Introduction
Osteosarcoma is the most common type of primary bone
tumor worldwide and exhibits a peak incidence in the second and
third decades of life (1).
Osteosarcoma can arise in any bone, however, it is most common in
the metaphyses of long bones (1).
Although survival rates have increased between 20 and- 75% due to
the combination of radical surgery and neoadjuvant chemotherapy
(2–4), for patients who present with
metastatic disease or present with tumor recurrence, the survival
rates remain <30 and <20%, respectively (5). This emphasizes the requirement for
the development of novel therapeutic targets and approaches for the
treatment of osteosarcoma.
Sex determining region Y-box 18 (SOX18) is a member
of the sex-determining region of the Y chromosome-related high
mobility group box (SOX) family of transcription factors. It
selectively interacts with the common SOX target sequence
(A/T)ACAA(A/T)G, and activates transcription via a transactivation
domain adjacent to the high mobility group domain (6,7).
Previous studies (8,9) have suggested that the expression
level of SOX18 may affect tumor growth. It has been reported that
the expression levels of SOX18 are increased in gastric cancer
tissues, compared with normal gastric tissues (10). Furthermore, the expression of SOX18
is correlated with poor survival rates (10). The expression of SOX18 has also
been correlated with poor clinical outcome in patients with
non-small cell lung cancer (11),
ovarian cancer (12) and invasive
ductal breast carcinoma (13).
However, the expression pattern and biological functions of SOX18
in osteosarcoma remain to be fully elucidated.
The present study aimed to investigate the role of
SOX18 in osteosarcoma. Initially, the expression levels of SOX18
were analyzed using reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) analysis in osteosarcoma tissue samples
obtained from 25 patients. Subsequently, the biological function of
SOX18 in osteosarcoma cell lines was investigated using RNA
interference (RNAi). The present study also aimed to elucidate
whether SOX18 is involved in cell proliferation, cell cycle
progression, apoptosis, adhesion and invasion, and whether SOX18 is
involved in these processes by regulating the expression of
transforming growth factor-β1 (TGF-β1), platelet-derived growth
factor-A (PDGF-A), PDGF-B and Ras homolog family member A
(RhoA).
Materials and methods
Patients and tissue samples
Between 2010 and 2012, 25 patients (16 men and 9
women) with conventional (occurring in the metaphyses of the long
bones) osteosarcoma, who were admitted to the Department of
Orthopedics, Shanghai Tenth People's Hospital (Tongji University,
Shanghai, China), were enrolled in the present study. Complete
clinical and pathological follow-up data were obtained for all
patients. The patients ranged in age from 7 to 49 years with a
median age of 18 years. Osteosarcoma tissues (0.1–0.2 g) were
obtained from femur or tibia of these 25 patients and normal bone
tissues were also collected as negative controls. These normal bone
tissues were resected within at least 5 cm of the tumor margin when
the patients underwent definitive surgery. Ethical approval for the
present study was provided by the independent ethics committee of
Shanghai Tenth People's Hospital, Tongji University (Shanghai,
China). Informed and written consent was obtained from all patients
or their advisers, according to the ethics committee
guidelines.
Antibodies
The following primary antibodies were used in the
present study: Mouse polyclonal SOX18 (Ab66145; 1:1,000), rabbit
polyclonal TGF-β1 (Ab92486; 1:400) and rabbit polyclonal RhoA
(Ab68826; 1:2,000) (Abcam, Cambridge, MA, USA), rabbit polyclonal
PDGF-A (BA0408; 1:200) and rabbit polyclonal PDGF-B (BA0519-2;
1:200) (Wuhan Boster Biological Technology, Ltd. (Wuhan, China) and
rabbit monoclonal glyceraldehyde 3-phosphate dehydrogenase (GAPDH;
#5174; 1:2,000), Cell Signaling Technology, Inc. (Danvers, MA,
USA). Horseradish peroxidase-conjugated goat anti-mouse and goat
anti-rabbit secondary antibodies were purchased from Beyotime
Institute Biotechnology (Shanghai, China).
Cell culture
MG63, HOS, 143B, Saos2, U2OS and HEK293T cells were
purchased from the American Type Culture Collection (Rockville, MD,
USA). The MG63, HOS, Saos2, 143B and HEK293T cells were grown in
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo
Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Thermo
Fisher Scientific, Inc.). The U2OS cells were grown in RPMI-1640
medium with 10% FBS (Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin. All cell lines were maintained at 37°C in
a 5% CO2 atmosphere.
Vector construction and virus
transduction
Three shRNAs targeting human SOX18 mRNA (SOX18-Ri-3,
AGGAAGCCGAACGGCTGCGTT; SOX18-Ri-2, AGGCTGCCTTCTTCCCTCCTT; and
SOX18-Ri-3, TACCACGTGGCACTGGCCATT; Generay Biotech Co., Ltd.,
Shanghai, China) were cloned into a lentiviral vector (PLKO.1;
Addgene, Inc., Cambridge, MA, USA). A non-specific scramble shRNA
sequence (TTCTCCGAACGTGTCACGTTT) was used as the negative control
(NC). The constructs were then transfected into the HEK293T cells
with lentiviral packaging vectors using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The viruses were collected 48 h subsequent
to transfection and used to transduce the U2OS cells and MG63
cells. After 48 h, the cells were processed for RT-qPCR and western
blotting.
RT-qPCR
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Total RNA (1 µg) was reverse
transcribed using a First Strand cDNA Synthesis kit (K1612; Thermo
Fisher Scientific Inc.), according to the manufacturer's protocol.
RT-qPCR was performed using a SYBR Green PCR kit (Thermo Fisher
Scientific, Inc.) on an ABI 7300 Real-time PCR machine (Applied
Biosystems; Thermo Fisher Scientific, Inc., Foster City, CA, USA)
using the following cycling parameters: 95°C for 10 min, followed
by 40 cycles of 95°C for 15 sec and 60°C for 45 sec. GAPDH served
as an internal control. The gene expression was calculated using
the ΔΔCt method (14). All data
represent the average of three replicates. The primers used
(Generay Biotech Co., Ltd.) were as follows: SOX18 (NM_018419.2),
forward 5′-CGCGTGTATGTTTGGTTC-3′ and reverse
5′-ATGTAACCCTGGCAACTC-3′; TGF-β1 (NM_000660.4), forward
5′-GACTACTACGCCAAGGAGGTC-3′ and reverse 5′-GAGAGCAACACGGGTTCAG-3′;
PDGF-A (NM_002607.5), forward 5′-CGTAGGGAGTGAGGATTCTTTG-3′ and
reverse 5′-AAATGACCGTCCTGGTCTTG-3′; PDGF-B (NM_002608.2), forward
5′-CTCGATCCGCTCCTTTGATG-3′ and reverse 5′-AGGAAGTTGGCGTTGGTG-3′;
RhoA (NM_001664.2), forward 5′-GAGTGTTCAGCAAAGACCAAAG-3′ and
reverse 5′-TTGCAGCAAGGTTTCACAAG-3′; GAPDH (NM_001256799.1), forward
5′-CACCCACTCCTCCACCTTTG-3′ and reverse
5′-CCACCACCCTGTTGCTGTAG-3′.
Western blotting
The treated and untreated MG63 and U2OS cells were
harvested and washed twice with phosphate-buffered saline (PBS) and
lysed in ice-cold radio immunoprecipitation assay buffer (JRDUN
Biotechnology Co., Ltd., Shanghai, China) with freshly added 0.01%
protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA).
The cells were then incubated on ice for 30 min. The cell lysates
were centrifuged at 16,000 × g for 10 min at 4°C. Protein
concentration was measured using the Bicinchoninic Acid Assay kit
(Thermo Fisher Scientific, Inc.) and the supernatant (20–30
µg protein) was run on a 15% SDS-PAGE gel and transferred
electrophoretically onto a nitrocellulose membrane (EMD Millipore,
Billerica, MA, USA). Subsequent to blocking with 5% skimmed milk,
the membranes were incubated with the primary antibodies, followed
by the corresponding horseradish peroxidase-conjugated secondary
antibodies (Beyotime Institute of Biotechnology). The blots were
then visualized using enhanced chemiluminescence (EMD
Millipore).
Cell proliferation assay
Cell proliferation was measured using a Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) according to manufacturer's protocol. In brief,
the U2OS and MG63 cells (~1–5×103) were seeded into
96-well plates. At 0, 24, 48 and 72 h, CCK-8 solution (10 µl
in 100 µl DMEM) was added into each well, followed by
incubation at 37°C for 1 h. The optical density values were
measured at a wavelength of 450 nm using a microplate reader (Model
550; Bio-Rad Laboratories, Inc., Hercules, CA, USA). All
experiments were performed in triplicate and repeated a minimum of
three times.
Cell cycle distribution analysis
Propidium iodide (PI; Sigma-Aldrich) staining was
performed to analyze the DNA content in the cells to determine cell
cycle distribution. The cells were harvested 48 h following
transduction, and were labeled with PI, as previously described
(15). In brief, the cells were
resuspended in PBS and fixed with 70% ethanol. Cells were washed
twice with PBS and then suspended at a concentration of
1×106 cells/ml. Following treatment with ribonuclease
(Sigma-Aldrich) for 15 min at 37°C, PI (0.05 mg/ml) was added to
the cells, followed by incubation at room temperature in the dark
for 30 min. DNA content was then analyzed using a FACScan
instrument equipped with FACStation running CellQuest software,
version 3.3 (BD Biosciences, San Jose, CA, USA).
Cell apoptosis assay
The percentages of the cells actively undergoing
apoptosis were determined by double staining with annexin
V-fluorescein isothiocyanate (FITC) and PI. The adherent and
floating virally transduced or control cells were harvested after
48 h, and double-labeled with annexin V-FITC and PI (BD
Biosciences), according to the manufacturer's protocol. The cells
were analyzed using a FACScan instrument equipped with FACStation
running CellQuest software.
Cell adhesion assay
To determine cell adhesion, the assay was performed
in 12-well plates. The plates were pre-coated with 1 ml fibronectin
(5 µg/ml) for 2 h at room temperature. The cells were
transduced, as described above, 48 h prior to the assay. The cells
were seeded into the coated plates at a density of 105
cells/well and allowed to adhere at 37°C for 1 h. Non-adherent
cells were washed off with PBS and the cells were fixed in 4%
paraformaldehyde (Beijing Solarbio Science & Technology Co.,
Ltd., Beijing, China) and stained with 0.2% crystal violet (Beijing
Solarbio Science & Technology Co., Ltd.). The number of
adherent cells was determined in five randomly selected fields
under a microscope (Eclipse E600; Nikon Corporation, Tokyo, Japan),
as previously described (16).
In vitro invasion assay
The upper well of a Transwell chamber (Corning
Incorporated, NY, USA) was coated with Matrigel (BD Biosciences) at
37°C in a 5% CO2 incubator for 1 h. The virus-treated
and untreated cells were serum starved for 24 h, then 500 µl
cell suspension containing 105 cells/ml were placed in
the upper compartment of the chamber. Culture medium supplemented
with 10% FBS (750 µl) was added into the lower well of the
chamber. The cells were allowed to invade through the Matrigel
membrane for 48 h, and non-invasive cells were removed from the
upper membrane. The invasive cells on the underneath were washed
with PBS, fixed in 4% paraformaldehyde and stained with 0.2%
crystal violet. The invading cells were observed under a
microscope. Cells were counted in the central field of the
membranes in triplicate.
Statistical analysis
All data are presented as the mean ± standard
deviation. Statistical significance was determined using Student's
two-tailed t-test with SPSS software, version 13.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
SOX18 is overexpressed in
osteosarcoma
The mRNA levels of SOX18 were measured in the
osteosarcoma and adjacent normal tissues of 25 patients using
RT-qPCR. As presented in Fig. 1A,
SOX18 was overexpressed in 88% (22/25) of the osteosarcoma tissues
assessed. Statistical analysis using Student's t-test indicated
that SOX18 mRNA was significantly overexpressed in osteosarcoma
tissues, compared with normal tissues (P<0.001).
 | Figure 1SOX18 is overexpressed in osteosarcoma
tissues, and the knockdown of SOX18 suppresses the proliferation of
osteosarcoma cells. (A) mRNA expression levels of SOX18 were
significantly increased in the osteosarcoma tissues (n=25),
compared with the levels in the normal tissues (n=25), obtained
from patients admitted to the Shanghai Tenth People's Hospital
(Shanghai, China) between 2009 and 2012. In the graph, a positive
log2 Tumor/Normal ratio on the y-axis indicates increased
expression levels of SOX18 in the tumor tissue, whereas a negative
log2 Tumor/Normal ratio indicates reduced expression levels of
SOX18 in the tumor tissue. (B) Expression levels of SOX18 in five
osteosarcoma cell lines were analyzed using western blotting (upper
panel) and RT-qPCR (lower panel). Data are representative of three
independent experiments. (C and D) Expression levels of SOX18 in
U2OS and MG63 cells was analyzed using western blotting (upper
panel) and RT-qPCR (lower panel). (E and F) Cell proliferation was
detected 24, 48 and 72 h subsequent to viral transduction of the
U2OS and MG63 cells. Data are representative of three independent
experiments and are presented as the mean ± standard deviation
(**P<0.01, vs. NC). SOX18, sex-determining region
Y-box 18; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; WT,
wild-type; NC, scrambled shRNA; RNAi-1, SOX18-shRNA-1 virus
transduction; SOX18-Ri-3, SOX18-shRNA-3 virus transduction; OD,
optical density; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction. |
Knockdown of SOX18 suppresses the
proliferation of osteosarcoma cells
The expression levels of SOX18 in five osteosarcoma
cell lines, Saos2, U2OS, HOS, MG63 and 143B, were assessed using
RT-qPCR and western blotting. The results demonstrated that two of
these cell lines, MG63 and U2OS, exhibited higher mRNA and protein
expression levels of SOX18, compared with the remaining Saos2, HOS
and 143B cell lines, which exhibited lower mRNA and protein
expression levels of SOX18 (Fig.
1B).
To investigate the effect of SOX18 on osteosarcoma,
SOX18 was knocked down in osteosarcoma cells using RNAi. U2OS and
MG63 cells were selected for the RNAi experiment due to the fact
that they expressed higher levels of SOX18. Three pairs of shRNA
(SOX18-Ri-3, SOX18-Ri-2 and SOX18-Ri-3) targeting human SOX18, and
a non-specific scramble shRNA (NC) were designed and cloned into a
lentiviral plasmid. The recombinant lentivirus was then packaged
into the HEK293T cells and used to transduce the U2OS and MG63
cells. The silencing effect of the shRNA was evaluated by western
blotting and RT-qPCR (Fig. 1C and
D). The results indicated that SOX18-Ri-3 was the most
efficient shRNA, with a knockdown efficiency of ~70%. Therefore,
SOX18-Ri-3 was selected for the following assays.
The effect of SOX18 RNAi on the proliferation of
osteosarcoma was then assessed. Knockdown of SOX18 by transduction
of the SOX18-shRNA virus into the U2OS or MG63 cells resulted in a
reduced cell growth rate, compared with the corresponding control
(Fig. 1E and F), whereas a similar
growth rate was observed between the WT cells and the NC cells.
These results indicated that SOX18 may promote the proliferation of
osteosarcoma cells.
Silencing of SOX18 induces S-phase arrest
and apoptosis in osteosarcoma cells
The potential effects of SOX18 knockdown on cell
cycle progression were then investigated. PI staining and flow
cytometry analysis revealed that knockdown of SOX18 in the U2OS
(Fig. 2A) and MG63 cells (Fig. 2B) resulted in an increase in the
number of cells in the S-phase and a corresponding reduction in the
number of cells in the G0/G1-phase. These
results suggested that silencing of SOX18 prevented the
osteosarcoma cells from entering the G2/M-phase.
The apoptotic function of SOX18 in U2OS and MG63
cells was also assessed using the annexin V-FITC/PI staining assay.
As shown in Fig. 2C and D, flow
cytometric analysis demonstrated that knockdown of SOX18 in the
U2OS or MG63 cells significantly induced cell apoptosis, compared
with the corresponding scramble shRNA (U2OS cells, 23.03±0.46, vs.
3.80±0.23%; MG63 cells, 34.43±1.32, vs. 3.93±0.18%). These results
indicated that the proliferation-promoting function of SOX18 may be
mediated via the promotion of cell cycle progression between the
S-phase and G2/M-phase, inhibiting apoptosis.
Knockdown of SOX18 inhibits the
metastasis of osteosarcoma cells
Metastasis begins with the invasion of tumor cells
into the surrounding host tissue. The invasive tumor cells must
first alter cell-to-cell adhesion and adhesion to the extracellular
matrix (17). The effects of SOX18
on the adherent ability of osteosarcoma cells were evaluated in the
present study (Fig. 3A and B). The
number of adherent SOX18-Ri-3 cells was 29.5% of that of the NC
cells when U2OS cells were used. Similar results were obtained with
the MG63 cells. These data suggested that the adherent ability to
fibronectin was significantly inhibited in osteosarcoma cells by
SOX18 knockdown.
Whether SOX18 affected the invasive ability of
osteosarcoma cells was also investigated using a Transwell assay.
As shown in Fig. 3C and D,
transduction of the SOX18-shRNA virus into U2OS or MG63 cells
significantly reduced the cell invasion ability, compared with the
scramble shRNA (NC). These data suggested that SOX18 promoted
osteosarcoma cell invasion.
Expression levels of TGF-β1, PDGF-A,
PDGF-B and RhoA are downregulated by SOX18 RNAi
A previous study demonstrated that TGF-β1 is a
promoter of tumor progression and invasion (18). The classic PDGFs, PDGF-A and
PDGF-B, are regarded to be associated with metastasis in various
types of human cancer (19–21).
It is well known that small GTPase RhoA promotes the invasion of
tumor cells (22–24). In order to investigate the
molecular mechanisms underlying the role of SOX18 in osteosarcoma
cells, the mRNA and protein expression levels of TGF-β1, PDGF-A,
PDGF-B and RhoA were determined (Fig.
4). The expression levels of all the genes examined were
markedly reduced following the downregulation in the expression of
SOX18, which suggested that the biological function of SOX18 in
osteosarcoma may be associated with these genes.
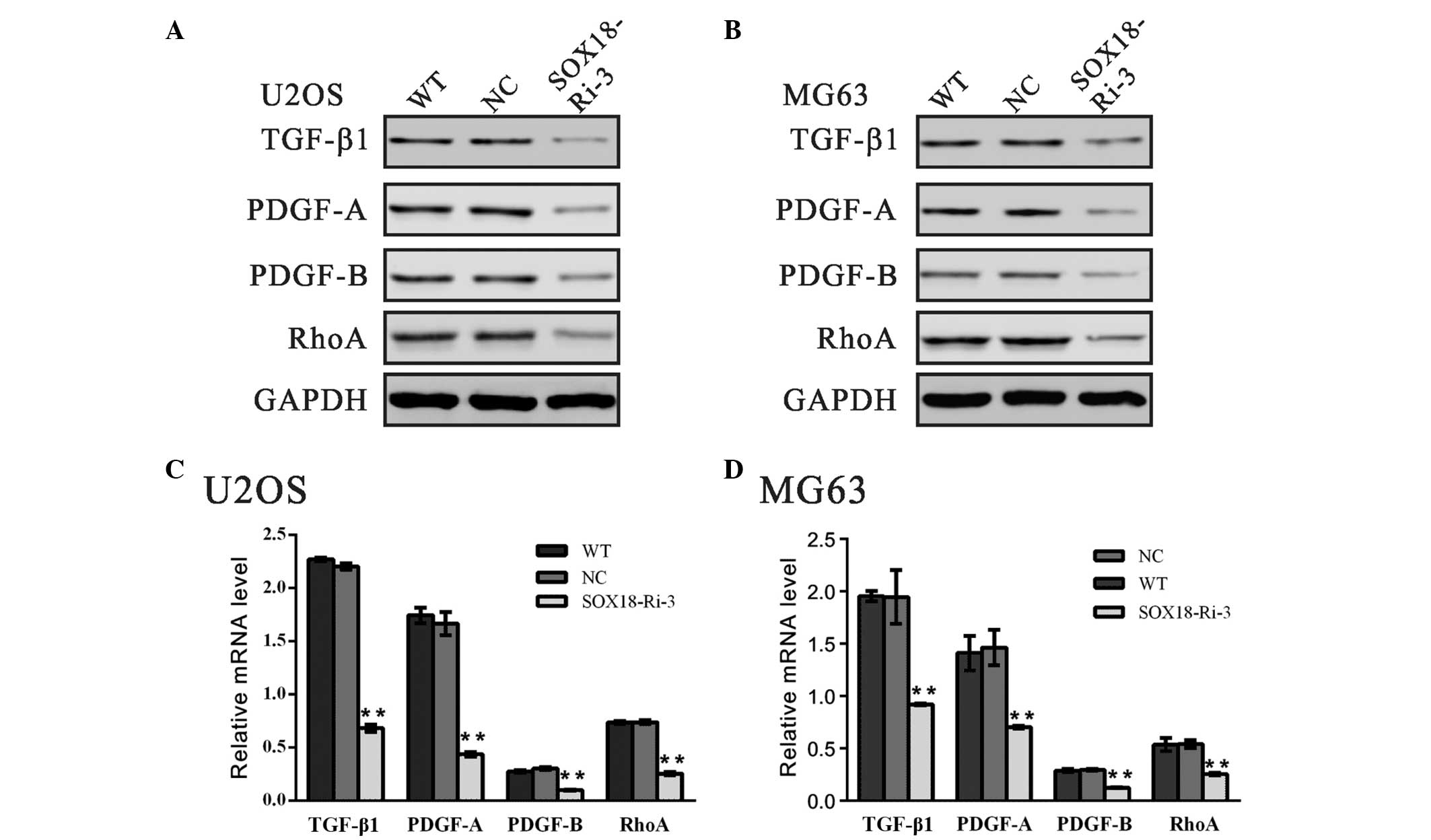 | Figure 4Expression levels of TGF-β1, PDGF-A,
PDGF-B and RhoA are downregulated by SOX18 RNAi. The protein and
mRNA levels of the indicated genes were evaluated using (A and B)
western blotting and (C and D) reverse transcription-quantitative
polymerase chain reaction in the U2OS and MG63 cells. Data are
presented as the mean ± standard deviation (**P<0.01,
vs. NC). TGF-β1, transforming growth factor-β1; PDGF-A,
platelet-derived growth factor-A; RhoA, Ras homolog family member
A; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; SOX18,
sex-determining region Y-box 18; WT, wild-type; NC, scrambled shRNA
virus transduction; SOX18-Ri-3, SOX18-shRNA-3 virus
transduction. |
Discussion
In the present study, it was found that SOX18 was
overexpressed in osteosarcoma. Knockdown of the expression of SOX18
markedly inhibited the transforming ability of osteosarcoma cells.
These data indicated the diagnostic and therapeutic value of SOX18
in osteosarcoma.
The involvement of SOX18 in several types of cancer
has been an area of investigation, and it has been reported that
SOX18 is overexpressed in several types of cancer tissue (10–13),
and that SOX18 may promote cellular proliferation (9,25).
Garcia-Ramirez et al (25)
found that SOX18 is co-localized with the proliferating cell
nuclear antigen protein in vascular smooth muscle cells of human
coronary atherosclerotic lesions, and that inhibiting the
expression of SOX18 results in a reduced proliferation rate in
these cells. The expression of dominant-negative SOX18 also reduces
the proliferation of human MCF-7 breast cancer cells (9). In the present study, the knockdown of
SOX18 in U2OS and MG63 osteosarcoma cells significantly reduced the
cell growth rate (Fig. 1). In
addition, cell cycle analysis revealed that SOX18 knockdown induced
S-phase arrest and apoptosis of osteosarcoma cells (Fig. 2), which may explain the inhibited
proliferation of the SOX18-knockdown cells.
Previously, SOX18 was reported to be associated with
cell migration and tumor metastasis, and dominant-negative SOX18
was reported to impair the migration of MCF-7 cells (9). Duong et al (26) reported that tumor metastasis is
inhibited in SOX18-deficient mice. In line with these observations,
the present study found that reduction in the expression of SOX18
in osteosarcoma cells by RNAi significantly reduced their adhesive
and invasive capabilities (Fig.
3), indicating that SOX18 may be important in promoting
metastasis of osteosarcoma.
The exact pathway that SOX18 may regulate in
osteosarcoma remains unclear. TGF-β1 has been considered as a
promoter of tumor progression and invasion (18). Additionally, PDGFs have been found
to induce tumor growth (27,28),
and their expression may be useful as a diagnostic and prognostic
marker for several types of cancer (29–31).
The classic PDGFs, PDGF-A and PDGF-B, are associated with the
metastasis of various types of human cancer (19–21).
In the present study, SOX18 RNAi significantly downregulated the
expression levels of TGF-β1, PDGF-A and PDGF-B, which indicated
that SOX18 may execute its functions through regulating the
expression of these genes.
It is well known that small GTPase RhoA promotes the
invasion of tumor cells (22–24).
The expression levels of RhoA may be positively correlated with the
progression of carcinoma, suggesting that RhoA may be important in
tumorigenesis and tumor progression (32–35).
The malignant phenotype in gastric cancer cells (36) and breast cancer cells (37) can be reversed by inhibiting the
expression of RhoA. In the present study, it was observed that
SOX18 knockdown impaired the expression of RhoA (Fig. 4). Therefore, it was hypothesized
that SOX18 may perform its biological function through regulating
the expression of RhoA.
In conclusion, the present study demonstrated high
expression levels of SOX18 in osteosarcoma, which suggested that
SOX18 may be a diagnostic marker for osteosarcoma. The present
study suggested for the first time, to the best of our knowledge,
that SOX18 is key in the proliferation, apoptosis and metastasis of
osteosarcoma cells. In addition, SOX18 may regulate these
biological processes through TGF-β1, PDGF-A, PDGF-B and RhoA, thus
providing potentially useful information for the targeted therapy
of osteosarcoma.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar
|
|
2
|
Provisor AJ, Ettinger LJ, Nachman JB,
Krailo MD, Makley JT, Yunis EJ, Huvos AG, Betcher DL, Baum ES,
Kisker CT and Miser JS: Treatment of nonmetastatic osteosarcoma of
the extremity with preoperative and postoperative chemotherapy: A
report from the children's cancer group. J Clin Oncol. 15:76–84.
1997.PubMed/NCBI
|
|
3
|
Bacci G, Ferrari S, Bertoni F, Ruggieri P,
Picci P, Longhi A, Casadei R, Fabbri N, Forni C, Versari M and
Campanacci M: Long-term outcome for patients with nonmetastatic
osteosarcoma of the extremity treated at the istituto ortopedico
rizzoli according to the istituto ortopedico rizzoli/osteosarcoma-2
protocol: An updated report. J Clin Oncol. 18:4016–4027.
2000.PubMed/NCBI
|
|
4
|
Rytting M, Pearson P, Raymond AK, Ayala A,
Murray J, Yasko AW, Johnson M and Jaffe N: Osteosarcoma in
preadolescent patients. Clin Orthop Relat Res. 39–50. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferguson WS and Goorin AM: Current
treatment of osteosarcoma. Cancer Invest. 19:292–315. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hosking BM, Muscat GE, Koopman PA, Dowhan
DH and Dunn TL: Trans-activation and DNA-binding properties of the
transcription factor, Sox-18. Nucleic Acids Res. 23:2626–2628.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hosking BM, Wyeth JR, Pennisi DJ, Wang SC,
Koopman P and Muscat GE: Cloning and functional analysis of the
Sry-related HMG box gene, Sox18. Gene. 262:239–247. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Darby IA, Bisucci T, Raghoenath S, Olsson
J, Muscat GE and Koopman P: Sox18 is transiently expressed during
angiogenesis in granulation tissue of skin wounds with an identical
expression pattern to Flk-1 mRNA. Lab Invest. 81:937–943. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Young N, Hahn CN, Poh A, Dong C, Wilhelm
D, Olsson J, Muscat GE, Parsons P, Gamble JR and Koopman P: Effect
of disrupted SOX18 transcription factor function on tumor growth,
vascularization and endothelial development. J Natl Cancer Inst.
98:1060–1067. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eom BW, Jo MJ, Kook MC, Ryu KW, Choi IJ,
Nam BH, Kim YW and Lee JH: The lymphangiogenic factor SOX 18: A key
indicator to stage gastric tumor progression. Int J Cancer.
131:41–48. 2012. View Article : Google Scholar
|
|
11
|
Jethon A, P ula B, Olbromsk i M, Wer ynska
B, Muszczynska-Bernhard B, Witkiewicz W, Dziegiel P and
Podhorska-Okolow M: Prognostic significance of SOX18 expression in
non-small cell lung cancer. Int J Oncol. 46:123–132. 2015.
|
|
12
|
Pula B, Kobierzycki C, Solinski D,
Olbromski M, Nowak-Markwitz E, Spaczynski M, Kedzia W, Zabel M and
Dziegiel P: SOX18 expression predicts response to platinum-based
chemotherapy in ovarian cancer. Anticancer Res. 34:4029–4037.
2014.PubMed/NCBI
|
|
13
|
Pula B, Olbromski M, Wojnar A,
Gomulkiewicz A, Witkiewicz W, Ugorski M, Dziegiel P and
Podhorska-Okolow M: Impact of SOX18 expression in cancer cells and
vessels on the outcome of invasive ductal breast carcinoma. Cell
Oncol (Dordr). 36:469–483. 2013. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Henry MK, Lynch JT, Eapen AK and Quelle
FW: DNA damage-induced cell-cycle arrest of hematopoietic cells is
overridden by activation of the PI-3 kinase/Akt signaling pathway.
Blood. 98:834–841. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Silletti S, Paku S and Raz A: Autocrine
motility factor and the extracellular matrix. I. Coordinate
regulation of melanoma cell adhesion, spreading and migration
involves focal contact reorganization. Int J Cancer. 76:120–128.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mareel M, Oliveira MJ and Madani I: Cancer
invasion and metastasis: Interacting ecosystems. Virchows Arch.
454:599–622. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wakefield LM and Roberts AB: TGF-beta
signaling: Positive and negative effects on tumorigenesis. Curr
Opin Genet Dev. 12:22–29. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kodama M, Kitadai Y, Sumida T, Ohnishi M,
Ohara E, Tanaka M, Shinagawa K, Tanaka S, Yasui W and Chayama K:
Expression of platelet-derived growth factor (PDGF)-B and
PDGF-receptor β is associated with lymphatic metastasis in human
gastric carcinoma. Cancer Sci. 101:1984–1989. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Donnem T, Al-Saad S, Al-Shibli K, Busund
LT and Bremnes RM: Co-expression of PDGF-B and VEGFR-3 strongly
correlates with lymph node metastasis and poor survival in
non-small-cell lung cancer. Ann Oncol. 21:223–231. 2010. View Article : Google Scholar
|
|
21
|
Jechlinger M, Sommer A, Moriggl R, Seither
P, Kraut N, Capodiecci P, Donovan M, Cordon-Cardo C, Beug H and
Grünert S: Autocrine PDGFR signaling promotes mammary cancer
metastasis. J Clin Invest. 116:1561–1570. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Struckhoff AP, Rana MK and Worthylake RA:
RhoA can lead the way in tumor cell invasion and metastasis. Front
Biosci (Landmark Ed). 16:1915–1926. 2011. View Article : Google Scholar
|
|
23
|
Yan G, Zou R, Chen Z, Fan B, Wang Z, Wang
Y, Yin X, Zhang D, Tong L, Yang F, et al: Silencing RhoA inhibits
migration and invasion through Wnt/β-catenin pathway and growth
through cell cycle regulation in human tongue cancer. Acta Biochim
Biophys Sin (Shanghai). 46:682–690. 2014. View Article : Google Scholar
|
|
24
|
Yoshioka K, Nakamori S and Itoh K:
Overexpression of small GTP-binding protein RhoA promotes invasion
of tumor cells. Cancer Res. 59:2004–2010. 1999.PubMed/NCBI
|
|
25
|
Garcia-Ramírez M, Martinez-González J,
Juan-Babot JO, Rodríguez C and Badimon L: Transcription factor
SOX18 is expressed in human coronary atherosclerotic lesions and
regulates DNA synthesis and vascular cell growth. Arterioscler
Thromb Vasc Biol. 25:2398–2403. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duong T, Proulx ST, Luciani P, Leroux JC,
Detmar M, Koopman P and Francois M: Genetic ablation of SOX18
function suppresses tumor lymphangiogenesis and metastasis of
melanoma in mice. Cancer Res. 72:3105–3114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ostman A: PDGF receptors-mediators of
autocrine tumor growth and regulators of tumor vasculature and
stroma. Cytokine Growth Factor Rev. 15:275–286. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uehara H, Kim SJ, Karashima T, Shepherd
DL, Fan D, Tsan R, Killion JJ, Logothetis C, Mathew P and Fidler
IJ: Effects of blocking platelet-derived growth factor-receptor
signaling in a mouse model of experimental prostate cancer bone
metastases. J Natl Cancer Inst. 95:458–470. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sulzbacher I, Birner P, Trieb K, Träxler
M, Lang S and Chott A: Expression of platelet-derived growth
factor-AA is associated with tumor progression in osteosarcoma. Mod
Pathol. 16:66–71. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peterson JE, Zurakowski D, Italiano JE Jr,
Michel LV, Connors S, Oenick M, D'Amato RJ, Klement GL and Folkman
J: VEGF, PF4 and PDGF are elevated in platelets of colorectal
cancer patients. Angiogenesis. 15:265–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ariad S, Seymour L and Bezwoda WR:
Platelet-derived growth factor (PDGF) in plasma of breast cancer
patients: Correlation with stage and rate of progression. Breast
cancer Res Treat. 20:11–17. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abraham MT, Kuriakose MA, Sacks PG, Yee H,
Chiriboga L, Bearer EL and Delacure MD: Motility-related proteins
as markers for head and neck squamous cell cancer. Laryngoscope.
111:1285–1289. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Horiuchi A, Imai T, Wang C, Ohira S, Feng
Y, Nikaido T and Konishi I: Up-regulation of small GTPases, RhoA
and RhoC, is associated with tumor progression in ovarian
carcinoma. Lab Invest. 83:861–870. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kamai T, Arai K, Tsujii T, Honda M and
Yoshida K: Overexpression of RhoA mRNA is associated with advanced
stage in testicular germ cell tumour. BJU International.
87:227–231. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kamai T, Tsujii T, Arai K, Takagi K, Asami
H, Ito Y and Oshima H: Significant association of Rho/ROCK pathway
with invasion and metastasis of bladder cancer. Clin Cancer Res.
9:2632–2641. 2003.PubMed/NCBI
|
|
36
|
Liu N, Bi F, Pan Y, Sun L, Xue Y, Shi Y,
Yao X, Zheng Y and Fan D: Reversal of the malignant phenotype of
gastric cancer cells by inhibition of RhoA expression and activity.
Clin Cancer Res. 10:6239–6247. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pillé JY, Denoyelle C, Varet J, Bertrand
JR, Soria J, Opolon P, Lu H, Pritchard LL, Vannier JP, Malvy C, et
al: Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and
invasiveness of MDA-MB-231 breast cancer cells in vitro and in
vivo. Mol Ther. 11:267–274. 2005. View Article : Google Scholar : PubMed/NCBI
|


















