Introduction
The transformation of normal stem cells into
lymphoma is a multistep process comprising accumulated genetic and
epigenetic aberrations, including mutations of the genome of
proto-oncogenes, tumor suppressor genes and other genes associated
with important cellular processes, such as differentiation and cell
proliferation. Alterations in the methylation patterns of various
genes have been observed in almost all cancer types, including
hematological malignancies and solid tumors (1,2).
Numerous studies have shown that hypermethylation of CpG islands of
tumor-suppressor genes, which are unmethylated under normal
conditions, is associated with transcriptional silencing of the
respective genes, which therefore has a critical role in tumor
development and progression (1–4).
The inhibitor of DNA binding (ID) family is an
important methylation site associated with non-Hodgkin lymphoma
(NHL), which is utilized for clinical diagnosis. The functions of
ID proteins comprise cell cycle control, lymphocyte development and
cellular senescence (5,6). By forming hetero-dimers with
transcription factors, ID proteins act as dominant-negative
inhibitors of gene transcription and negatively regulate the
function of basic-helix-loop-helix (bHLH) transcription factors to
affect the balance between cell growth and differentiation
(5,7). The ID protein family comprises four
members, ID1-4, among which ID4 was first discovered in 2004
(8). However, its expression and
function in various tumor types have remained controversial:
Kuzontkoski et al (9) and
Zeng et al (10) showed
that ID4 is highly expressed in glioblastoma multiforme (GBM), in
which it promotes angiogenesis and growth, while other studies
showed that ID4 protein expression was decreased in several types
of human cancer (11,12). A study utilizing a mouse model of
acute lymphoblastic leukemia of the T/natural killer cell lineage
showed that ID4 protein expression was downregulated by promoter
methylation and identified the ID4 gene as a putative tumor
suppressor gene (8). The ID4 gene
has also been confirmed to have an increased degree of methylation
in a variety of human tumors (13,14),
including gastric adenocarcinoma (15), tumors of haematopoietic and
lymphoid tissues (8,16,17),
breast carcinoma (18,19), esophageal adenocarcinomas (20) and prostate cancers (12). ID4 methylation was also shown to be
significantly correlated with World Health Organization sub-types
and risk groups of cancer determined by the International
Prognostic Scoring System. Multivariate analysis indicated that the
ID4 methylation status was an independent prognostic factor for
leukemia-free survival (21).
Thus, the present study hypothesized that ID4 may be a therapeutic
target in NHL. The demethylating reagent 5-aza-2′-deoxycytosine
(CdR) inhibits DNA methyltransferases and reverses DNA methylation.
It has been reported that CdR inhibits cancer cell growth,
particularly that of leukemia cells. It has been applied for the
treatment of myelodysplastic syndromes (MDS) and in preliminary
experimental cancer treatments (14,20,22),
and the most clinically advanced agents, 5-azacytidine and CdR,
provide a promising approach for the treatment of hematopoietic
malignancies, including MDS and acute myeloid leukemia (21,23).
However, the mechanisms of action of CdR in NHL have largely
remained elusive.
The present study explored the expression of ID4
protein and the status of ID4 gene methylation in the Raji human
Burkitt's lymphoma cell line as well as their modulation by
demethylating agent CdR. Furthermore, the effects of CdR on cell
cycle distribution and apoptosis of Raji cells were investigated.
The results demonstrated that ID4 was hypermethylated and its
protein expression was low in Raji cells, while CdR reversed the
abnormal DNA methylation and induced re-expression of the ID4
protein; furthermore, CdR enhanced apoptosis and induced cell-cycle
arrest in Raji cells.
Materials and methods
Drugs and reagents
CdR was purchased from Sigma-Aldrich (St. Louis, MO,
USA) and was dissolved in phosphate-buffered saline (PBS). ID4
rabbit polyclonal antibody (cat no. SC-B047) was purchased from
Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). One step modified
base kit (cat. no. E112885) was purchased from Epigentek
(Farmingdale, NY, USA). Ex Taq enzyme was purchased from Takara
(Otsu, Japan).
Cell line and culture
The CCL-86 Raji human Burkitt's lymphoma cell line
was purchased from the Chinese Academy of Sciences Shanghai
Institute of Cell Biology (Shanghai, China) and was grown in
RPMI-1640 culture medium (HyClone; GE Healthcare, Little Chalfont,
UK)containing 10% fetal bovine serum, 100 U/ml penicillin and 100
mg/ml streptomycin (Harbin Pharmaceutical Group Holding Co.,
Harbin, China) in an incubator with a humidified atmosphere
containing 5% CO2 at 37°C. Cells were passaged and
inoculated at 2×105/ml in the logarithmic growth
phase.
Cell growth study
Raji cells in the logarithmic growth phase were
treated with CdR at 0, 0.1, 0.5, 1.0, 2.5 or 5.0 µmol/l. The
total numbers of live cells in each group were counted every day
for seven days under a light microscope and growth curves were
drawn.
Treatment groups
On the basis of the growth curves, CdR at
concentrations of 0, 0.5 and 5.0 µmol/l with incubation for
24, 48 or 72 h were selected as the experimental conditions of the
present study. In the control group, cells were treated with medium
only. Cells were then subjected to analysis using the
immunocytochemical streptavidin-peroxidase (SP) method, flow
cytometry and methylation-specific polymerase chain reaction
(MS-PCR) technology to assess the expression of ID4 protein,
apoptosis and cell cycle and the ID4 gene methylation status of
Raji cells, respectively.
Immunocytochemical SP method and
semiquantitative analysis of ID4 protein expression
Immunocytochemistry was performed according to a
previously described method (24).
Cells were seeded onto cover slips and peroxidase was blocked by
incubating the cells with 3% H2O2 (Maixin
Biotech Co., Fuzhou, China) for 30 min. Protein was blocked by
incubation with normal goat serum (Maixin Biotech Co.) for 30 min.
Following incubation with ID4 rabbit polyclonal antibody at 1:100
dilution in PBS overnight at 4°C, cells were treated with
biotinylated secondary antibody (rat polyclonal; 1:50 dilution;
cat. no. KIT-9710; Maixin Biotechnology) and peroxidase.
Peroxidase-conjugated streptavidin was used with the Dako Real
Detection System and diaminobenzidine (Dako, Glostrup, Denmark)
according to the manufacturer's instructions. As a negative
control, the primary antibody was replaced with PBS.
The intensity of ID4 staining in the cytoplasm was
evaluated by two independent, experienced pathologists blinded to
the experimental groups using a scale of 0–12. The correlation
coefficient for the ID4 scoring determined by the two observers was
r=0.93–0.96.
Flow cytometric analysis
In each group, 1×106 Raji cells were
harvested by centrifugation at 447 × g, for 5 min and washed with
PBS twice. The apoptosis assay was performed on Raji cells of each
group using the Annexin V-FITC Apoptosis Detection Kit I (BD
Biosciences, Franklin Lakes, NJ, USA) and analyzed by
fluorescence-activated cell sorting.
For cell cycle analysis, cells were fixed by adding
700 ml/l ethanol dropwise on ice and incubation at 4°C overnight.
Following two washes with PBS, cells were incubated with propidium
iodide (PI; Bender Medsystems, Vienna, Austria) for 20 min in the
dark and analyzed by flow cytometry (FACSCalibur; BD Biosciences)
with ModFit LT™ software (Verity Software House, Topsham, ME, USA)
was used for quantification of cells in each phase of the cell
cycle.
MS-PCR
For MS-PCR, sodium bisulfite-treated DNA was
amplified using either a methylation-specific or a
non-methylation-specific primer set, synthesized by AuGCT Co.
(Beijing, China). The sequences of the methylation-specific primers
were 5′-TTT TAT AAA TAT AGT TGC GCG GC-3′ (forward) and 5′-GAA ACT
CCG ACT AAA CCC GAT-3′ (reverse). Sequences of the
non-methylation-specific primers were 5′-GTT TTA TAA ATA TAG TTG
TGT GGT GG-3′ (forward) and 5′-AAA ACT CCA ACT AAA CCC AAT CT-3′
(reverse) (24). DNA extracted
from the Raji cell line using the QlAamp DNA Mini kit (Qiagen,
Hilden, Germany) and then methylated at CpG sites using CpG
methylase enzyme (New England Biolabs, Ipswich, MA, USA) according
to the manufacturer's instructions. DNA derived from the human
breast cancer cell line MDA-MB231 (Chinese Academy of Sciences
Shanghai Institute of Cell Biology) was used as a positive control,
whose DNA was extracted using a procedure identical to that of the
Raji cells, and distilled water was treated with bisulfide and CpG
methylase to serve as a negative control (17). MSP was performed with the following
cycling conditions: 95°C for 5 min; 39 cycles of denaturation at
95°C for 1 min; specific annealing at 59°C for 1 min and extension
for 72°C for 1 min; and a final extension of 7 min at 72°C. Takara
Taq™ Hot Start Version (Takara) was used in the experiment. The PCR
mixture contained 50 ng bisulfite-treated DNA, 4 µl (2.5 mM)
deoxynucleoside triphosphate mixture, 0.5 µl (20 M) of each
primer, 10X PCR buffer and 1.25 units of Takara Taq enzyme in a
total volume of 50 µl. PCR was performed in a PTC-200 cycler
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). The amplification
products were analyzed on 2.2% agarose gels with 50-bp DNA Ladder
Maker (Takara) and visualized under ultraviolet illumination
(Ultra-Violet Products Ltd., Cambridge, UK).
Statistical analysis
All experiments were repeated three times. All
values are expressed as the mean ± standard deviation and analyzed
using SPSS 19.0 software (International Business Machines, Armonk,
NY, USA). Significance of comparisons between experimental groups
was tested using Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
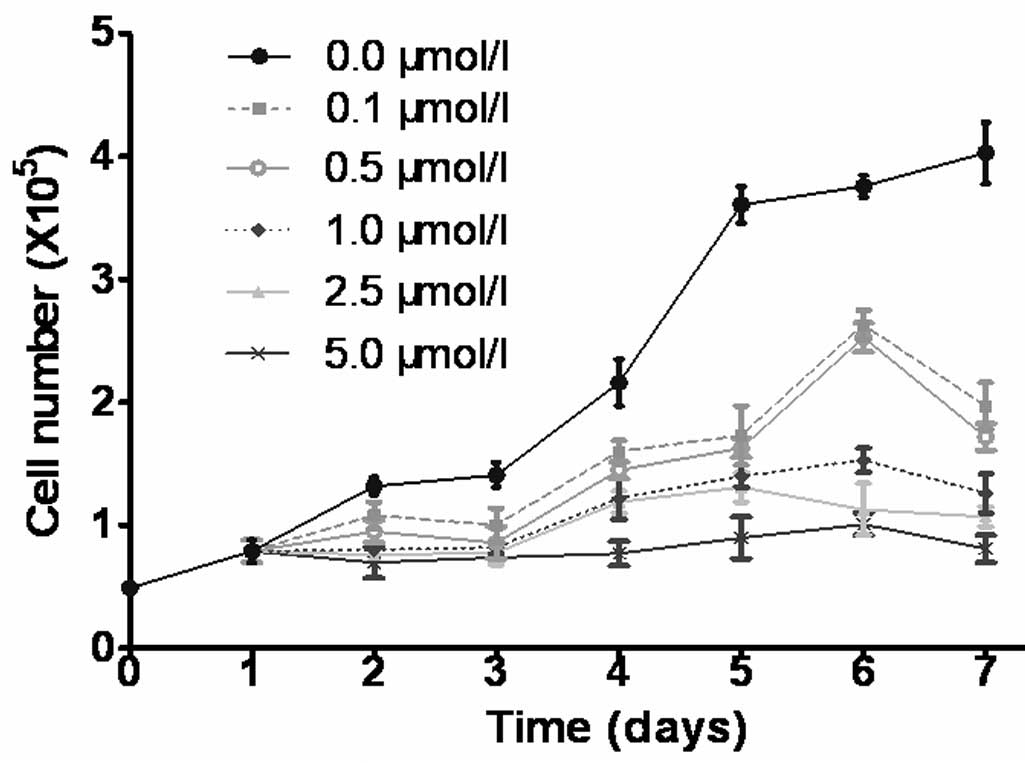
CdR inhibits cell growth
The growth curves indicated that CdR inhibited the
growth of Raji cells in a time- and dose-dependent manner (Fig. 1). As marked differences between the
growth curves of cells treated with 0, 0.5 and 5.0 µmol/l
CdR were present, these conditions were used in subsequent
experiments.
CdR enhances the expression of ID4
protein
In untreated Raji cells, ID4 protein expression was
low and only present in the cytoplasm. However, a marked increase
in ID4 protein expression was observed following CdR treatment, and
ID4 was present in the cytoplasm and in the nucleus (Fig. 2A). The expression of ID4 was
highest after 72 h of incubation with 5.0 µmol/l CdR. CdR
significantly increased the protein expression of ID4 in a
concentration-dependent manner with constant incubation time
(P<0.05); furthermore, CdR at a constant concentration of 5.0
µmol/l significantly increased the expression of ID4 in a
time-dependent manner (P<0.05) (Fig. 2B).
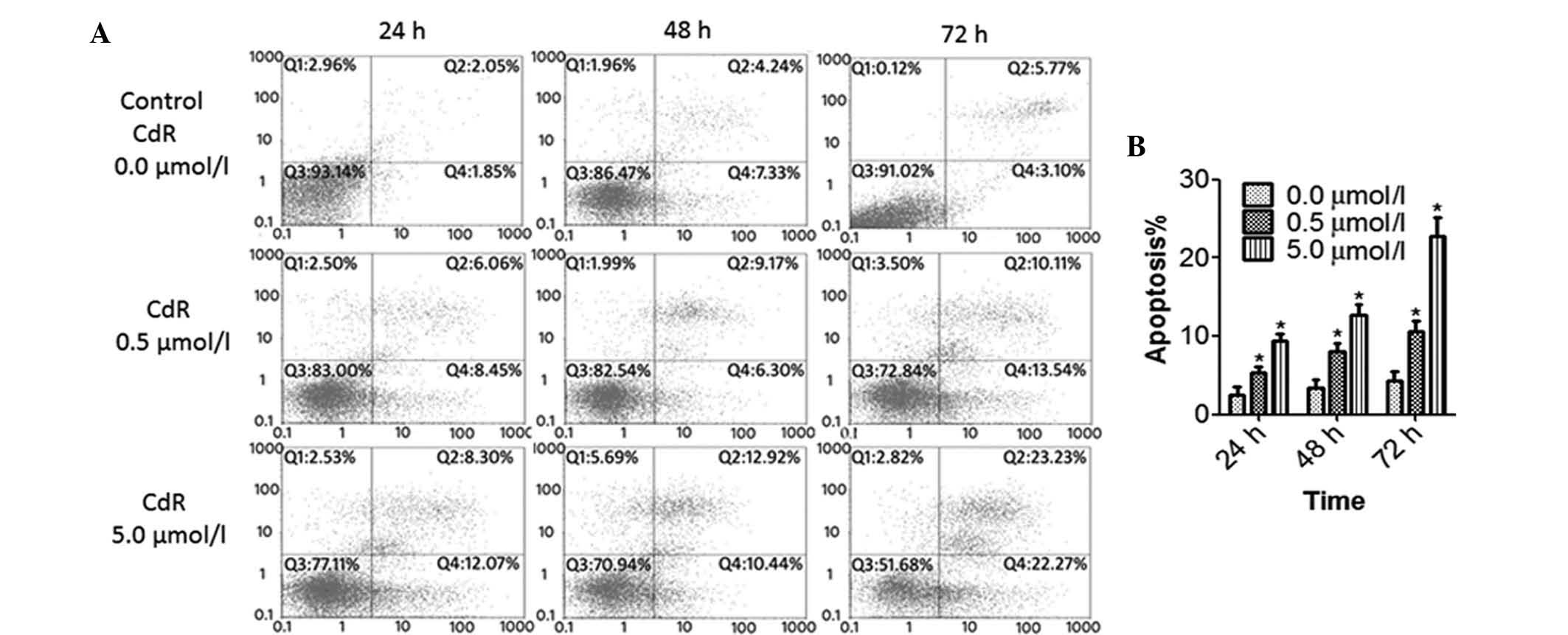
CdR causes apoptosis and S-phase arrest
in Raji cells
The effects if CdR on the apoptotic rate and cell
cycle of Raji cells were detected by flow cytometry. The number of
apoptotic cells was significantly increased by CdR treatment in a
concentration- and time-dependent manner (P<0.05) (Fig. 3). Furthermore, CdR significantly
enhanced the S-phase population of Raji cells in a concentration-
and time-dependent manner (P<0.05), indicating that CdR causes
cell cycle arrest in S phase in Raji cells (Fig. 4).
CdR reverses the hypermethylation of ID4
in Raji cells
Untreated Raji cells exhibited methylated ID4 only,
indicating that ID4 is hypermethylated in Raji cells. Of note, in
Raji cells treated with 5.0 µmol/l CdR for 72 h, only
unmethylated ID4 was present, indicating that CdR had the capacity
to fully demethylate the gene. In the group treated with 0.5
µmol/l for 48 h, methylated as well as unmethylated ID4 was
present, indicating that the gene was partially methylated
(Fig. 5).
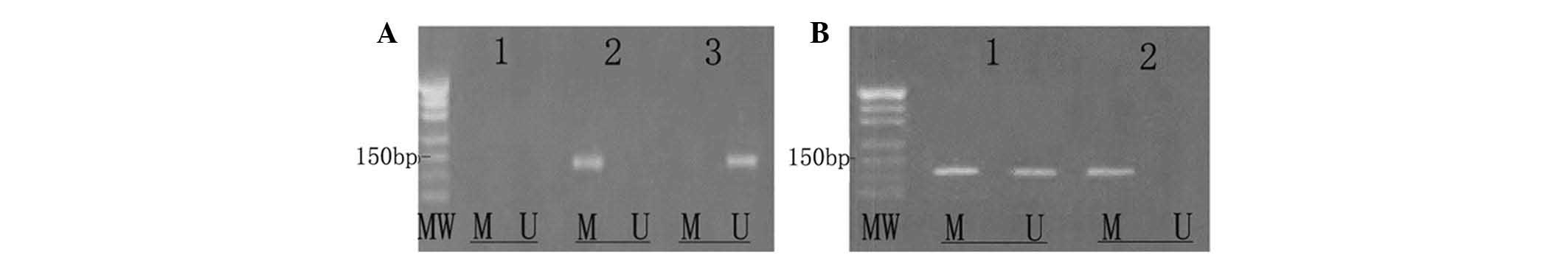 | Figure 5CdR reverses abnormal DNA methylation.
Raji cells were treated with CdR (0, 0.5 or 5.0 µmol/l) for
24, 48 or 72 h. Methylation-specific polymerase chain reaction was
used to test the ID4 gene methylation status in each group. (A)
Lanes: 1, blank control group (DNA replaced with distilled water);
2, Raji cells treated with medium only - the methylated gene as the
sole amplification product indicated that the ID4 gene is
completely methylated in Raji cells; 3, Raji cells treated with 5.0
µmol/l CdR for 72 h - amplification of the unmethylated gene
only indicated that the ID4 gene was fully demethylated by CdR. (B)
Lanes: 1, Raji cells treated with 5.0 µmol/l CdR for 48 h -
amplification of methylated and unmethylated gene indicated that
the ID4 gene was partially demethylated by CdR; 2, in untreated
Raji cells - only the methylated gene was amplified. MW, molecular
weight marker; M, methylated gene fragment; U, unmethylated gene
fragment; CdR, 5-aza-2′-deoxycytosine; ID4, inhibitor of DNA
binding 4. |
Discussion
ID4 has become a hot spot in cancer research due to
its heterogeneous roles in various cancer types (25). ID4 has been observed to be involved
in human neoplasia, including lymphoma, GBM and breast cancer, and
also represents a therapeutic target. However, as studies on the
function of ID4 in various cancer types display inconsistencies,
further clarification of its roles and the underlying mechanisms is
required. ID4 protein has been reported to enhance tumor
angiogenesis and to be is highly expressed in GBM (10,26);
however, ID4 protein expression was shown to be decreased in
several types of human cancer (7,11)
and ID4 is a putative tumor suppressor gene (8,26).
Therefore, the present study was performed to identify the role of
ID4 in Bukitt's lymphoma.
As ID4 has been suggested to be a crucial factor
controlling cell differentiation, the present study hypothesized
that epigenetic regulation of the ID4 gene may affect
differentiation and progression of Burkitt's lymphoma. In the
present study, the link between ID4 promoter hypermethylation,
protein expression levels and apoptosis was determined.
First, immunocytochemistry and semi-quantitative
analysis of ID4 protein expression proved that the expression of
ID4 protein in Raji cells was inhibited, which was consistent with
the results of previous studies (2,23,24).
The detection of ID4 protein may aid in the diagnosis of Burkitt's
lymphoma, which requires confirmation by future study. In addition,
ID4 protein was upregulated by CdR in a dose- and time-dependent
manner. These results suggested that demethylation of the promoter
region of ID4 silenced through hypermethylation in lymphoma may
represent a novel treatment approach of high therapeutic value and
good prospects.
Furthermore, flow cytometric analysis performed in
the present study showed that CdR enhanced apoptosis in Raji cells
and caused cell cycle arrest in S phase. To the best of our
knowledge, the present study was the first to report that CdR
induces cell apoptosis. It can be concluded that hypermethylated
ID4 promoted the proliferation of Burkitt's lymphoma cells, which
is in agreement with the results of Qu et al (27). Of note, CdR was able to demethylate
ID4, which may be of therapeutic significance and improve the
outcome of Burkitt's lymphoma.
The MS-PCR results of the present study were
consistent with those of previous studies (27,28),
suggesting that specific methyltransferase inhibitor CdR reversed
the abnormal DNA methylation and induced the re-expression of ID4
protein, thereby effectively inhibiting the proliferation and
promoting the differentiation and apoptosis of NHL cells. Abnormal
DNA methylation has also been shown to be reversed by other drugs,
including arsenic trioxide (27);
therefore, DNA demethylating agents may represent a novel clinical
treatment approach. In general, the treatment of ID4 gene
methylation in NHL is of great therapeutic significance.
In conclusion, the present study indicated that the
ID4 gene was hypermethylated and its protein expression was low in
Burkitt's lymphoma cells, while CdR reversed the abnormal DNA
methylation and induced re-expression of the ID4 protein.
Hypermethylation of tumor suppressor gene ID4 promotes the
proliferation of Burkitt's lymphoma cells, which can be reversed by
demethylating drug CdR, which represents a promising therapeutic
approach for Burkitt's lymphoma.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. U1204814). The
authors would like to thank the Henan Key Laboratory for Tumor
Pathology (Department of Pathology, The First Affiliated Hospital
of Zhengzhou University, Zhengzhou, China) for their technical
assistance, including Miss Dong-Ling Gao and Miss Lan Zhang for
assistance with flow cytometric analysis, Miss Shuang Xue for
MS-PCR, as well as Dr Sheng-Lei Li and Miss Ming Huang.
References
|
1
|
Sandoval J, Mendez-Gonzalez J, Nadal E,
Chen G, Carmona FJ, Sayols S, Moran S, Heyn H, Vizoso M, Gomez A,
et al: A prognostic DNA methylation signature for stage I
non-small-cell lung cancer. J Clin Oncol. 31:4140–4147. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Esteller M: Dormant hypermethylated tumour
suppressor genes: Questions and answers. J Pathol. 205:172–180.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sato T, Arai E, Kohno T, Tsuta K, Watanabe
S, Soejima K, Betsuyaku T and Kanai Y: DNA methylation profiles at
precancerous stages associated with recurrence of lung
adenocarcinoma. PLoS One. 8:594442013. View Article : Google Scholar
|
|
4
|
Guillaumet-Adkins A, Richter J, Odero MD,
Sandoval J, Agirre X, Catala A, Esteller M, Prósper F, Calasanz MJ,
Buño I, et al: Hypermethylation of the alternative AWT1 promoter in
hematological malignancies is a highly specific marker for acute
myeloid leukemias despite high expression levels. J Hematol Oncol.
7:42014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zebedee Z and Hara E: Id proteins in cell
cycle control and cellular senescence. Oncogene. 20:8317–8325.
2001. View Article : Google Scholar
|
|
6
|
Rivera R and Murre C: The regulation and
function of the Id proteins in lymphocyte development. Oncogene.
20:8308–8316. 2001. View Article : Google Scholar
|
|
7
|
Yu L, Liu C, Vandeusen J, Becknell B, Dai
Z, Wu YZ, Raval A, Liu TH, Ding W, Mao C, et al: Global assessment
of promoter methylation in a mouse model of cancer identifies Id4
as a putative tumor-suppressor gene in human leukemia. Nat Genet.
37:265–274. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilson JW, Deed RW, Inoue T, Balzi M,
Becciolini A, Faraoni P, Potten CS and Norton JD: Expression of Id
helix-loop-helix proteins in colorectal adenocarcinoma correlates
with p53 expression and mitotic index. Cancer Res. 61:8803–8810.
2001.PubMed/NCBI
|
|
9
|
Kuzontkoski PM, Mulligan-Kehoe MJ, Harris
BT and Israel MA: Inhibitor of DNA binding-4 promotes angiogenesis
and growth of glioblastoma multiforme by elevating matrix GLA
levels. Oncogene. 29:3793–3802. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeng W, Rushing EJ, Hartmann DP and Azumi
N: Increased inhibitor of differentiation 4 (id4) expression in
glioblastoma: A tissue microarray study. J Cancer. 1:1–5. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arnold JM, Mok SC, Purdie D and
Chenevix-Trench G: Decreased expression of the Id3 gene at 1p36.1
in ovarian adenocarcinomas. Br J Cancer. 84:352–359. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deleu S, Savonet V, Behrends J, Dumont JE
and Maenhaut C: Study of gene expression in thyrotropin-stimulated
thyroid cells by cDNA expression array: ID3 transcription
modulating factor as an early response protein and tumor marker in
thyroid carcinomas. Exp Cell Res. 279:62–70. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lasorella A, Uo T and Iavarone A: Id
proteins at the cross-road of development and cancer. Oncogene.
20:8326–8333. 2001. View Article : Google Scholar
|
|
14
|
Hagiwara K, Nagai H, Li Y, Ohashi H, Hotta
T and Saito H: Frequent DNA methylation but not mutation of the ID4
gene in malignant lymphoma. J Clin Exp Hematop. 47:15–18. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Umetani N, Mori T, Koyanagi K, Shinozaki
M, Kim J, Giuliano AE and Hoon DS: Aberrant hypermethylation of ID4
gene promoter region increases risk of lymph node metastasis in T1
breast cancer. Oncogene. 24:4721–4727. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cen J, Shen J, Wang X, Kang H, Wang L, Sun
L, Li Y and Yu L: Association between lymphoma prognosis and
aberrant methylation of ID4 and ZO-1 in bone marrow and
paraffin-embedded lymphoma tissues of treatment-naive patients.
Oncol Rep. 30:455–461. 2013.PubMed/NCBI
|
|
17
|
Smith E, De Young NJ, Pavey SJ, Hayward
NK, Nancarrow DJ, Whiteman DC, Smithers BM, Ruszkiewicz AR,
Clouston AD, Gotley DC, et al: Similarity of aberrant DNA
methylation in Barrett's esophagus and esophageal adenocarcinoma.
Mol Cancer. 7:752008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Noetzel E, Veeck J, Horn F, Hartmann A,
Knüchel R and Dahl E: Promoter methylation of ID4. A marker for
recurrence-free survival in human breast cancer. Pathologe.
29(Suppl 2): S319–S327. 2008.In German. View Article : Google Scholar
|
|
19
|
Yang Q, Shan L, Yoshimura G, Nakamura M,
Nakamura Y, Suzuma T, Umemura T, Mori I, Sakurai T and Kakudo K:
5-aza-2′-deoxycytidine induces retinoic acid receptor beta 2
demethylation, cell cycle arrest and growth inhibition in breast
carcinoma cells. Anticancer Res. 22:2753–2756. 2002.
|
|
20
|
Wang H, Wang XQ, Xu XP and Lin GW: ID4
methylation predicts high risk of leukemic transformation in
patients with myelodysplastic syndrome. Leuk Res. 34:598–604. 2010.
View Article : Google Scholar
|
|
21
|
Rüter B, Wijermans PW and Lübbert M: DNA
methylation as a therapeutic target in hematologic disorders:
Recent results in older patients with myelodysplasia and acute
myeloid leukemia. Int J Hematol. 80:128–135. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Umetani N, Takeuchi H, Fujimoto A,
Shinozaki M, Bilchik AJ and Hoon DS: Epigenetic inactivation of Id4
in colorectal carcinomas correlates with poor differentiation and
unfavorable prognosis. Clin Cancer Res. 10:7475–7483. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vinarskaja A, Goering W, Ingenwerth M and
Schulz WA: ID4 is frequently downregulated and partially
hypermethylated in prostate cancer. World J Urol. 30:319–325. 2012.
View Article : Google Scholar
|
|
24
|
Xu RR, Liu F, Cui X, Zhang XW and Wang Y:
ID4 promoter methylation in acute myeloid leukemia. J Exp Hematol.
19:582–584. 2011.In Chinese.
|
|
25
|
Dell'Orso S, Ganci F, Strano S, Blandino G
and Fontemaggi G: ID4: A new player in the cancer arena.
Oncotarget. 1:48–58. 2010. View Article : Google Scholar
|
|
26
|
Chan AS, Tsui WY, Chen X, Chu KM, Chan TL,
Chan AS, Li R, So S, Yuen ST and Leung SY: Downregulation of ID4 by
promoter hypermethylation in gastric adenocarcinoma. Oncogene.
22:6946–6953. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qu F, Zhao CH, Diao YQ, Zhu XL, Chen J, Li
M, Liu CP, Jiang L and Jin J: Methylation of Id4 gene and
inhibitive effect of arsenic trioxide on it in Raji cells. Clin J
Hematol. 31:821–825. 2010.In Chinese.
|
|
28
|
Vandeputte DA, Troost D, Leenstra S,
Ijlst-Keizers H, Ramkema M, Bosch DA, Baas F, Das NK and Aronica E:
Expression and distribution of id helix-loop-helix proteins in
human astrocytic tumors. Glia. 38:329–338. 2002. View Article : Google Scholar : PubMed/NCBI
|