Introduction
Kidney cancer is the 14th most common cancer in the
world, and its incidence and mortality has plateaued in North
America and Europe in previous years and continues to increase in
incidence in developing countries (1,2). The
5-year survival rate of renal cell carcinoma (RCC) is significantly
higher for stage I disease than stage III disease, underscoring the
importance of early detection and treatment of RCC. However, early
detection and treatment is difficult, since RCC is characterized by
a lack of early-warning signs, protean clinical manifestations, and
resistance to radiotherapy and chemotherapy (3,4).
Therefore, the identification of biomarkers for early-stage RCC is
important. However, no accurate biomarker for RCC currently exists
(5).
Kidney cancer is not a single disease but a number
of different types of cancer that occur in the kidney, each caused
by a different gene with a distinct histology and clinical course
that responds differently to therapy (6). These known kidney cancer genes,
VHL, MET, FLCN, TSC1, TSC2,
FH and SDH, are all possibly regulated by microRNAs
(miRNAs) through translational inhibition and/or mRNA degradation
(7). There have been an increasing
number of studies focusing on the role of miRNAs in RCC and their
clinical and pathogenetic significance (8–10).
Mutations in the miRNA processing machinery as well as deregulated
miRNA expression are observed in a number of different human cancer
types, including RCC (8).
Dysregulated miRNAs identified by array-based miRNA profiling not
only discriminate RCC from normal kidney tissue, but also
distinguish different RCC histological subtypes. Specific miRNAs
implicated in the pathogenesis of RCC provide a possible new target
or tool for clear-cell renal cell carcinoma (ccRCC) therapy
(9,10).
Previous microarray studies suggested that
microRNA-106b (miR-106b) was significantly upregulated in RCC
tissues compared with paired normal kidney tissues (11–14).
Slaby et al identified a prognostic significance of
miR-106b, which, following validation on a larger group of
patients, may be useful as a promising biomarker for the prediction
of early metastasis following nephrectomy (14). It has been reported that miR-106b
is widely upregulated in various types of human malignancy,
including head and neck (15),
ovarian (16), prostate (17), colon (18), gastric (19), laryngeal (20) and brain (21) cancer by comprehensive miRNA
profiling. However, the expression of miR-106b requires
quantification and the function of miR-106b in RCC cells requires
elucidation. The aim of present study was to determine the
upregulation of miR-106b in RCC tissues and cell lines compared
with the controls and describe the function of miR-106b in RCC
tumorigenesis.
Materials and methods
Clinical specimens, cell culture and RNA
isolation
RCC tissues and adjacent normal tissues were
obtained from the Department of Urology, Peking University Shenzhen
Hospital (Shenzhen, China) and the Anhui Medical University First
Affiliated Hospital (Anhui, China) between February 2006 and
February 2011, in accordance with Institutional Review
Board-approved protocol for human specimen collection and for the
use of these materials and associated clinical information for
research purposes. The adjacent normal tissues, ~5 cm from the edge
of the tumor, were confirmed by two experienced pathologists. Once
dissected, all samples were stored at −80°C in RNAlater (Qiagen,
Valencia, CA, USA) for further research. No treatment was
administered prior to surgery. All the specimens were evaluated
according to the 2010 American Joint Committee on Cancer staging
system (22). The
clinicopathological information of all the patients is summarized
in Table I.
 | Table IClinicopathological features in renal
cell carcinoma patients. |
Table I
Clinicopathological features in renal
cell carcinoma patients.
| Characteristic | Number of
cases |
|---|
| Mean age range
(years) | 50 (27–70) |
| Gender |
| Male | 25 |
| Female | 10 |
| Histological
type |
| Clear cell | 26 |
| Papillary | 9 |
| pT stage |
| T1 | 20 |
| T2 | 14 |
| T3+T4 | 1 |
| Fuhrman grade |
| I | 10 |
| II | 15 |
| III | 8 |
| IV | 2 |
| AJCC clinical
stages |
| I | 19 |
| II | 14 |
| III+IV | 2 |
RCC cell lines (786-O and ACHN) and human embryonic
kidney 293T (HEK-293T) cells were obtained from the Guangdong and
Shenzhen Key Laboratory of Male Reproductive Medicine and Genetics
(Shenzhen, China). All cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 1% antibiotics [100 U/ml penicillin and
100 mg/ml streptomycin sulfates (Gibco; Thermo Fisher Scientific,
Inc.)] and 1% glutamate (Gibco; Thermo Fisher Scientific, Inc.),
and then incubated at 37°C in a humidified chamber containing 5%
CO2.
Total RNA, including miRNA was extracted from 35
paired RCC samples and adjacent normal tissue and cells using
TRIzol® (Invitrogen: Thermo Fisher Scientific, Inc.) and
purified using the RNeasy® Maxi kit (Qiagen), according
to the manufacturer's instructions. The RNA samples with 260/280
ratios of 1.8–2.0 were used for further experiments.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
To quantify the expression level of miR-106b in
tissues and cells, RT-qPCR was performed. cDNA was obtained from
RNA by using the miScript II RT kit (Qiagen). cDNA templates were
diluted 10 times and then used for RT-qPCR. RT-qPCR was performed
with the miScript SYBR® Green PCR kit (Qiagen, Valencia,
CA, USA) according to the manufacturer's instructions on the Roche
LightCycler 480 Real-Time PCR System (Roche Applied Science,
Indianapolis, IN, USA). The cycling conditions were as follows:
95°C for 15 min; and 40 cycles of 94°C for 15 sec, 55°C for 30 sec
and 72°C for 30 sec. The expression levels are shown as fold
differences relative to the U6, which were calculated using the
following formula: Relative quantification (RQ) = 2−ΔΔCq
[ΔΔCq = (meanCqtumor − meanCqcontrol) −
(meanCqnormal − meanCqcontrol)]. The primer
sequences are summarized in Table
II.
 | Table IISequences of primers and miRNAs. |
Table II
Sequences of primers and miRNAs.
| Name | Sequence |
|---|
| miR-106b | Forward:
5′-TAAAGTGCTGACAGTGCAGAT-3′
Reverse: Universal Primer, miScript SYBR® Green PCR
kit |
| U6 | Forward:
5′-CTCGCTTCGGCAGCACA-3′
Reverse: 5′-ACGCTTCACGAATTTGCGT-3′ |
| miR-106b
inhibitor |
5′-AUCUGCACUGUCAGCACUUUA-3′ |
| NC |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
Cell transfection
For upregulation of miR-106b, the cancer cell lines
were transfected with synthesized miR-106b mimics (Shanghai
GenePharma Co., Ltd., Shanghai, China) using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), which were mixed in
Opti-MEM® I Reduced Serum Medium (Gibco; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions
following plating for 24 h. The miR-106b inhibitor (200, 100 and 5
pmol) or a negative control (NC) was transfected into RCC cells in
6-well, 12-well and 96-well plates, respectively, for 4–6 h. The
sequences of the miR-106b inhibitor and NC are summarized in
Table II.
Cell scratch assay
The migratory ability of 786-O and ACHN cells was
assessed by wound scratch assay in vitro. Approximately
3×105 cells were seeded per 12-well dish. After 6 h of
transfection, a sterile 200 µl pipette tip was used to
scrape a clear line through the cell layer. The cells were then
rinsed with phosphate-buffered saline (PBS) and cultured in
serum-free DMEM. Images of the scratches were acquired, using a
digital camera system, 0 and 48 h after the scratches were made
(IX71; Olympus Corporation, Tokyo, Japan). The experiments were
performed in triplicate and repeated more than three times.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
The MTT (5 mg/ml) assay (Sigma-Aldrich, St. Louis,
MO, USA) was used to analyze cell proliferation. 786-O and ACHN
cells (5,000 cells/well) were plated into 96-well plates with five
replicate wells of each condition. Each well was assessed at 0, 24,
48 or 72 h post-transfection. Prior to measurement, 20 µl
MTT was added to cells and incubated at 37°C in a humidified
chamber containing 5% CO2 for 6 h. Subsequently, the MTT
medium mixtures were discarded and 120 µl dimethyl sulfoxide
(Sigma-Aldrich, Shanghai, China) was added. Following agitation for
30 min at room temperature, the absorbance was measured using a
Model 680 enzyme-linked immunosorbent assay microplate reader
(Bio-Rad, Hercules, CA, USA) at a wave length of 490 nm (with 630
nm as the reference wave length).
Cell apoptosis assay
786-O or ACHN (3×105) cells were plated
in 6-well plates for the cell apoptosis assay. At 48 h
post-transfection, the cells, including floating cells, were
harvested, washed twice with 4°C PBS and resuspended in 100
µl 1X binding buffer and stained with 3 µl propidium
iodide and 5 µl Annexin V-fluorescein isothiocyanate
(Invitrogen; Thermo Fisher Scientific, Inc.) for 15 min at room
temperature. Flow cytometry (EPICS Xl-4; Beckman Coulter, Inc.,
Brea, CA, USA) was used to quantify the percentage of apoptotic
cells within 30 min after staining and 400 µl 1X binding
buffer was added to each sample prior to measurement. Each
experiment was performed at least three times.
Statistical analysis
All data are presented as the mean ± standard
deviation from three independent experiments. Statistical analysis
was performed with the SPSS 19.0 statistical software package
(SPSS, Inc., Chicago, IL, USA). Statistical significance was
determined using Student's t-test. For the comparison of miR-106b
expression levels in matched tumor/normal samples, paired t-test
was used. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-106b is highly expressed in human RCC
clinical specimens and RCC cell lines
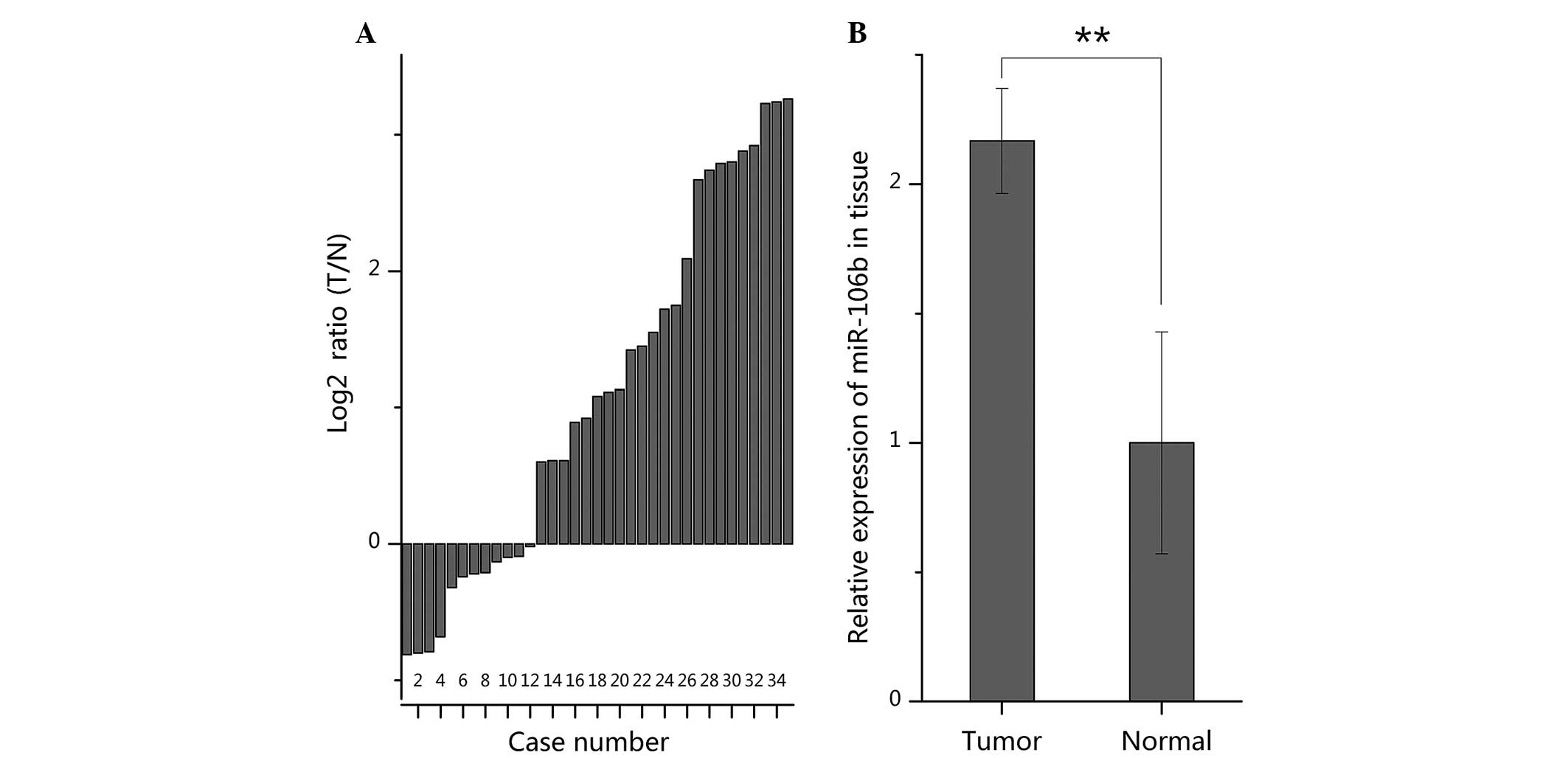
RT-qPCR was used to quantify the expression of
miR-106b in 35 paired ccRCC specimens and normal specimens. The
results demonstrated that miR-106b was significantly upregulated in
23/35 of RCC specimens (Fig. 1A),
with an average of 2.17-fold increase in expression (P<0.01;
Fig. 1B). The high expression of
miR-106b was in accordance with the results of previous miRNA
profiling in RCC (11–14).
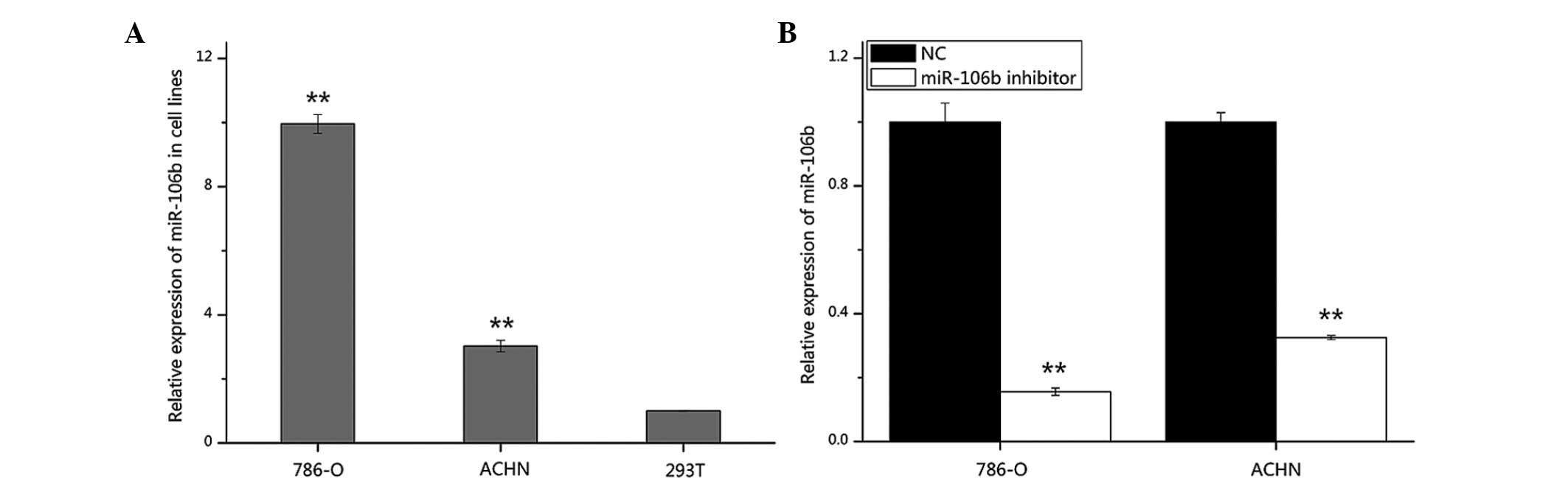
The expression of miR-106b was also analyzed in RCC
cell lines (786-O and ACHN) and HEK-293T cells by RT-qPCR. miR-106b
expression was significantly higher in 786-O and ACHN cells
(P<0.01 and P<0.01) compared with that in 293T cells, which
is in accordance with the expression pattern of miR-106b in tissues
(Fig. 2A). In addition, in order
to validate the transfection efficiency of the miR-106b inhibitor
in 786-O and ACHN cells, RT-qPCR was performed. As shown in
Fig. 2B, the expression of
miR-106b was suppressed by 84.4 and 67.5% in 786-O and ACHN cells,
respectively.
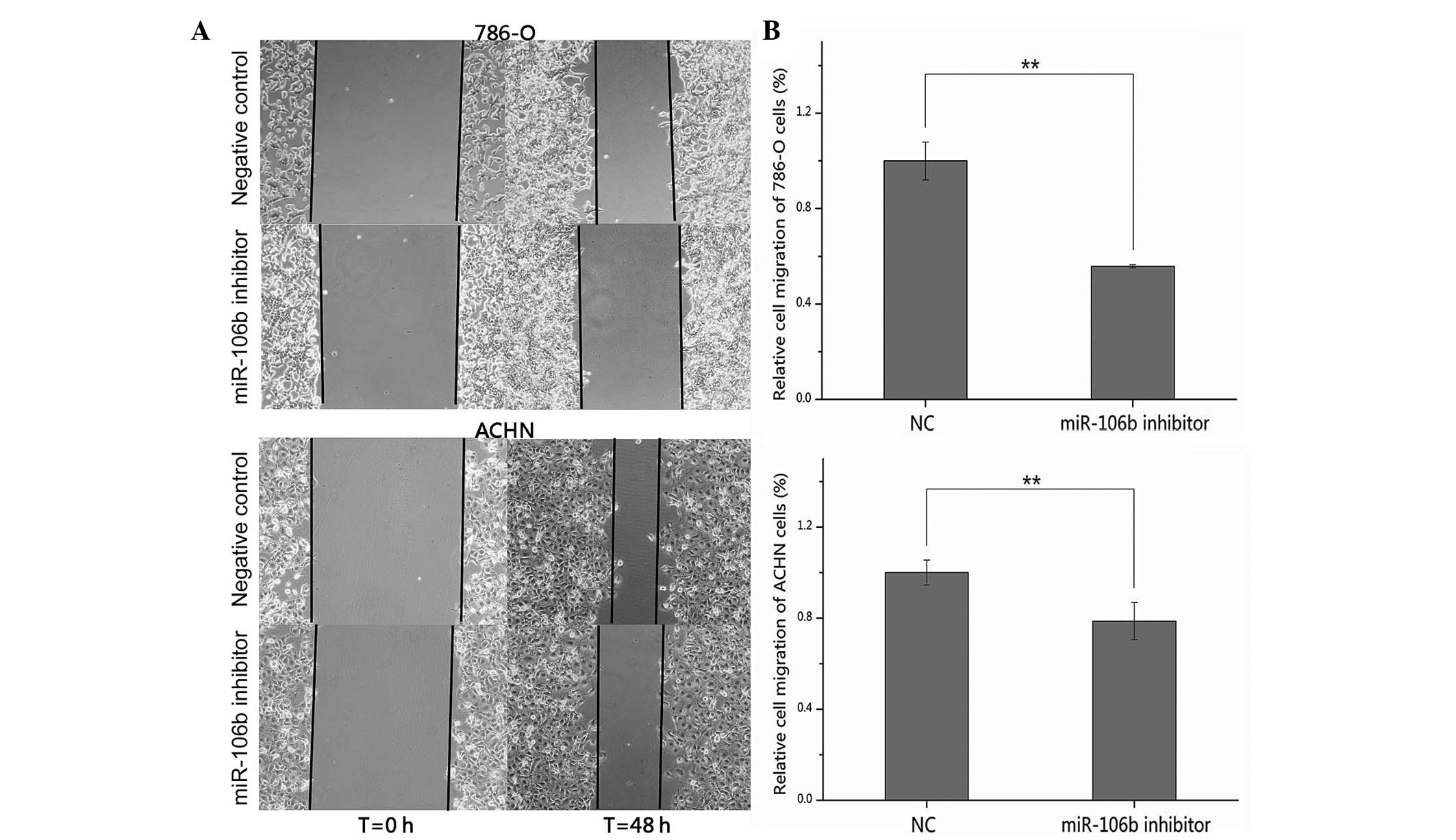
Suppression of miR-106b inhibits 786-O
and ACHN cell migration
The effects of suppression of miR-106b expression on
cell migration in RCC cells were assessed by wound scratch assay.
As shown in Fig. 3A, the wound
widths of the cells transfected with the miR-106b inhibitor were
wider than cells transfected with the NC after 48 h. Statistical
analysis demonstrated that the migratory distances of the miR-106b
inhibitor group were significantly decreased by 44.2 (P<0.01)
and 21.3% (P<0.01) in 786-O and ACHN cells, compared with the NC
group (Fig. 3B). These results
suggested that down-regulation of miR-106b inhibited the migratory
ability of RCC cells.
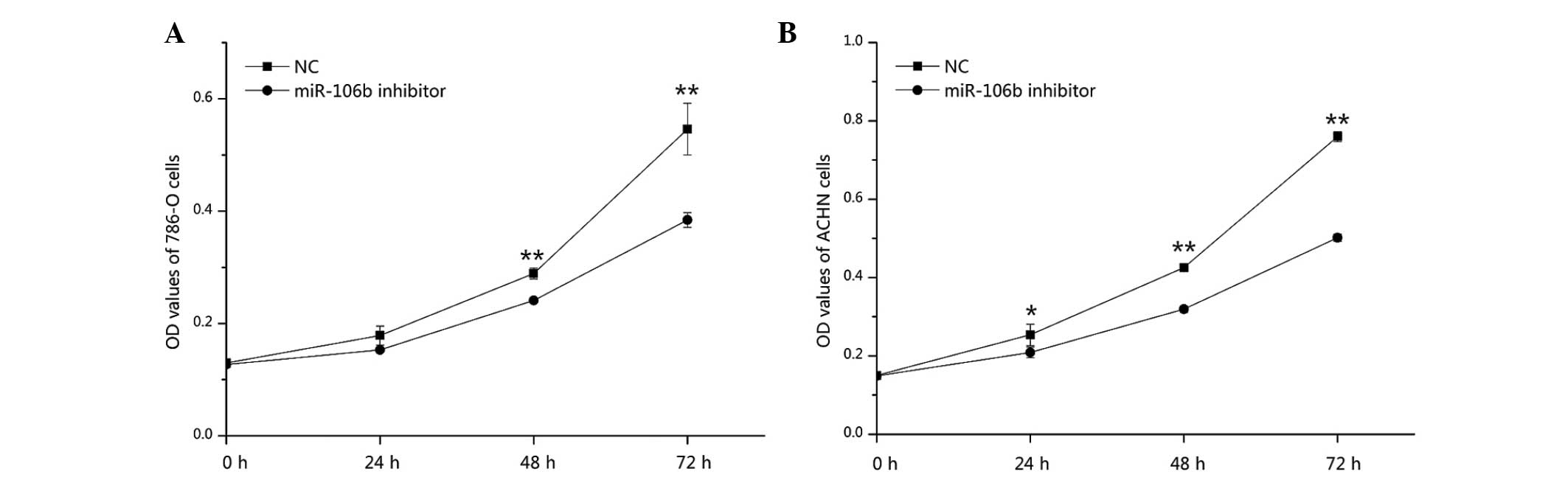
Downregulation of miR-106b suppresses
786-O and ACHN cell proliferation
The effect of downregulation of miR-106b on cell
proliferation in RCC was analyzed by an MTT assay. Optical density
values of the miR-106b inhibitor group and NC group were measured
at 0, 24, 48 and 72 h after transfection. The results revealed that
the proliferation of 786-O cells was decreased by 14.37 (P=0.09),
16.70 (P<0.01) and 29.61% (P<0.01), and the proliferation of
ACHN cells was inhibited by 17.74 (P<0.05), 24.86 (P<0.01)
and 33.96% (P<0.01) at 24, 48 and 72 h after transfection of the
miR-106b inhibitor, respectively, as compared with the NC group.
The results suggested that downregulation of miR-106b significantly
suppressed the proliferation of renal cancer cells (Fig. 4).
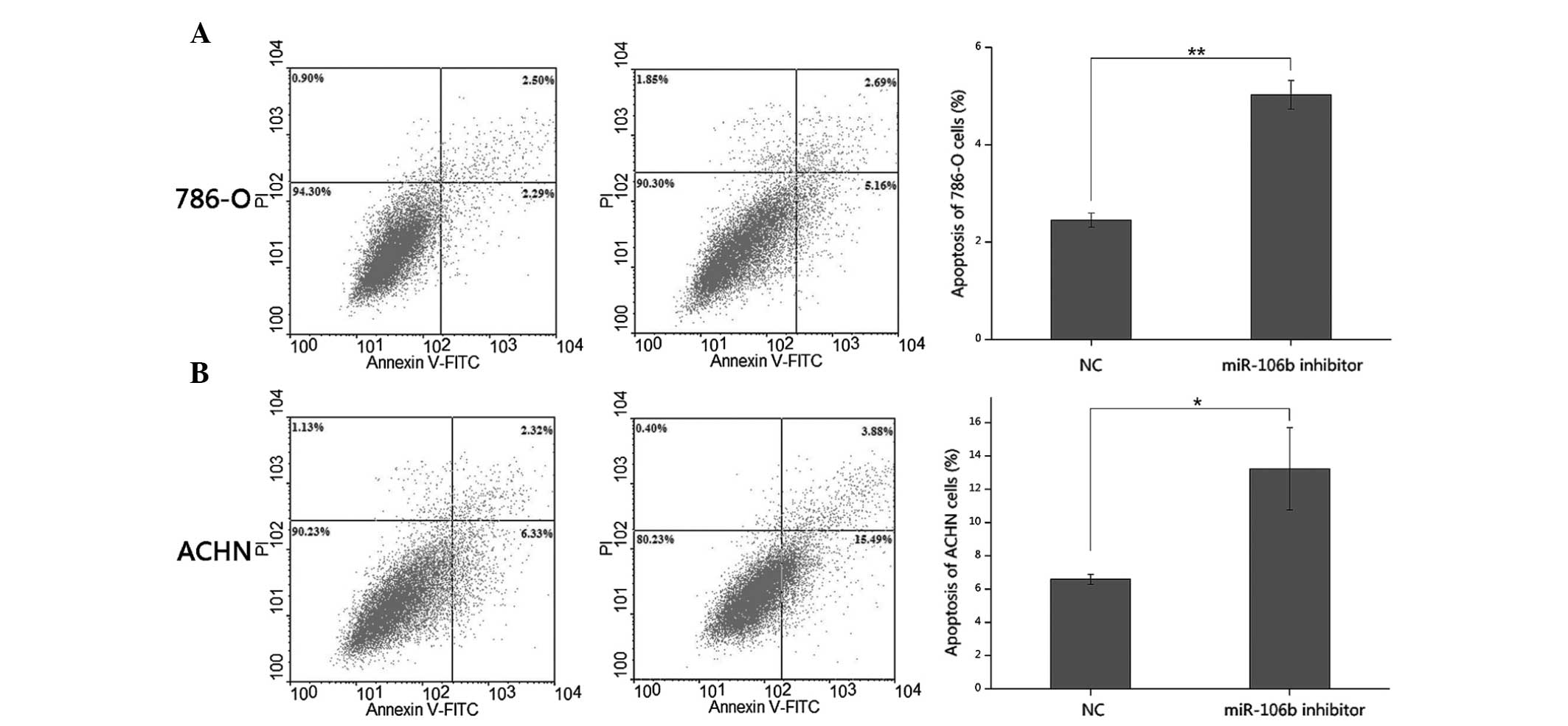
miR-106b inhibitor induces apoptosis of
786-O and ACHN cells
The effects of the miR-106b inhibitor on apoptosis
were determined by flow cytometric analysis. Following transfection
of either the miR-106b inhibitor or NC for 48 h, 786-O and ACHN
cells were harvested, stained and measured. The results revealed
that the average apoptotic rate of 786-O cells transfected with the
miR-106b inhibitor or NC was 5.03 and 2.45% (P<0.01), while the
average apoptotic rate of ACHN cells in the miR-106b inhibitor or
NC group was 13.24 vs. 6.59% (P<0.05), suggesting that the
miR-106b inhibitor induced apoptosis in RCC cells (Fig. 5).
Discussion
miRNAs are ~22 nt, single-strand, non-coding RNAs
that bind to the 3′-untranslated region of target genes and
regulate gene expression by translation inhibition or inducing mRNA
degradation (9). There are
increasing studies on the crucial role of miRNAs in cancer
initiation, progression, metastasis and invasion (7,23).
miRNAs are important regulators of all hallmarks of cancer,
including cell growth and cell cycle control, evasion of apoptosis,
tissue invasion and metastasis, angiogenesis and unlimited
replicative potential (7). There
are two essential implications of miRNAs: Firstly, the expression
profile studies of miRNAs have demonstrated that upregulated or
downregulated miRNAs in RCC, including RCC subtypes may serve as
biomarkers for diagnostic, prognostic, predictive and disease
monitoring purposes, and secondly, the function and mechanism
studies of specific miRNAs could contribute to the development of
novel therapies (23).
miR-106b has been reported to be upregulated in
tumors of numerous organs (15–21)
by comprehensive miRNA profiling. Previous microarray profiles in
RCC and adjacent normal tissues have demonstrated a high expression
of miR-106b (11–14). However, the expression pattern of
miR-106b in RCC has not been verified by RT-PCR and the
tumor-associated function of miR-106b remains to be elucidated. In
the present study, RT-qPCR was performed to validate the
upregulation of miR-106b in RCC tissues and cells, as compared with
adjacent normal tissues and HEK-293T cells. In addition, the
effects of miR-106b on cell migration, proliferation and apoptosis
in RCC cells were examined by wound scratch assay, MTT assay and
flow cytometry. The results demonstrated that miR-106b was
significantly higher in RCC tissues and cells compared with normal
tissues and cells, and the reduction of miR-106b by the synthesized
miR-106b inhibitor significantly suppressed cell migration,
proliferation and evasion of apoptosis. These results suggested
that miR-106b may be a biomarker for RCC diagnosis, disease
monitoring and prognosis and could be a target for therapeutic
purposes.
miRNAs exert their functions on cancer cells mainly
depending on their target genes. The various molecular mechanisms
of how miR-106b affects tumor initiation, development, metastasis
and invasion have been reported in several different types of
cancer. In brain cancer, through downregulation of p21/WAF1/Cip1,
miRNA-106b was verified to be important in melanoma growth, and
grape seed proanthocyanidins acted as an inhibitor of miR-106b
thereby inhibiting melanoma growth in vitro and in
vivo models (24). Another
study identified that overexpression of miR-106b-5p in glioma tumor
cells significantly promoted cell proliferation and inhibited cell
apoptosis, while knockdown of miR-106b-5p significantly inhibited
cell proliferation and enhanced cell apoptosis in vitro and
in vivo. Furthermore, the authors found that miR-106b-5p
affected cell proliferation by targeting retinoblastoma-like 1
(RBL1) and RBL2 and regulated cell apoptosis by decreasing
caspase-8 (25). The miR-106b-25
miRNA was also found to be associated with prostate cancer
progression and disease outcome and may do so by altering
apoptosis- (caspase-7, apoptosis-related cysteine peptidase) and
focal adhesion-associated pathways (24,26).
Epithelial-mesenchymal transition (EMT) describes a transcriptional
mechanism that ensures tissue remodeling during embryonic
morphogenesis and is considered to be an important step in cancer
cell dissemination and metastasis (27). By targeting TWIST1, miR-106b
modulates EMT in invasive endometrial cancer cell lines (28). In laryngeal cancer, miR-106b was
reported to target RUNX3 and RB is important in proliferation and
invasion of laryngeal carcinoma cells (29,30).
Stromal interactions with cancer cells in the microenvironment
determine whether cancer cells remain stable or progress into
invasive and metastatic tumors (31). miRNA-106b in cancer-associated
fibroblasts from gastric cancer function as an oncogene, promoting
cell migration and invasion by targeting PTEN (32). One study demonstrated that AAAGUGC
seed-containing miRNAs, including miR-106b promote cell expansion,
replating capacity and signaling in hematopoietic cells by
interference with quantitative proteomics identified sequestosome
1-regulated pathways (33). In
hepatocellular carcinoma, miR-106b targeted adenomatous polyposis
coli (APC) and downregulation of APC caused accumulation of
β-catenin, which is a major cellular effector of Wnt signaling
(34). miR-106b was necessary for
hepatocellular carcinoma cell proliferation, anchorage-independent
growth, apoptosis (targeting F2F1) and contributed to metastasis
and migration by activating EMT (35,36).
In clinical application research, miR-106b in plasma
was significantly higher in gastric cancer patients compared with
in the control, suggesting that miR-106b may be a new tumor marker
for gastric cancer (37–39) and laryngeal cancer (20). Urinary cell-free miRNA-106b has
been verified as a novel biomarker for the detection of bladder
cancer and the level of urinary miR-106b was associated with
advanced tumor stage (40).
Increased tumor miR-106b expression was also reported to be
associated with disease recurrence in prostate cancer (26), adverse outcomes and a higher risk
of systemic spread in osteosarcomas (41).
In conclusion, to the best of our knowledge, the
present study is the first to reveal the relative expression and
cellular function of miR-106b in RCC. The results in the present
study demonstrated that miR-106b was significantly upregulated in
RCC tissues and cell lines compared with controls, and inhibition
of miR-106b suppressed cell migration and proliferation, although
induced cell apoptosis. In addition, the data suggested that
miR-106b may not only be a promising biomarker for diagnosis,
disease monitoring and prognosis, but may also be a therapeutic
target for the treatment of RCC. Further studies are required to
examine the roles and target genes of miR-106b in RCC.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81101922), Science
and Technology Development Fund Project of Shenzhen (grant nos.
JCYJ20130402114702124, JCYJ20130402113131201 and
JCYJ20150403091443329) and the fund of Guangdong Key Medical
Subject.
References
|
1
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
3
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iwamoto H, Kanda Y, Sejima T, Osaki M,
Okada F and Takenaka A: Serum miR-210 as a potential biomarker of
early clear cell renal cell carcinoma. Int J Oncol. 44:53–58.
2014.
|
|
5
|
Ljungberg B, Cowan NC, Hanbury DC, Hora M,
Kuczyk MA, Merseburger AS, Patard JJ, Mulders PF and Sinescu IC;
European Association of Urology Guideline Group: EAU guidelines on
renal cell carcinoma: The 2010 update. Eur Urol. 58:398–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Linehan WM, Srinivasan R and Schmidt LS:
The genetic basis of kidney cancer: A metabolic disease. Nat Rev
Urol. 7:277–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fendler A, Stephan C, Yousef GM and Jung
K: MicroRNAs as regulators of signal transduction in urological
tumors. Clin Chem. 57:954–968. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oulas A, Karathanasis N, Louloupi A and
Poirazi P: Finding cancer-associated miRNAs: Methods and tools. Mol
Biotechnol. 49:97–107. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maher ER: Genomics and epigenomics of
renal cell carcinoma. Semin Cancer Biol. 23:10–17. 2013. View Article : Google Scholar
|
|
10
|
Rydzanicz M, Wrzesiński T, Bluyssen HA and
Wesoly J: Genomics and epigenomics of clear cell renal cell
carcinoma: Recent developments and potential applications. Cancer
Lett. 341:111–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chow TF, Youssef YM, Lianidou E, Romaschin
AD, Honey RJ, Stewart R, Pace KT and Yousef GM: Differential
expression profiling of microRNAs and their potential involvement
in renal cell carcinoma pathogenesis. Clin Biochem. 43:150–158.
2010. View Article : Google Scholar
|
|
12
|
Wulfken LM, Moritz R, Ohlmann C,
Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel
G, von Ruecker A, Müller SC and Ellinger J: MicroRNAs in renal cell
carcinoma: Diagnostic implications of serum miR-1233 levels. PLoS
One. 6:e257872011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Muller S and Nowak K: Exploring the
miRNA-mRNA regulatory network in clear cell renal cell carcinomas
by next-generation sequencing expression profiles. Biomed Res Int.
2014:9484082014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Slaby O, Jancovicova J, Lakomy R, Svoboda
M, Poprach A, Fabian P, Kren L, Michalek J and Vyzula R: Expression
of miRNA-106b in conventional renal cell carcinoma is a potential
marker for prediction of early metastasis after nephrectomy. J Exp
Clin Cancer Res. 29:1756–9966. 2010. View Article : Google Scholar
|
|
15
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B,
Waldron J, et al: Comprehensive MicroRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji T, Zheng ZG, Wang FM, Xu LJ, Li LF,
Cheng QH, Guo JF and Ding XF: Differential microRNA expression by
Solexa sequencing in the sera of ovarian cancer patients. Asian Pac
J Cancer Prev. 15:1739–1743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe
TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al:
Genomic profiling of microRNA and messenger RNA reveals deregulated
microRNA expression in prostate cancer. Cancer Res. 68:6162–6170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen
XM and Gao HJ: Initial study of microRNA expression profiles of
colonic cancer without lymph node metastasis. J Dig Dis. 11:50–54.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni
L, Zhang WG, Nan KJ, Song TS and Huang C: MicroRNA profiling of
human gastric cancer. Mol Med Rep. 2:963–970. 2009.PubMed/NCBI
|
|
20
|
Yu X, Wu Y, Liu Y, Deng H, Shen Z, Xiao B
and Guo J: miR-21, miR-106b and miR-375 as novel potential
biomarkers for laryngeal squamous cell carcinoma. Curr Pharm
Biotechnol. 15:503–508. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Zhang H, Zhang A, Han L, Wang K,
Liu R, Yang S, Pu P, Shen C, Kang C and Yu C: Upregulation of
miR-20a and miR-106b is involved in the acquisition of malignancy
of pediatric brainstem gliomas. Oncol Rep. 28:1293–1300.
2012.PubMed/NCBI
|
|
22
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer; New York, NY: pp. 479–489. 2010
|
|
23
|
Schaefer A, Stephan C, Busch J, Yousef GM
and Jung K: Diagnostic, prognostic and therapeutic implications of
microRNAs in urologic tumors. Nat Rev Urol. 7:286–297. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Prasad R and Katiyar SK: Down-regulation
of miRNA-106b inhibits growth of melanoma cells by promoting
G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1
protein. Oncotarget. 5:10636–10649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu F, Gong J, Huang W, Wang Z, Wang M,
Yang J, Wu C, Wu Z and Han B: MicroRNA-106b-5p boosts glioma
tumorigenesis by targeting multiple tumor suppressor genes.
Oncogene. 33:4813–4822. 2014. View Article : Google Scholar
|
|
26
|
Hudson RS, Yi M, Esposito D, Glynn SA,
Starks AM, Yang Y, Schetter AJ, Watkins SK, Hurwitz AA, Dorsey TH,
et al: MicroRNA-106b-25 cluster expression is associated with early
disease recurrence and targets caspase-7 and focal adhesion in
human prostate cancer. Oncogene. 32:4139–4147. 2013. View Article : Google Scholar
|
|
27
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong P, Kaneuchi M, Watari H, Sudo S and
Sakuragi N: MicroRNA-106b modulates epithelial-mesenchymal
transition by targeting TWIST1 in invasive endometrial cancer cell
lines. Mol Carcinog. 53:349–359. 2014. View
Article : Google Scholar
|
|
29
|
Xu Y, Wang K, Gao W, Zhang C, Huang F, Wen
S and Wang B: MicroRNA-106b regulates the tumor suppressor RUNX3 in
laryngeal carcinoma cells. FEBS Lett. 587:3166–3174. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai K, Wang Y and Bao X: MiR-106b promotes
cell proliferation via targeting RB in laryngeal carcinoma. J Exp
Clin Cancer Res. 30:1756–9966. 2011. View Article : Google Scholar
|
|
31
|
Rubin H: Contact interactions between
cells that suppress neoplastic development: Can they also explain
metastatic dormancy? Adv Cancer Res. 100:159–202. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang TS, Yang XH, Chen X, Wang XD, Hua J,
Zhou DL, Zhou B and Song ZS: MicroRNA-106b in cancer-associated
fibroblasts from gastric cancer promotes cell migration and
invasion by targeting PTEN. FEBS Lett. 588:2162–2169. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meenhuis A, van Veelen PA, de Looper H,
van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru
AH, van Adrichem AJ, et al: MiR-17/20/93/106 promote hematopoietic
cell expansion by targeting sequestosome 1-regulated pathways in
mice. Blood. 118:916–925. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen G, Jia H, Tai Q, Li Y and Chen D:
miR-106b downregulates adenomatous polyposis coli and promotes cell
proliferation in human hepatocellular carcinoma. Carcinogenesis.
34:211–219. 2013. View Article : Google Scholar
|
|
35
|
Yau WL, Lam CS, Ng L, Chow AK, Chan ST,
Chan JY, Wo JY, Ng KT, Man K, Poon RT and Pang RW: Over-expression
of miR-106b promotes cell migration and metastasis in
hepatocellular carcinoma by activating epithelial-mesenchymal
transition process. PLoS One. 8:e578822013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Tan W, Neo TW, Aung MO, Wasser S,
Lim SG and Tan TM: Role of the miR-106b-25 microRNA cluster in
hepatocellular carcinoma. Cancer Sci. 100:1234–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsujiura M, Ichikawa D, Komatsu S,
Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi
K, Fujiwara H, et al: Circulating microRNAs in plasma of patients
with gastric cancers. Br J Cancer. 102:1174–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang R, Wang W, Li F, Zhang H and Liu J:
MicroRNA-106b~25 expressions in tumor tissues and plasma of
patients with gastric cancers. Med Oncol. 31:2432014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cai H, Yuan Y, Hao YF, Guo TK, Wei X and
Zhang YM: Plasma microRNAs serve as novel potential biomarkers for
early detection of gastric cancer. Med Oncol. 30:4522013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou X, Zhang X, Yang Y, Li Z, Du L, Dong
Z, Qu A, Jiang X, Li P and Wang C: Urinary cell-free microRNA-106b
as a novel biomarker for detection of bladder cancer. Med Oncol.
31:1972014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Arabi L, Gsponer JR, Smida J, Nathrath M,
Perrina V, Jundt G, Ruiz C, Quagliata L and Baumhoer D:
Upregulation of the miR-17–92 cluster and its two paraloga in
osteosarcoma - reasons and consequences. In: Genes Cancer. 5. pp.
56–63. 2014
|