Introduction
Echinococcosis, also termed cystic echinococcosis,
is a type of zoonotic parasitic disease, which is caused by
infection with Echinococcus larvae. Echinococcosis is a
severely harmful disease affecting humans and animals, and is
associated with high rates of mortality and disability worldwide
(1), particularly in developing
countries. China is one of the countries with the highest incidence
of echinococcosis. Echinococcosis is predominantly endemic in
pasture regions, including Xinjiang, Qinghai, Gansu and Ningxia, as
well as in semi-pasture regions. At present, surgery is considered
the primary therapeutic strategy for the treatment of
echinococcosis, whereas drug therapy is supplementary and less
efficient. In recent years, vaccination against Echinococcus
has attracted increased attention (2).
Ferritin is a multifunctional and multimeric
protein, which is widely distributed amongst organisms and has a
significant role in the regulation of immune function (3–6).
Previous studies have identified Echinococcus granulosus
(Eg.) ferritin as an antigenic molecule, which is associated with a
certain immunological protection. In the 1990s, Eresfeld and Craig
(7) cloned the Eg. ferritin gene,
and determined that it exhibits immunogenicity and can be used to
diagnose echinococcosis. Therefore, the Eg. ferritin gene has
attracted increasing attention. The present study aimed to predict
the T-cell and B-cell antigen epitopes of Eg. ferritin and perform
sequence analysis, in order to diagnose and treat hepatic
echinococcosis. The present study may provide novel evidence
supporting the development of an epitope vaccine against
echinococcosis.
Materials and methods
Primary reagents
TRIzol®, Taq enzyme and an AMV
First Strand cDNA Synthesis kit were purchased from Invitrogen Life
Technologies (Carlsbad, CA, USA). The DL2000 DNA Marker was
purchased from Takara Biotechnology Co., Ltd. (Dalian, China).
Specimen collection of Echinococcus
granulosus
Fresh livers of sheep which had naturally contracted
Echinococcus granulosus, as identified by vesicae in the
liver, were obtained from Xinjiang Slaughterhouse (Urumqi, China)
and cystic fluid was extracted using a 50 ml sterile syringe. The
fluid was transferred into a centrifuge tube, and the protoscolex
were allowed to naturally precipitate. Following rinsing three
times with sterile saline, the protoscolex were collected and
stored at 4°C for further analysis. The study was approved by the
ethics committee (ZACUS-20130425002) of Xinjiang Medical University
(Urumqi, China).
Primer design and synthesis
According to the Eg. ferritin gene sequence (GenBank
ID: Z31712; http://www.ncbi.nlm.nih.gov/nuccore/Z31712), the
following primer was designed using DNAman software (LynnonBiosoft
Corp., San Ramon, CA, USA): Eg. ferritin, forward
5′-CGGAATTCATGAGGAATGCGAACGTG-3′ and reverse
5′-CGCAAGCTTTGATAAAAAATTATTTGT-3′. The primer was synthesized by
Sangon Biotech Co., Ltd. (Shanghai, China).
Analytical software
The Self-Optimized Prediction Method with Alignment
(SOPMA) server (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.%20.html)
was used to predict the secondary structure of Eg. ferritin; the
Immune Epitope Database(IEDB; http://tools.immuneepi-tope.org/tools/bcell/iedb_input)
and Linear Epitope Prediction Based on Propensity Scale and SVM
(LEPS; http://leps.cs.ntou.edu.tw/index.php) tool were used
to predict the B-cell epitope; and the SYFPEITHI (http://www.syfpeithi.de) database and IEDB tools were
used to predict the T-cell epitope. The online software, 3D
Ligandsite (http://www.sbg.bio.ic.ac.uk/~3dligandsite/) and RasMol
(http://www.rasmol.org/) were used to predict the
three dimensional (3D) structure of Eg. ferritin.
Extraction of total RNA from Echinococcus
granulosus protoscolex and synthesis of cDNA
Echinococcus protoscolices were ground
between three and six times in liquid nitrogen (provided by The
State Key Laboratory Incubation Base of Xinjiang Major Diseases
Research, The First Affiliated Hospital of Xinjiang Medical
University, Urumqi, China) prior to RNA extraction in a sterile
mortar. A total of 1 ml TRIzol® (Invitrogen Life
Technologies, Inc.) was then added per 100 ml sample, and ground
between three and six times. Total RNA was extracted using
TRIzol® according to the manufacturer's instructions.
The RNA concentration was determined using an ultraviolet
spectrophotometer (ND1000; NanoDrop; Thermo Fisher Scientific,
Waltham, MA, USA) and was dissolved in 50 µl water treated
with diethylpyrocarbonate (Tianjing Fuyu Chemical Co., Ltd.,
Tianjing, China). The samples (5 µl) were then run on a 1.2%
3-(N-morpholino) propanesulfonic acid (MOPS)-formaldehyde
denaturing gel (Tianjing Fuyu Chemical Co., Ltd). RNA was
reverse-transcribed into cDNA using a RevertAid™ First strand cDNA
Synthesis kit (Thermo Fisher Scientific) according to the
manufacturer's instructions. Reactions were performed using 2
µl cDNA in a 20-µl reaction volume and the following
thermocycling profile: 10 min of denaturation at 95°C, 40 cycles of
denaturation at 95°C for 15 sec and 60 sec of extension at
60°C.
Cloning and identification of the Eg.
ferritin gene
The Eg. ferritin gene was cloned from the
protoscolex cDNA. Amplification of Eg95 was performed in a
20-µl mixture containing 1 µl cDNA template, 2
µl 10X buffer, 0.5 µl of each primer, 0.5 µl
10 mm dNTP, 0.5 µl Taq enzyme and 15.5 µl pure
water (2XTaq PCR Master Mix or Maxima SYBR Green/ROX qPCR Master
Mix; Invitrogen Life Technologies, Inc.). The cycling conditions of
the polymerase chain reaction (PCR) were as follows: Initial
denaturation at 95°C for 6 min, followed by 30 consecutive cycles
of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec
and extension at 72°C for 1 min, and a final extension step at 72°C
for 5 min. The PCR products were subsequently detected using 1.2%
agarose gel electrophoresis (Sigma-Aldrich, St. Louis, MO,
USA).
Amino acid sequences coded by Eg.
Ferritin
Validation of gene sequence analysis was performed
and the corresponding amino acid sequences were identified using
the DNAman software program. Multiple sequences were detected,
according to the Eg. ferritin gene sequence obtained from
GenBank.
Prediction of secondary protein
structure
Prediction of the secondary structure of the Eg.
ferritin protein was performed using the online SOPMA server
(http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.%20.html).
B cell epitope prediction software
Predictions of B cell hydrophilicity, antigenicity
and flexibility were made using the IEDB (http://tools.immuneepitope.org/tools/bcell/iedb_input)
and LEPS (http://leps.cs.ntou.edu.tw/index.php) online
prediction software.
T-cell epitope prediction software
Prediction of the potential major histocompatibility
complex (MHC)-I type human leukocyte antigen (HLA)-A 0201
restrictive T-cell epitope was made using the SYFPEITHI (http://www.syfpeithi.de) and IEDB (http://tools.immuneepitope.org/tools/bcell/iedb_input)
online resources.
Prediction of the tertiary structure of
Eg. Ferritin
The tertiary structure of Eg. ferritin was predicted
using the 3DLigandsite (http://www.sbg.bio.ic.ac.uk/~3dligandsite/) online
server, combined with RasMol software, in order to analyze and
determine different models of presentation, including Cartoon,
Structure and Group.
Results
Extraction of total RNA from Echinococcus
granulosus protoscolex
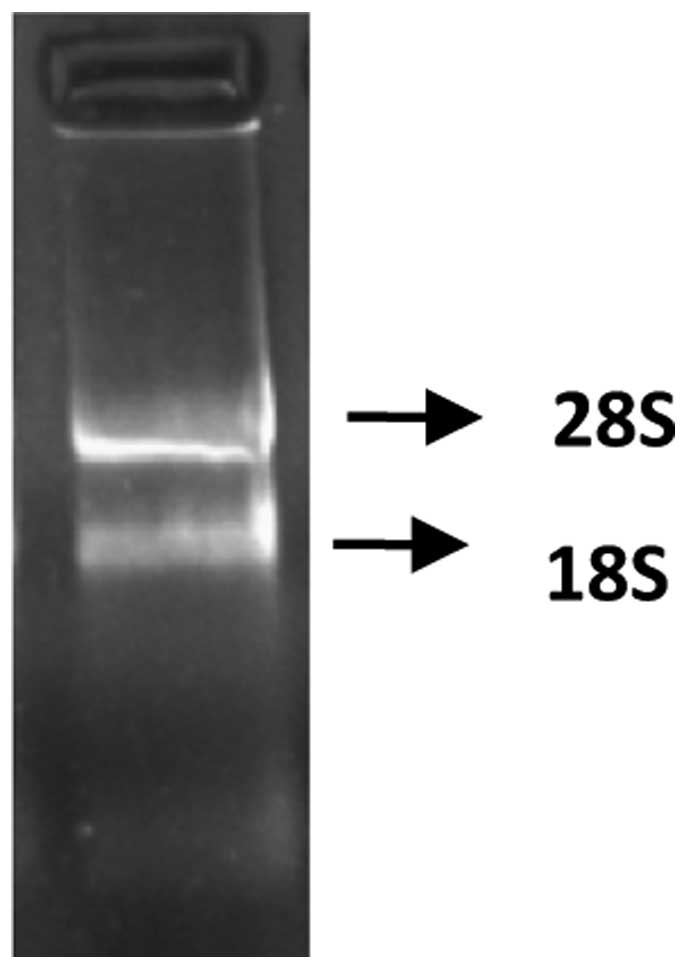
The absorption value of the extracted total RNA was
detected at 260 and 280 nm wavelengths, using a nucleic acid
analyzer. The value of protoscolex RNA was 1.94; demonstrating that
the RNA was extracted successfully. The resulting MOPS-formaldehyde
denaturing gel electrophoresis of Eg. ferritin is shown in Fig. 1.
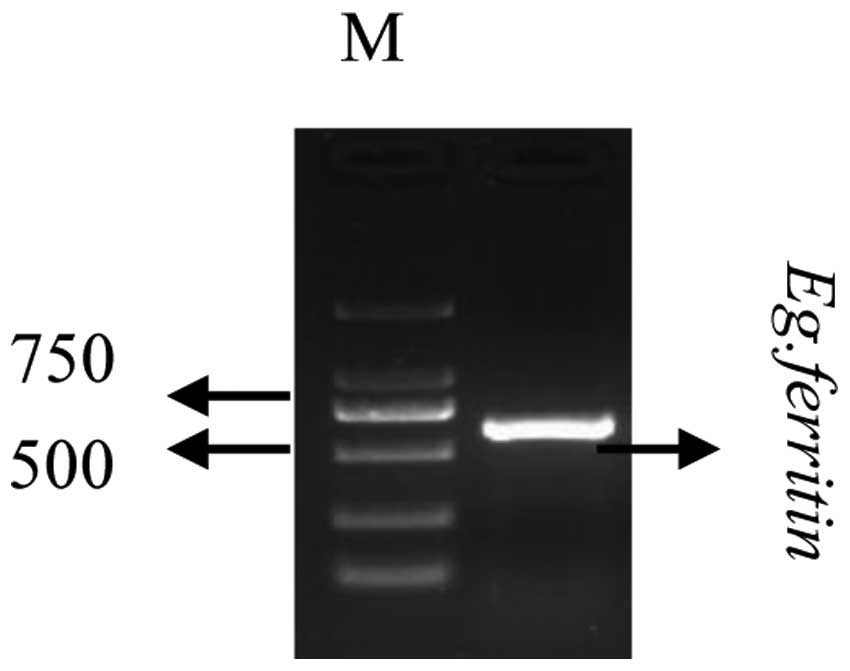
Cloning of the Eg. ferritin gene
cDNA was subsequently used as a template for PCR
amplification using an Eg. ferritin primer. The PCR products were
verified using 1.2% agarose gel electrophoresis. As shown in
Fig. 2. the Eg. ferritin PCR
products resulted in a specific band at 653 bp, whereas no such
band was detected in the negative control group, in which water was
used instead of template. This indicated that a specific PCR
fragment had been successfully amplified from the cDNA.
Amino acid sequence coded by Eg.
Ferritin
The corresponding amino acid sequence to be
translated from the Eg. ferritin gene was predicted using DNAman
online software. The following 176 amino acid residue was
identified:
MSLVRQNFHEECERGINRQINMELYASYLYLAMSQHFDRDDVALPGFREFFAKASEEEREHAIKLMRYQCGRGGRIVYQDIAKPQTTEWASGLEAMEMALKIEREVNESLLALRGVANKNNDSQFCEFLEGEFLGEQVSDIKKLAGYVTNLKRCGPGLGEYIFDKETLQGGEK.
Prediction of the secondary structure of
Eg. ferritin anti-genic protein
Prediction of the secondary structure of Eg.
ferritin antigenic protein was made using the online software,
SOPMA. α-helix structures accounted for 73.41% of the total amino
acid sequence, and β-fold structures and random coils accounted for
4.05 and 16.76% of the total amino acid sequence, respectively. The
distribution of the different structures of Eg. ferritin antigenic
protein is shown in Fig. 3.
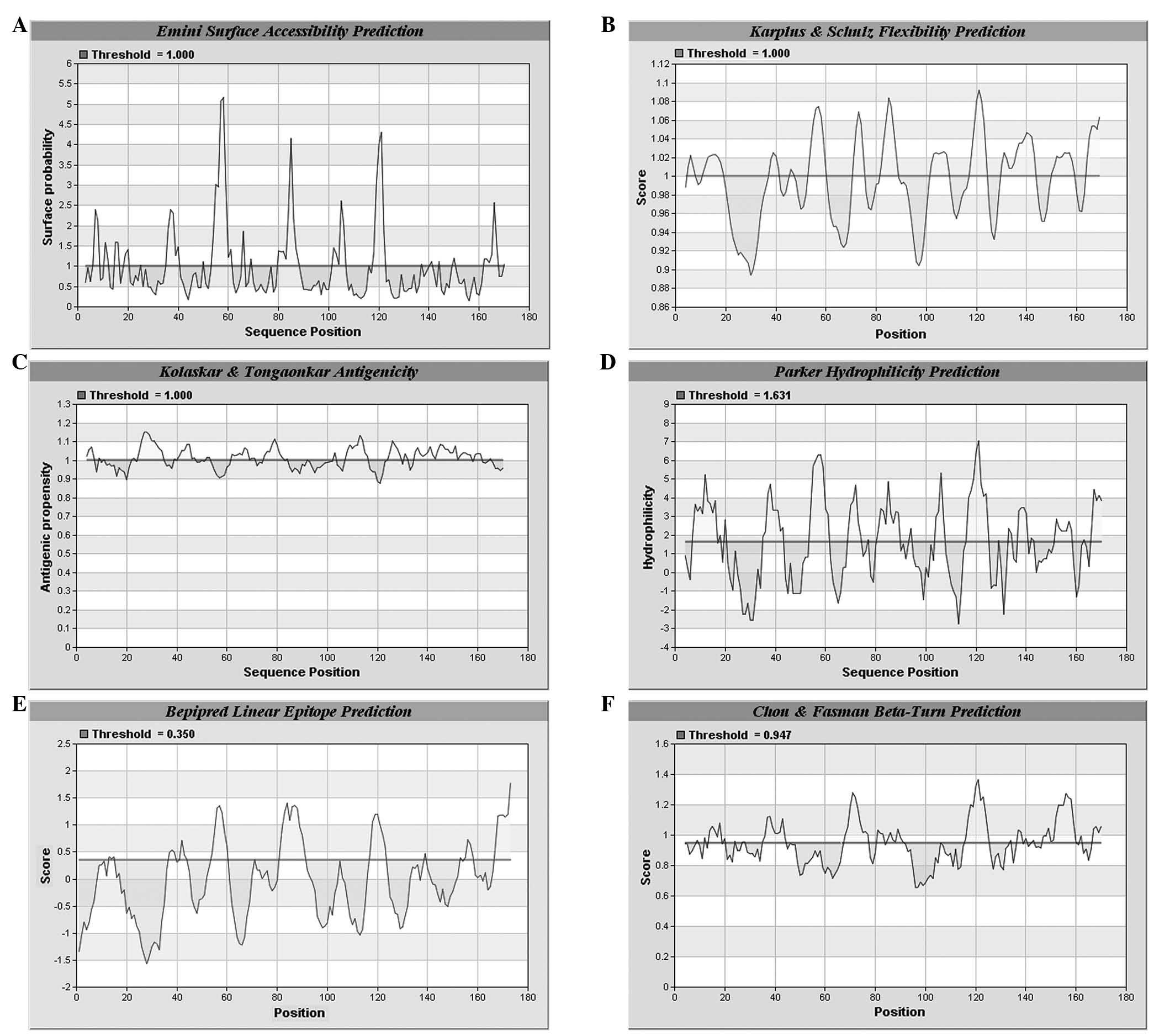
B-cell antigen epitope prediction of Eg.
Ferritin
A prediction was made on the combined
hydrophilicity, antigenicity and flexibility of Eg. ferritin using
IEDB and LEPS (http://leps.cs.ntou.edu.tw/index.php) online software.
Regions with high values are considered to be potential B-antigen
epitopes. According to the predicted results, several high value
amino acid sequences were identified (Figs. 4 and 5). Combining the results of the two
software analyses, seven potential B-antigen epitopes were
identified, comprising the 8–16, 54–61, 70–75, 80–90, 103–109,
117–124 and 167–173 amino acid sequence regions.
T-cell antigen epitope prediction of Eg.
Ferritin
In order to obtain the most accurate results, the
SYFPEITHI (http://www.syfpeithi.de) database and
IEDB (http://tools.immuneepitope.org/tools/bcell/iedb_input)
online prediction tool were used to predict the MHC-I type HLA-A
0201 restrictive T-antigen epitope. These each identified 15
regions with high values. The results of the analyses are shown in
Table I. Combining the results of
the two software analyses, four potential T-cell antigen epitopes,
including the 85–93, 105–113, 133–141 and 157–165 amino acid
sequence regions, were identified.
 | Table IPredicted major histocompatibility
complex-I type human leukocyte antigen-A 0201 restrictive T-cell
epitopes using SYFPEITHI and IEDB. |
Table I
Predicted major histocompatibility
complex-I type human leukocyte antigen-A 0201 restrictive T-cell
epitopes using SYFPEITHI and IEDB.
| Order | Initiation site | Amino acid
sequence | Score |
|---|
| SYFPEITHI | | | |
| 1 | 143 | KLAGYVTNL | 30 |
| 2 | 133 | FLGEQVSDI | 25 |
| 3 | 92 | GLEAMEML | 21 |
| 4 | 150 | NLKRCGPGL | 21 |
| 5 | 105 | EVNESLLAL | 20 |
| 6 | 23 | ELYASYLYL | 19 |
| 7 | 109 | SLLALRGVA | 19 |
| 8 | 21 | NMELYASYL | 18 |
| 9 | 98 | MALKIEREV | 18 |
| 10 | 157 | GLGEYIFDK | 18 |
| 11 | 110 | LLALRGVAN | 17 |
| 12 | 112 | ALRGVANKN | 17 |
| 13 | 140 | DIKKLAGYV | 17 |
| 14 | 85 | QTTEWASGL | 16 |
| 15 | 102 | IEREVNESL | 16 |
| IEDB | | | |
| 1 | 9 | HEECERGIN | 100 |
| 2 | 11 | ECERGINRQ | 100 |
| 3 | 13 | ERGINRQIN | 100 |
| 4 | 56 | EEEREHAIK | 100 |
| 5 | 164 | DKETLQGGE | 99 |
| 6 | 158 | LGEYIFDKE | 96 |
| 7 | 52 | AKASEEERE | 95 |
| 8 | 60 | EHAIKLMRY | 94 |
| 9 | 70 | CGRGGRIVY | 94 |
| 10 | 134 | LGEQVSDIK | 94 |
| 11 | 58 | EREHAIKLM | 93 |
| 12 | 113 | LRGVANKNN | 93 |
| 13 | 103 | EREVNESLL | 91 |
| 14 | 86 | TTEWASGLE | 90 |
| 15 | 40 | DDVALPGFR | 90 |
Using multiple sequence alignment with DNAman
software and comparing the potential T-cell and B-cell epitopes,
the highly overlapped amino acid sequence 105–109 was predicted as
a T- and B-combined epitope (Table
II).
 | Table IIT- and B-combined epitope. |
Table II
T- and B-combined epitope.
| Epitope | Predicted
region | Amino acid
sequence |
|---|
| B-cell | 103–109 | EREVNES |
| T-cell | 105–113 | EVNESLLAL |
| B- and
T-combined | 105–109 | EVNES |
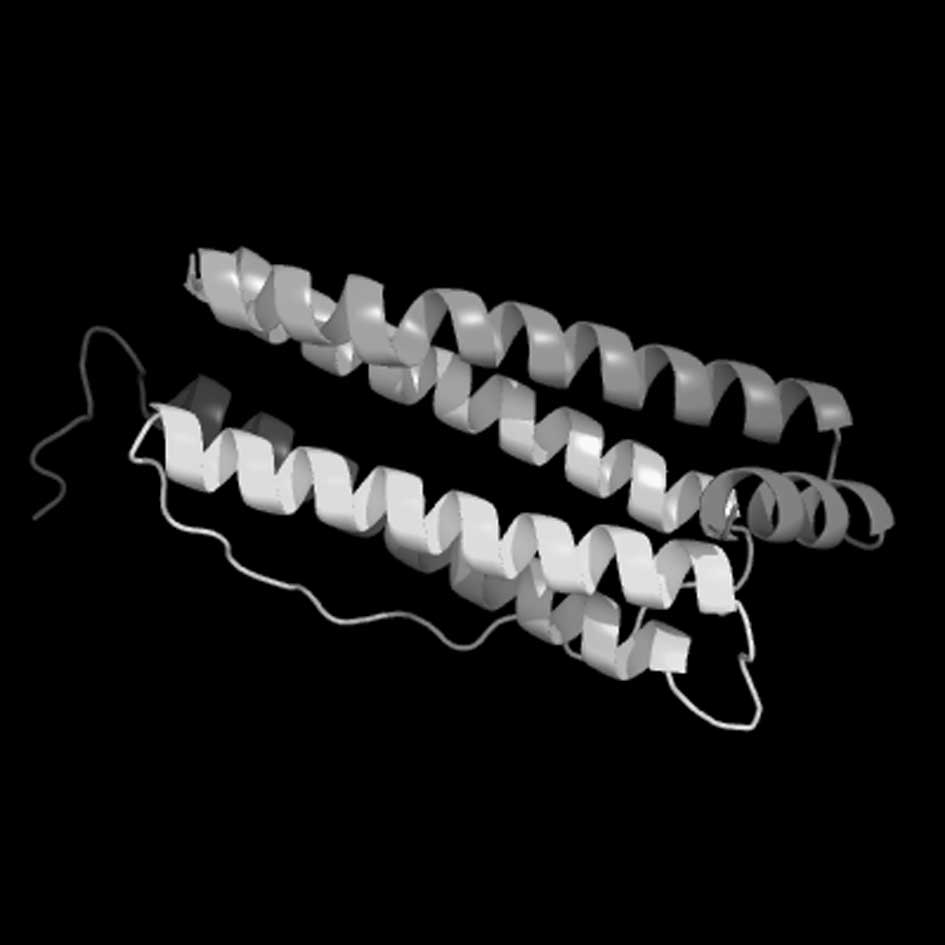
Analyses of Eg. ferritin tertiary
structure
Segments of the Eg. ferritin amino acid code were
submitted to the 3DLigandsite server (http://www.sbg.bio.ic.ac.uk/~3dligandsite/), in order
to predict and analyze the 3D structure of the protein (Fig. 6). Different demonstration models
were applied, including the Structure and Group (Fig. 7A–C), to determine the specific
position of each amino acid on the tertiary structure of Eg.
ferritin using RasMol and 3D Ligandsite analysis software. A marked
similarity was observed between an area in the structural model and
the flexible area predicted by the secondary structure analysis.
The Group structure model detected that this assembled area and was
distributed at the surface of the structure, indicating that it is
most likely the combined epitope of antigen and antibody.
Discussion
China remains one of the countries with a high
incidence of echinococcosis, which is a significant factor
affecting economic development and public health in western China.
Therefore, the identification of an antigen with high specificity
and high sensitivity is important for the diagnosis and treatment
of echinococcosis. It has previously been demonstrated that Eg.
ferritin attains a protective immunity of 85.6% in animals;
therefore, it can be considered as a potential antigen for
investigation (8,9).
Early investigation by Schuler (10) identified the FMDV immune locus,
which is an antigenic epitope and specific chemical group among
antigen molecules for determining antigenic specificity, also
termed an antigen determinant. Epitopes can be divided into B-cell
epitopes and T-cell epitopes. B-cell epitopes are located on the
surface of the antigenic molecule and induce the humoral immune
response and production of specific antibodies from B-cells. T-cell
epitopes are linear peptides, which, following the
antigen-presenting cell process, delivers antigens from the MHC
molecule to the T-cell receptor and is connected to the cellular
immune response (11). Epitope
vaccines are produced according to epitope amino acid sequences
(12) and epitope vaccines have
become an important focus of molecular vaccine investigations;
Kouguchi et al (13)
demonstrated that the EmY162 recombinant antigen can induce 74.3%
immune protection in mice. The most important process of epitope
vaccine production is the identification of a sequence with a
highly specific epitope location (14).
In the prediction of the secondary protein
structure, random coils and β-folds are considered the prominent
structural features, the majority of which appear predominantly on
the surface of the protein antigens and are beneficial for the
recognition of antigens, and are therefore likely to be an
antigenic epitope (15). The
present study demonstrated that random coils accounted for 16.76%
and β-folds accounted for 4.05% of the total antigen protein
structure of Eg. ferritin. These results indicated that these
structures are within the distribution region of the antigenic
epitope with marked immunogenicity (16,17).
The higher the level flexibility, the easier it is to form an
antigenic epitope. Antigen accessibility is made possible through
contact between amino acid residues and solvent molecules,
indicating the distribution of internal and external antigen
residues. In the 3D protein structure, globular or oval structures,
which are formed by peptide coils and folds, always form a
hydrophilic molecule and a hydrophobic nuclear molecule, enabling
stability of the 3D structure due to the presence of hydrophobic
and hydrogen bonds (11). With
prediction of the flexibility of the antigen protein and epitope
accessibility in the present study, further evidence were gained to
confirm this.
In the T-cell epitope prediction in the present
study, an MHC-I type antigenic epitope was predicted with high
accuracy. HLA-A 0201 restrictive T-cells are most common among the
Han Chinese population (11,18,19).
In the present study, nine peptides of HLA-A0201 MHC-I type
antigenic epitopes were predicted using online software, and four
regions with high values were identified: 85–93, 105–113, 133–141
and 157–165. The present study also identified a potential T- and
B-combined epitope, which possessed a highly overlapped region
(105–109). These results may provide evidence supporting the
production of a T- and B-antigen epitope vaccine, contributing to
the therapy of echinococcosis in terms of humoral and cellular
immunity.
Eg. ferritin has the potential to form T-cell and
B-cell epitopes (20,21). The present study used IEDB and LEPS
online software to predict the potential B-cell epitope of Eg.
ferritin. In total, seven amino acid sequence positions were
identified (8–16, 54–61, 70–75, 80–90, 103–109, 117–124 and
167–173), which readily form B-cell epitopes. Furthermore, the
present study also predicted the T-cell epitope using the SYFPEITHI
and IEDB online servers. This identified four amino acid sequences
(85–93, 105–113, 133–141 and 157–165), which readily form T-cell
epitopes. Following observation of the potential T-cell and B-cell
epitopes, a highly overlapped sequence was found (105–109). In
conclusion, the results of the present study provide evidence
supporting the production of a highly efficient and safer epitope
vaccine, and establishes the foundation for the treatment of
echinococcosis.
Acknowledgments
The present study was supported by the Scientific
Research Project of Science Department of Xinjiang Autonomous
Region (grant no. 2012211A034), and the National Natural Science
Foundation (grant nos. 31160194, 81260253, 30960358, 81160378;
31000411, 30901374, 81060135 and 30860263).
References
|
1
|
No authors listed. New influenza A (HINl)
virus: Global epidemiological situation, June 2009. Wkly Epidemiol
Rec. 84:249–257. 2009.
|
|
2
|
Wen H and Ding Z: Atlas of Echinococcosis.
2nd edition. Science Press; Shanghai: pp. 92008, In Chinese.
|
|
3
|
Ponka P: Recent advances in cellular iron
metabolism. J Trace Elem Exp Med. 16:201–217. 2003. View Article : Google Scholar
|
|
4
|
Wang QL, Kong B and Huang HQ: Progress in
structural and functional study of nanometer protein shell of the
ferritin. Prog Chem. 16:516–519. 2004.In Chinese.
|
|
5
|
Qadri F, Jonson G, Begum YA, Wennerås C,
Albert MJ, Salam MA and Svennerholm AM: Immune response to the
mannose-sensitive hemagglutinin in patients with cholera due to
Vibrio cholerae O1 and O139. Clin Diagn Lab Immunol. 4:429–434.
1997.PubMed/NCBI
|
|
6
|
Qadri F, Ryan ET, Faruque AS, Ahmed F,
Khan AI, Islam MM, Akramuzzaman SM, Sack DA and Calderwood SB:
Antigen-speeific immunoglobulin A antibodies secreted from
circulating B cells are an effective marker for recent local immune
responses in patients with cholera: Comparison to
antibody-secreting cell responses and other immunological markers.
Infect Immun. 71:4808–4814. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eresfeld K and Craig PS: Cloning and
immunological characterization of Echinococcus granulosus ferritin.
Parasitol Res. 81:382–387. 1995. View Article : Google Scholar
|
|
8
|
Wang YN, Li ZJ, Li ZY, Bo Y and Zhao W:
Recombinant ferritin protects mice against challenge with
Echinococcus granulosus. Acta Parasitologica. 54:335–340. 2009.
View Article : Google Scholar
|
|
9
|
Wang YN, Ding SQ, Wang J, Zhang Y, Wang J,
Wang S and Zhao W: High level expression and identification of
recombinant ferritin of Echinococcus granolosus. Chinese Journal of
Zoonoses. 22:399–406. 2006.
|
|
10
|
Schuler MM, Nastke MD and Stevanovikć S:
SYFPEITHI: Database for searching and T-cell epitope prediction.
Methods Mol Biol. 409:75–93. 2007. View Article : Google Scholar
|
|
11
|
Ma X, Zhou X, Zhu Y, Li Y, Wang H, Mamuti
W, Li Y, Wen H and Ding J: The prediction of T- and B-combined
epitope and tertiary structure of the Eg95 antigen of Echinococcus
granulosus. Exp Ther Med. 6:657–662. 2013.PubMed/NCBI
|
|
12
|
Ben-Yedidia T and Arnon R: Towards an
epitope-based human vaccine for influenza. Hum Vaccin. 1:95–101.
2005. View Article : Google Scholar
|
|
13
|
Kouguchi H, Matsumoto J, Katoh Y, Oku Y,
Suzuki T and Yagi K: The vaccination potential of EMY162 antigen
against Echinococcus multilocularis infection. Biochem Biophys Res
Commun. 363:915–920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
You L, Brusic V, Gallagher M and Bodén M:
Using Gaussian process with test rejection to detect T-Cell
epitopes in pathogen genomes. IEEE/ACM Trans Comput Biol Bioinform.
7:741–751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
LIU Xianfei, Wang Hongying, ZHOU Xiaotao,
et al: Cloning and bioinformatics prediction of EM18 in
Echinococcus multilocularis [J]. Chin J Zoonoses. 29:23–26.
2013.
|
|
16
|
Sikic K, Tomic S and Carugo O: Systematic
comparison of crystal and NMR protein structures deposited in the
protein data bank. Open Biochem J. 4:83–95. 2010. View Article : Google Scholar
|
|
17
|
Sy SM, Chen J and Huen MS: The
53BP1-EXPAND1 connection in chromatin structure regulation.
Nucleus. 1:472–474. 2010. View Article : Google Scholar
|
|
18
|
Yan C, Wang R, Li J, Deng Y, Wu D, Zhang
H, Zhang H, Wang L, Zhang C, Sun H, et al: HLA-A gene polymorphism
defined by high-resolution sequence-based typing in 161 Northern
Chinese Han people. Genomics Proteomics Bioinformatics. 1:304–309.
2003.
|
|
19
|
Lin L, Tan B, Pantapalangkoor P, Ho T,
Hujer AM, Taracila MA, Bonomo RA and Spellberg B: Acinetobacter
baumannii rOmpA vaccine dose alters immune polarization and
immunodominant epitopes. Vaccine. 31:313–318. 2013. View Article : Google Scholar :
|
|
20
|
Jonson G, Holmgren J and Svennerholm AM:
Identification of a mannose-binding pilus on Vibrio cholerae EI
Tor. Microb Pathog. 11:433–441. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jonson G, Lebens M and Holmgren J: Cloning
and sequencing of Vibrio cholerae mannose-sensitive haemagglutinin
pilin gene: Localization of mshA within a cluster of type 4 pilin
genes. Mol Microbiol. 13:109–118. 1994. View Article : Google Scholar : PubMed/NCBI
|