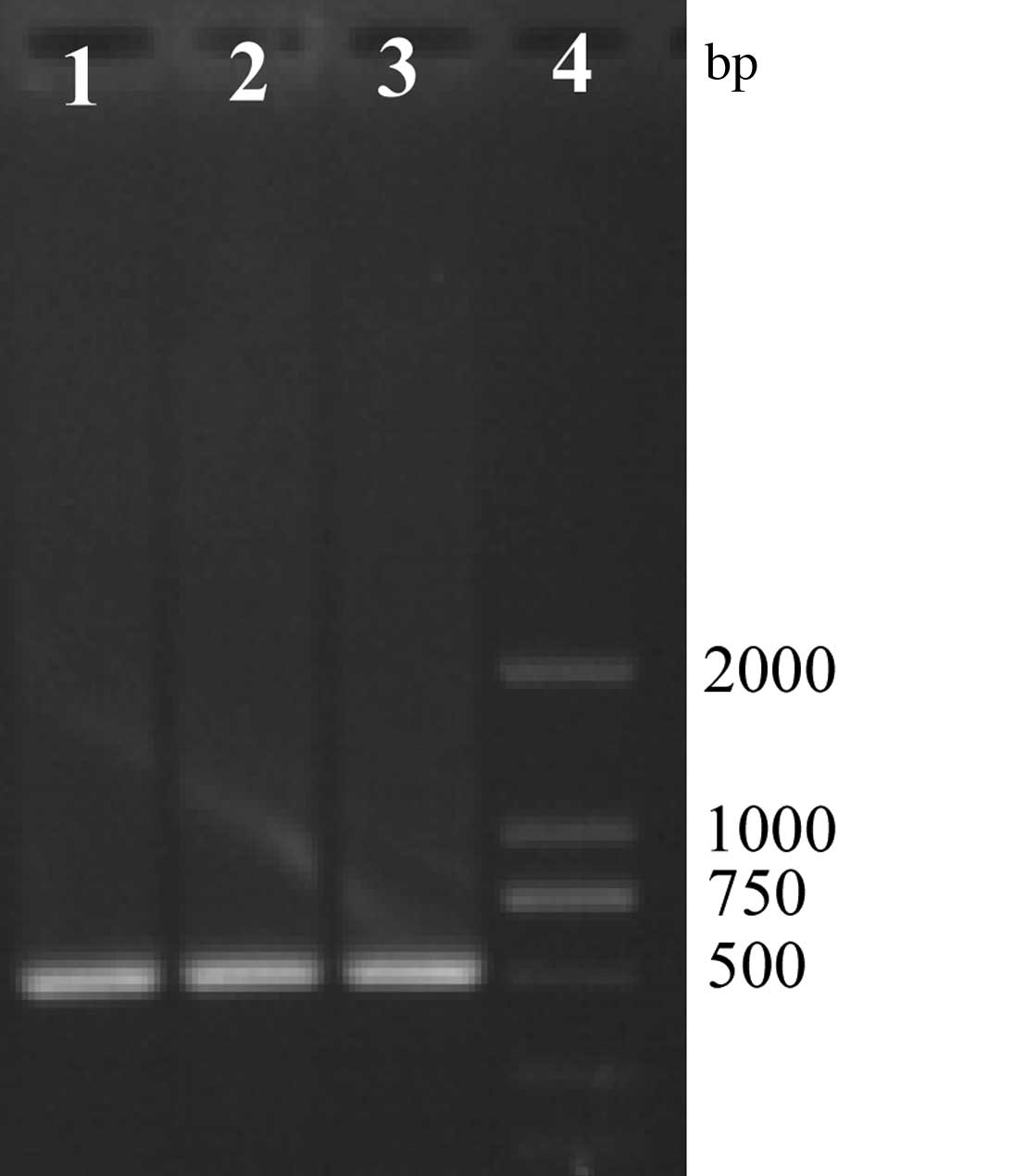
by GM F and GM R to produce a 1,000-bp product (Fig. 3). Amplification by luxS F and luxS
R produces two products at ~1,500 and 500 bp (Fig. 4). Bacteria were then subcultured a
third time, and amplified by luxS F and luxS R producing one
product at ~500 bp (Fig. 5).
Sequencing of the products indicated a 28-bp fragment was
successfully inserted into the luxS gene (Fig. 6). These findings indicate that
inactivation of the luxS and pfs genes had been
successful.
Effect of mutant strains on biofilm
formation
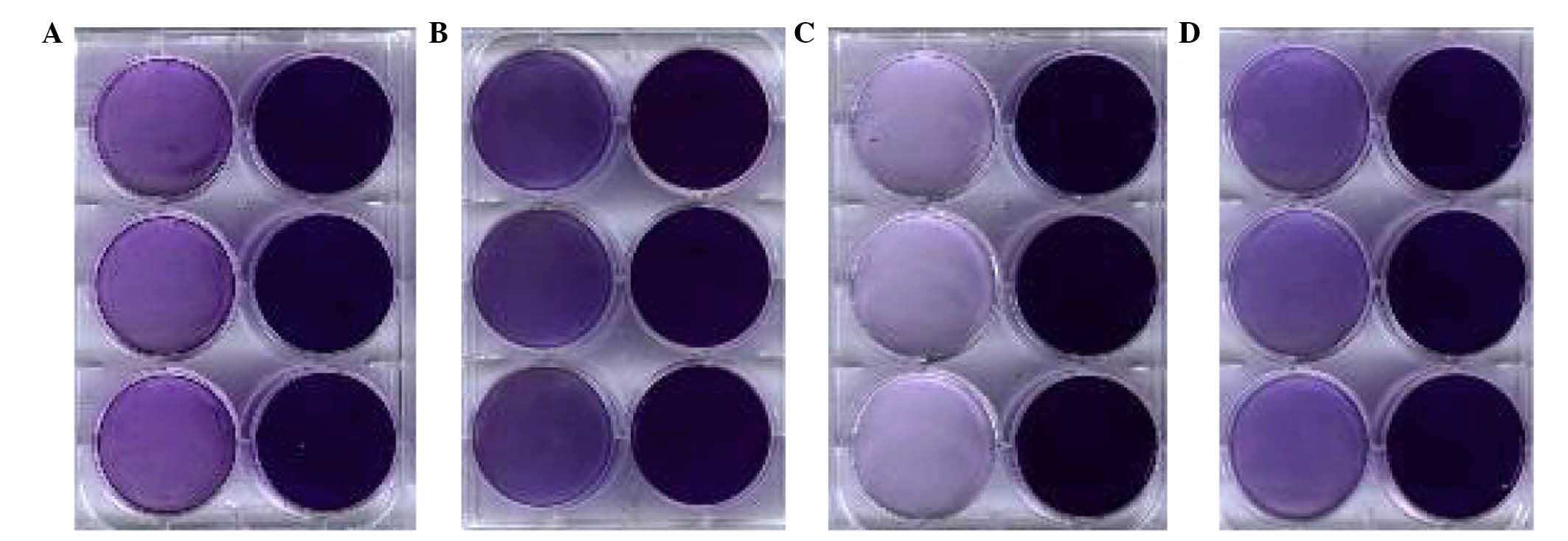
The ability to form biofilms was observed in four
strains of ETEC and is presented in Fig. 7. The ability to form biofilms was
detected by crystal violet staining in both the deletion plates and
the wild-type. It was demonstrated that the absorbance of the
deletion plates were significantly lower compared with the
wild-type, suggesting that deletion of the luxS gene
resulted in a decreased ability to form biofilms. Results of
ultraviolet spectrophotometry indicating absorption are presented
in Table III.
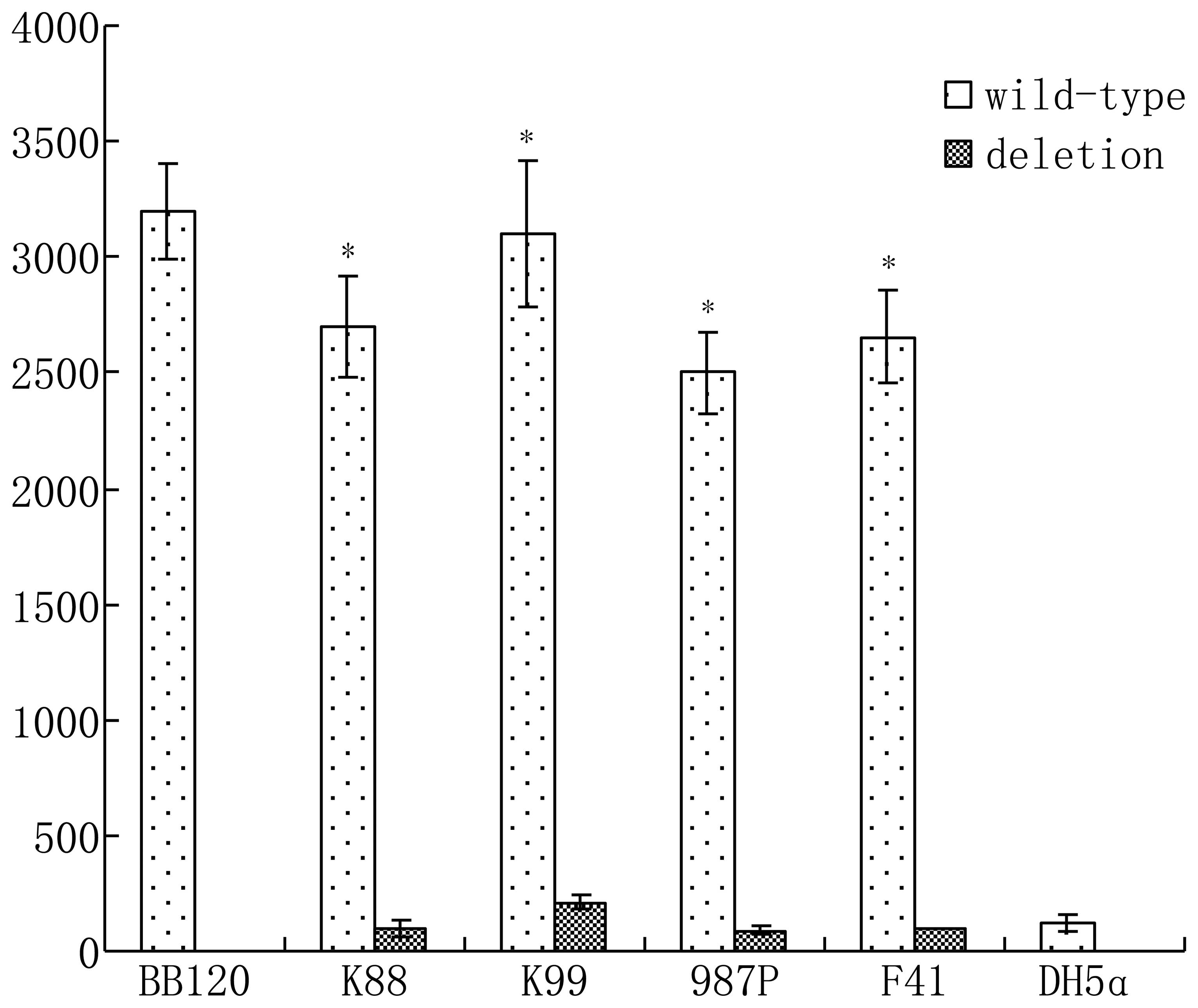
 | Table IIIAbsorbance of four strains of ETEC
bacteria. |
Table III
Absorbance of four strains of ETEC
bacteria.
| Absorbance l/(g·cm)
|
|---|
| Bacteria | K88 | K99 | 987P | F41 |
|---|
| Deletion | 0.21±0.04 | 0.44±0.11 | 0.16±0.09 | 0.22±0.08 |
| Wild-type |
1.17±0.11a | 1.21±0.13a | 1.57±0.19a | 1.39±0.21a |
AI-2 activity detection
V. harveyi BB170 was used to assay the AI-2
level of ETEC. Liquid supernatant of V. harveyi BB120 served
as the control group, E. coli DH5α served as the negative
control (Fig. 8). AI-2 secretion
was significantly decreased following deletion of the luxS
gene.
Discussion
The present study demonstrated a simple and rapid
technique for selectable marker gene deletion following gene
integration in bacteria, which should be applicable to all
prokaryotes with the ubiquitous Xer recombination system (15). The fragment, including a selectable
gene, dif sites and the chromosomal target gene was
amplified by PCR, or cloned into a plasmid, and then integrated
into the chromosome. ETECs that have undergone Xer recombination at
dif sites during continuous culture (and have, therefore,
lost the resistance gene) are identified by antibiotic sensitivity
testing and verified by PCR.
Competent cells of E. coli demonstrate a
sharp peak of transformation efficiency at an OD600
value of ~0.4 when the electroporation method is applied (16); Datsenko and Wanner (1) reported that the value was ~0.6,
however the present study indicated that a value of ~0.8–1.0
obtains greater transformation efficiency. This value varies in
previous studies, therefore, it is hypothesized that peak
transformation efficiency varies with the strains and should be
determined prior to experimentation (17). Datsenko and Wanner (1) used homologous arm fragments (length,
50 bp) to conduct recombination, however, the current study
observed that the efficiency of homologous recombination using a
fragment of ~200 bp was ten times greater than using a 50-bp
fragment. In the current study, the luxS or pfs genes
of ETEC were replaced with a GM resistance gene by homologous
recombination. As the chromosome mutation is irreversible, the
exogenous gene is highly stable in the bacteria. Throughout the
present study, the efficiency of recombination was observed to be
reduced when using fresh competent cells, however the efficiency
was increased when the competent cells were placed in the
refrigerator at −80°C for 24 h or longer. This was observed in the
four strains of ETEC that were investigated, whether the deleted
gene was luxS, pfs, or ack (data not shown).
The reason for this has not been determined in the current study,
however, is an important factor in recombination efficiency. The
Xer recombination system deletes selected genes with a homologous
arm fragment by recognizing the dif site of E. coli
after the homologous arm has been exchanged with the chromosome. In
the process of multiple gene deletion, the plasmid (such as pKD46)
used in λ Red recombination was maintained within the strain and
used to delete another gene, further simplifying the experimental
procedure. A previous study demonstrated that dif site
recombination is not affected by antibiotic use (3). The GM resistance gene can be removed
by Xer recombination, and upon addition of GM to the culture
medium, a 28-bp fragment remains in the luxS or pfs
gene. The recombination efficiency of the Xer system is high.
Fig. 4 presents results from the
first subculture, demonstrating production of two products at
~1,500 and 500 bp. The product at ~1,500 bp was the bacterial
chromosome fragment resulting from λ Red recombination, while the
~500-bp fragment had undergone Xer recombination. The bright bands
at ~500 bp suggests that the majority of bacteria used the Xer
system. Fig. 5 demonstrates the
third subculture, the presence of only the 500 bp product suggests
all bacteria had undergone λ Red and Xer recombination. Previous
research has speculated that multiple dif sites on a
chromosome may recombine again in the process of culture, leading
to unstable characteristics of gene mutant strains (18). In the current study, two dif
sites were used to mutate the luxS and pfs genes,
therefore the chromosome of the strains exhibited two dif
sites, however the sites did not recombine in subsequent culture.
The findings of the present study may enhance the methods of gene
mutation of ETEC. More than ten ETEC genes were deleted in the
present study and these strains lost the corresponding
characteristic during fermentation. For example, K88 and K99 have a
hemolysin toxin encoded by hlyA. The ability to destroy red blood
cells was significantly decreased after deletion of the hlyA gene.
The fliC gene was deleted which encoded the major flagellin protein
and thus, biofilm formation was reduced in the fliC gene mutant
strain. Finally, the EtpA gene was deleted, which encoded the
adhesin protein. This resulted in a reduction in biofilm
formation.
QS is a cell-to-cell signaling process in bacteria
enabling control of gene expression and synchronizing activities
that are beneficial only at a high population density (19). QS functions via the production,
secretion and detection of small molecules termed AIs; however
production and detection of the majority of AIs is restricted to a
single species. AI-2 (expressed by the luxS gene) is an
exception as it is widely distributed in different strains of
bacteria and controls distinct traits (5,20,21).
As a result of these characteristics, it is proposed that AI-2 is
used during interspecies communication. AI-2 was discovered in
V. harveyi and previous studies have demonstrated that
certain organisms use AI-2 in the regulation of genes, which
determine various functions, including expression of virulence
factors in Actinobacillus actinomycetemcomitans (22,23),
EHEC O157:H7 (24), P.
gingivalis (25,26), Streptococcus pyogenes
(27), Vibrio cholerae
(28–30), and Vibrio vulnificus
(31); motility in
Campylobacter jejuni (32),
EHEC O157:H7 and EPEC E. coli O127:H6 (33,34);
cell division in E. coli W3110 and EHEC O157:H7 (35,36);
production of antibiotics in Photorhabdus luminescens
(37); biofilm formation in
Streptococcus gordonii (38); and an AI-2 ATP binding
cassette-type transporter in Salmonella enterica serovar
Typhimurium (39). This previous
research demonstrates the wide variety of bacteria using AI-2 to
control a diverse range of function-specific genes.
Previous studies suggest that the expression of
virulence factors and metabolism of E. coli bacteria were
regulated by the luxS gene (40). Further analysis determined that
luxS performs a variety of functions, regulates the
expression of hundreds of genes, determines population density and
regulates biofilm formation by synthesis of AI-2 signal molecules.
In addition, exogenous AI-2 enhances biofilm formation and the
mechanism may function via the expression of flagella, which induce
bacterial activity (41–43); however, these previous studies
focused on EHEC and EPEC or other strains of E. coli. In the
current study, four strains of ETEC were used and it was
demonstrated that the ability to form biofilms was significantly
reduced without the luxS gene (Fig. 7 and Table III). The result suggests that the
effect of luxS on biofilm formation in ETEC is widespread,
with the largest impact observed on 987P and the smallest on K99.
Current research on ETEC is minimal, therefore, the aim of the
present study was to demonstrate that multiple gene inactivation to
delete the luxS gene (or other genes) may be used to
evaluate the influence of AI-2 on biofilm formation, the associated
drug resistance and virulence factors in order to contribute to the
prevention and treatment of piglet diarrhea.
In conclusion, gene inactivation by λ Red and Xer
recombination is a rapid, efficient and stable method for deletion
of ETEC genes. Deletion of luxS and pfs significantly
reduces the activity of AI-2 and the ability of ETEC to form
biofilms. However, further investigation is required to elucidate
the effect of QS on the expression of virulence genes and drug
resistance.
Acknowledgments
The present study was supported by the Chinese
Postdoctoral Science Foundation (grant no. 2014M552581).
References
|
1
|
Datsenko KA and Wanner BL: One-step
inactivation of chromosomal genes in Escherichia coli K-12 using
PCR products. Proc Natl Acad Sci USA. 97:6640–6645. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baba T, Ara T, Hasegawa M, Takai Y,
Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL and Mori H:
Construction of Escherichia coli K-12 in-frame, single-gene
knockout mutants: The Keio collection. Mol Syst Biol.
2:2006.00082006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bloor AE and Cranenburgh RM: An efficient
method of selectable marker gene excision by Xer recombination for
gene replacement in bacterial chromosomes. Appl Environ Microbiol.
72:2520–2525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Beeston AL and Surette MG: pfs-dependent
regulation of autoinducer 2 production in Salmonella enterica
serovar Typhimurium. J Bacteriol. 184:3450–3456. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xavier KB and Bassler BL: LuxS quorum
sensing: More than just a numbers game. Curr Opin Microbiol.
6:191–197. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costerton JW, Stewart PS and Greenberg EP:
Bacterial biofilms: A common cause of persistent infections.
Science. 284:1318–1322. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kozlova EV, Popov VL, Sha J, Foltz SM,
Erova TE, Agar SL, Horneman AJ and Chopra AK: Mutation in the
S-ribosylhomocysteinase (luxS) gene involved in quorum sensing
affects biofilm formation and virulence in a clinical isolate of
Aeromonas hydrophila. Microb Pathog. 45:343–354. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hancock V, Dahl M and Klemm P: Probiotic
Escherichia coli strain Nissle 1917 outcompetes intestinal
pathogens during biofilm formation. J Med Microbiol. 59:392–399.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Mey M, De Maeseneire S, Soetaert W and
Vandamme E: Minimizing acetate formation in E. coli fermentations.
J Ind Microbiol Biotechnol. 34:689–700. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hossain MM and Tsuyumu S:
Flagella-mediated motility is required for biofilm formation by
Erwinia carotovora subsp. carotovora. J Gen Plant Pathol. 72:34–39.
2006. View Article : Google Scholar
|
|
11
|
Bassler BL, Wright M, Showalter RE and
Silverman MR: Intercellular signalling in Vibrio harveyi: Sequence
and function of genes regulating expression of luminescence. Mol
Microbiol. 9:773–786. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bassler BL, Wright M and Silverman MR:
Multiple signalling systems controlling expression of luminescence
in Vibrio harveyi: Sequence and function of genes encoding a second
sensory pathway. Mol Microbiol. 13:273–286. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Surette MG and Bassler BL: Quorum sensing
in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci
USA. 95:7046–7050. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Surette MG and Bassler BL: Regulation of
autoinducer production in Salmonella typhimurium. Mol Microbiol.
31:585–595. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Recchia GD and Sherratt DJ: Conservation
of xer site-specific recombination genes in bacteria. Mol
Microbiol. 34:1146–1148. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sambrook J and Russell DW: Molecular
cloning: A laboratory Manual. 3rd Edn. New York: Cold Spring Harbor
Laboratory Press; 2001. pp. 1.199–1.122
|
|
17
|
Li Zhou, Dan-Dan Niu, Ning Li, Xian-Zhong
Chen, Gui-Yang Shi and Zheng-Xiang Wang: Multiple gene inactivation
approach in Escherichia coli mediated by a combination of red
recombination and Xer recombination. Microbiology China.
37:923–928. 2010.
|
|
18
|
Zhou L, Niu DD, Li N, Chen XZ, Shi GY and
Wand ZX: Multiple gene inactivation approach in Escherichia coli
mediated by a combination of red recombination and Xer
recombination. Microbiol China. 37:923–928. 2010.
|
|
19
|
Xavier KB and Bassler BL: Regulation of
uptake and processing of the quorum-sensing autoinducer AI-2 in
Escherichia coli. J Bacteriol. 187:238–248. 2005. View Article : Google Scholar :
|
|
20
|
Federle MJ and Bassler BL: Interspecies
communication in bacteria. J Clin Invest. 112:1291–1299. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Surette MG, Miller MB and Bassler BL:
Quorum sensing in Escherichia coli, Salmonella typhimurium, and
Vibrio harveyi: A new family of genes responsible for autoinducer
production. Proc Natl Acad Sci USA. 96:1639–1644. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fong KP, Chung WO, Lamont RJ and Demuth
DR: Intra-and interspecies regulation of gene expression by
Actinobacillus actinomycetemcomitans LuxS. Infect Immun.
69:7625–7634. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fong KP, Gao L and Demuth DR: luxS and
arcB control aerobic growth of Actinobacillus actinomycetemcomitans
under iron limitation. Infect Immun. 71:298–308. 2003. View Article : Google Scholar :
|
|
24
|
Sperandio V, Mellies JL, Nguyen W, Shin S
and Kaper JB: Quorum sensing controls expression of the type III
secretion gene transcription and protein secretion in
enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl
Acad Sci USA. 96:15196–15201. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Burgess NA, Kirke DF, Williams P, Winzer
K, Hardie KR, Meyers NL, Aduse-Opoku J, Curtis MA and Cámara M:
LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates
protease and haemagglutinin activities but is not essential for
virulence. Microbiology. 148:763–772. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chung WO, Park Y, Lamont RJ, McNab R,
Barbieri B and Demuth DR: Signaling system in Porphyromonas
gingivalis based on a LuxS protein. J Bacteriol. 183:3903–3909.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lyon WR, Madden JC, Levin JC, Stein JL and
Caparon MG: Mutation of luxS affects growth and virulence factor
expression in Streptococcus pyogenes. Mol Microbiol. 42:145–157.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lenz DH, Mok KC, Lilley BN, Kulkarni RV,
Wingreen NS and Bassler BL: The small RNA chaperone Hfq and
multiple small RNAs control quorum sensing in Vibrio harveyi and
Vibrio cholerae. Cell. 118:69–82. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miller MB, Skorupski K, Lenz DH, Taylor RK
and Bassler BL: Parallel quorum sensing systems converge to
regulate virulence in Vibrio cholerae. Cell. 110:303–314. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu J, Miller MB, Vance RE, Dziejman M,
Bassler BL and Mekalanos JJ: Quorum-sensing regulators control
virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci
USA. 99:3129–3134. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim SY, Lee SE, Kim YR, Kim CM, Ryu PY,
Choy HE, Choy SS and Rhee JH: Regulation of Vibrio vulnificus
virulence by the LuxS quorum-sensing system. Mol Microbiol.
48:1647–1664. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Elvers KT and Park SF: Quorum sensing in
Campylobacter jejuni: Detection of a luxS encoded signalling
molecule. Microbiology. 148:1475–1481. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Girón JA, Torres AG, Freer E and Kaper JB:
The flagella of enteropathogenic Escherichia coli mediate adherence
to epithelial cells. Mol Microbiol. 44:361–379. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sperandio V, Torres AG and Kaper JB:
Quorum sensing Escherichia coli regulators B and C (QseBC): A novel
two-component regulatory system involved in the regulation of
flagella and motility by quorum sensing in E. coli. Mol Microbiol.
43:809–821. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
DeLisa MP, Wu C-F, Wang L, Valdes JJ and
Bentley WE: DNA microarray-based identification of genes controlled
by autoinducer 2-stimulated quorum sensing in Escherichia coli. J
Bacteriol. 183:5239–5247. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sperandio V, Torres AG, Girón JA and Kaper
JB: Quorum sensing is a global regulatory mechanism in
enterohemorrhagic Escherichia coli O157:H7. J Bacteriol.
183:5187–5197. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Derzelle S, Duchaud E, Kunst F, Danchin A
and Bertin P: Identification, characterization, and regulation of a
cluster of genes involved in carbapenem biosynthesis in
Photorhabdus luminescens. Appl Environ Microbiol. 68:3780–3789.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McNab R, Ford SK, El-Sabaeny A, Barbieri
B, Cook GS and Lamont RJ: LuxS-based signaling in Streptococcus
gordonii: autoinducer 2 controls carbohydrate metabolism and
biofilm formation with Porphyromonas gingivalis. J Bacteriol.
185:274–284. 2003. View Article : Google Scholar :
|
|
39
|
Taga ME, Semmelhack JL and Bassler BL: The
LuxS-dependent autoinducer AI-2 controls the expression of an ABC
transporter that functions in AI-2 uptake in Salmonella
typhimurium. Molecular Microbiology. 42:777–793. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Walters M and Sperandio V: Quorum sensing
in Escherichia coli and Salmonella. Int J Med Microbiol.
296:125–131. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Beloin C, Roux A and Ghigo J-M:
Escherichia coli biofilms. Bacterial Biofilms. Romeo T: Springer;
Berlin Heidlberg: pp. 249–289. 2008, View Article : Google Scholar
|
|
42
|
Naves P, del Prado G, Huelves L, Gracia M,
Ruiz V, Blanco J, Dahbi G, Blanco M, Ponte MC and Soriano F:
Correlation between virulence factors and in vitro biofilm
formation by Escherichia coli strains. Microb Pathog. 45:86–91.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yamaguchi Y, Park JH and Inouye M: MqsR, a
crucial regulator for quorum sensing and biofilm formation, is a
GCU-specific mRNA interferase in Escherichia coli. J Biol Chem.
284:28746–28753. 2009. View Article : Google Scholar : PubMed/NCBI
|