Introduction
Interleukin (IL)-36RN was first discovered as an
IL-1 family cytokine, also known as IL-1F5, IL-1δ, IL-1Hy1, FIL-1δ,
IL-1H3, IL-1RP3 and IL-1L1 (1,2),
which is, together with classic IL-1 members, IL-37 and other IL-36
cytokines (IL-36α, IL-36β and IL-36γ) located in a 360-kb region of
chromosome 2q13 (3). IL-36RN was
identified to encode anti-inflammatory cytokine IL-36Ra, which is
52% homologous to the IL receptor antagonist (IL-1Ra) (4). IL-36Ra binds to IL-1Rrp2 and inhibits
IL-36α, IL-36β and IL-36γ in similar manner to IL-1Ra inhibiting
IL-1α and IL-1β (5). In spite of
its similar functions to those of IL-1Ra, IL-36Ra itself can induce
IL-4 expression in glial cells, while IL-4 is indispensable for the
anti-inflammatory activities of IL-36Ra in the brain; however,
IL-1Ra has not been found to induce any cytokines (6). To facilitate functional
investigations, IL-36 cytokines, including IL-36Ra, were re-named
in 2010 with the aim to distinguish them from the IL-1 cytokines
(2).
With regard the functions of IL-36, the perturbation
of the IL-36 signaling balance contributes to the pathogenesis of
immunological and inflammatory diseases (7). The balance can be disrupted by
aberrant expression of either agonists or antagonists of IL-36
signaling. The IL-36R signaling agonists IL-36α, -β and -γ are
highly expressed in several inflammatory diseases, including
chronic obstructive pulmonary disease (8), asthma (9), obesity (10), ankylosing spondylitis (11), rheumatoid arthritis (12) and allergic contact dermatitis
(13), and have a significant role
in these diseases. As an antagonist of IL-36 signaling, IL-36Ra is
also implicated in the pathogenesis of immunological and
inflammatory conditions. IL-36Ra expression is associated with
Kindler syndrome (14), brain
micromotion (15) and psoriasis
(16). It was recently shown that
mutations of IL36RN are closely associated with a serious disease
called general pustular psoriasis (GPP) (17–20).
Single-nucleotide polymorphisms (SNPs) in the IL-36RN gene can lead
to induction of a premature stop-codon, frame-shift mutation or an
amino acid substitution, resulting in a misfolded IL-36Ra protein
that is less stable and poorly expressed (17,18,20).
However, the roles of IL-36Ra in inflammation-associated tumors
have not been clearly elucidated, while IL-36 signaling has been
implicated in inflammatory diseases; therefore, an integrative
analysis of IL-36RN and its prognostic value in cancer is
required.
The present study assessed the IL-36RN gene in a
wide range of genomes using integrative genomic analyses.
Subsequently, functionally relevant SNP analysis and comparative
proteomic analysis of IL-36RN were conducted. The conserved
transcription-factor binding sites within the upstream region of
IL-36RN as well as the prognostic value of IL-36RN in cancer were
investigated.
Materials and methods
Identification of the IL-36RN gene in
vertebrate genomes and integrative genomic analyses
The nucleotide and amino acid sequences of IL-36RN
were obtained from the Ensembl database (www.ensembl.org), based on orthologous and paralogous
relationships. The IL-36RN gene sequences subjected to analysis
with the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi)
against the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) to confirm that
the best hits were the IL36RN genes for the selected species.
Conserved transcription-factor binding sites within promoter
regions of the human IL-36RN gene were obtained from the
DECipherment Of DNA Elements proprietary database (http://www.sabiosciences.com/chipqpcrsearch.php?app=TFBS)
of SABiosciences (Qiagen, Hilden, Germany), which combines text
mining with data from the genome browser of the University of
California, Santa Cruz (https://genome.ucsc.edu/).
Comparative proteomic analysis of the
IL-36RN protein
The ClustalW software implemented in MEGA 5.05
(http://www.megasoftware.net/) was used
to align the protein-coding sequences of IL-36RN. A maximum
likelihood tree of IL-36RN amino acid sequences was constructed
using MEGA 5.05 with the Kimura 2-parameter model (21). For the relative support of the
internal node, bootstrap analysis was performed with 1,000
replications for ML reconstructions. The positive selection of
IL-36RN during evolution (22) was
analyzed using the program CodeML implemented in the PAML4.7
software package (http://abacus.gene.ucl.ac.uk/software/paml.html).
Codon subsitution models M0 (one ratio), M1a (NearlyNeutral), M2a
(PositveSelection), M7 (β) and M8 (β and ω) were used. The
site-specific model was generated using likelihood ratio tests to
compare the models as previously described (23).
In silico expression analyses of the
human IL-36RN gene
The expression profiles of normal human tissues were
acquired from GeneAnnot (http://genecards.weizmann.ac.il/geneannot/index.shtml)
and ArrayExpress (https://www.ebi.ac.uk/array-express/). Using the human
IL-36RN gene (GenBank ID, NC_000002.12) as a query sequence,
expressed sequence tags (ESTs) derived from the human IL-36RN gene
were identified by BLAST as described previously (24). Virtual northern blot analysis was
also performed by searching the uniGene database of the National
Center of Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/unigene). In addition,
protein expression profiles of IL-36RN were obtained from the
Systematic Protein Investigative Research Environment (25) and the Model Organism Protein
Expression Database (26).
Evaluation of functionally relevant SNPs
of the human IL-36RN gene and identification of somatic mutations
in human cancer
Ensembl (http://www.ensembl.org/index.html) and the NCBI's
Database of SNPs (http://www.ncbi.nlm.nih.gov/snp/) were used to obtain
functionally relevant SNPs of the human IL-36RN gene as previously
described (24,27,28).
The SNPs that could disrupt exonic splicing enhancer (ESE)/exonic
splicing silencer (ESS) motifs or cause a missense mutation were
identified. Somatic mutations of the IL-36RN gene were identified
in human cancer types from the Catalogue Of Somatic Mutations In
Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cosmic/), which mines
complete cancer genomes (29).
Meta-analysis of the prognostic value of
the IL-36RN gene in cancer
The PrognoScan database (http://www.prognoscan.org/) (30) contains a large collection of
publicly available cancer microarray datasets with clinical
annotation, enabling it to also be used as an efficient tool for
assessing the association between gene expression and cancer
prognosis. During gene analysis, PrognoScan employed the minimum
P-value approach for grouping patients for survival analysis. Data
was collected for further analysis by searching the IL-36RN gene as
a query in PrognoScan.
Results
Comparative proteomic analysis of the
IL-36RN protein
All the IL-36RN gene and protein sequences were
collected from the Ensembl database and then confirmed by BLAST.
The complete IL-36RN genes were identified in human, chimpanzee,
gibbon, orangutan, olive baboon, vervet-African green monkey,
marmoset, bush baby, tarsier, rabbit, pika, rat, mouse, elephant,
cat, dog, panda, ferret, horse, cow, dolphin, guinea pig, sheep,
opossum, tasmanian devil, armadillo and tree shrew genomes. The
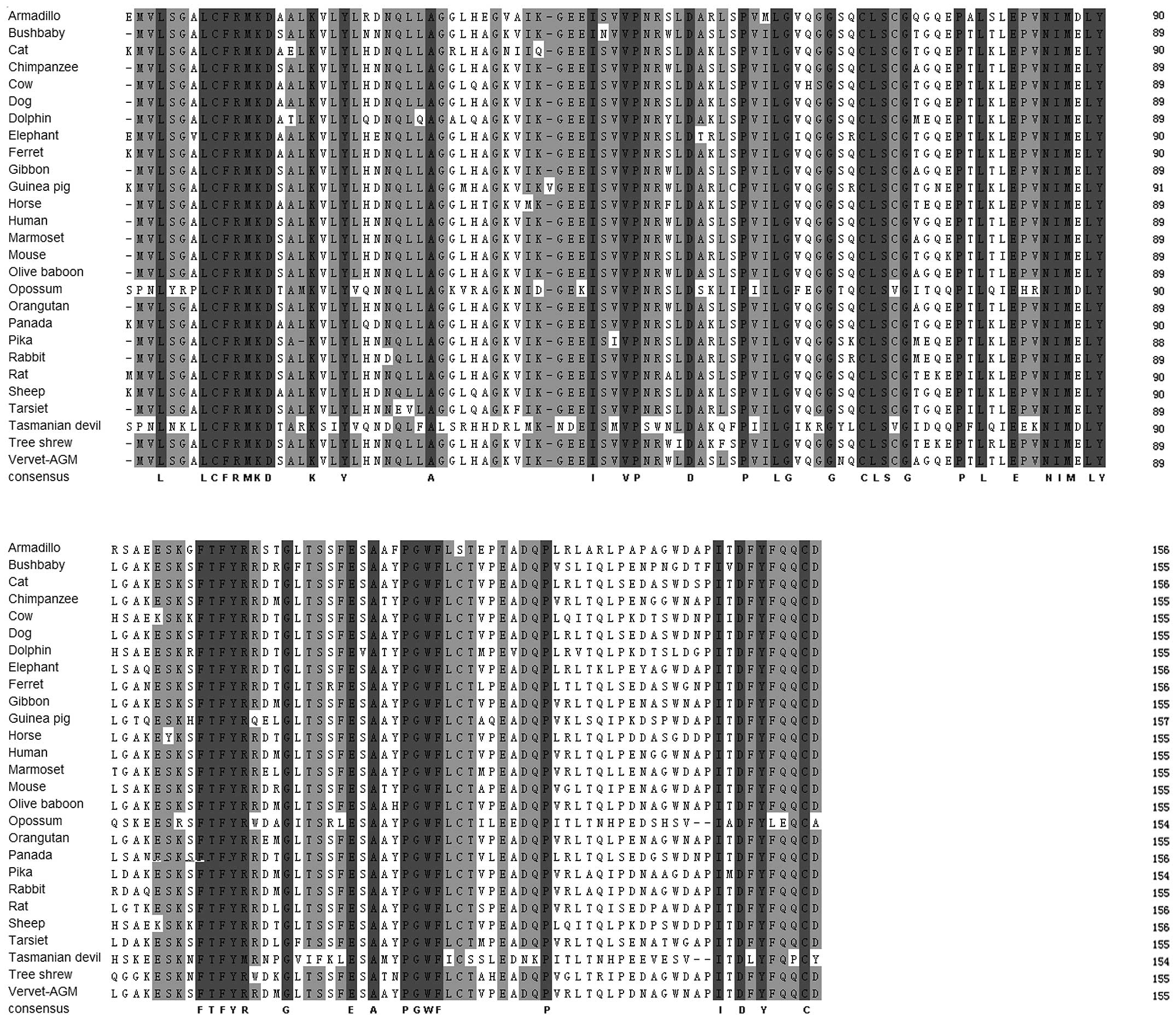
sequence and structural alignment of IL-36RN is illustrated in
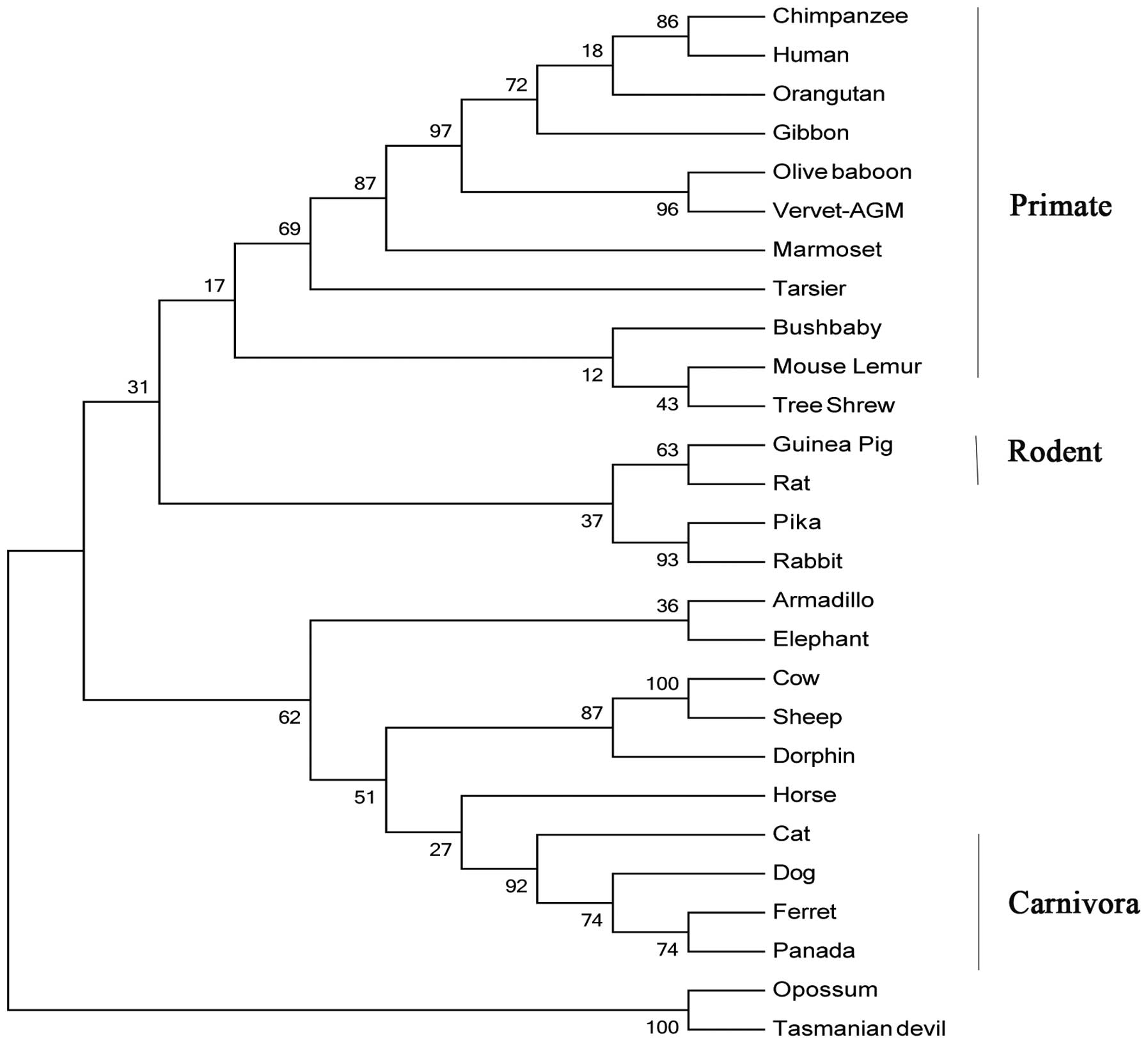
Fig. 1. Refined phylogentic trees
generated using the identified IL-36RN protein amino acid sequences
by ML and neighbor-joining (NJ) methods were almost identical;
therefore, only the results of the ML method are presented
(Fig. 2). It appeared that the
IL-36RN protein from the primate lineage forms a species-specific
cluster. Site-specific analysis for positive selection was
performed for primate, rodent, carnivora, mammalian and mammalian
excluding primate lineages. By using any of the six models in the
IL-36RN proteins, no positive selection site was identified.
Instead, purifying selection was observed among the proteins (data
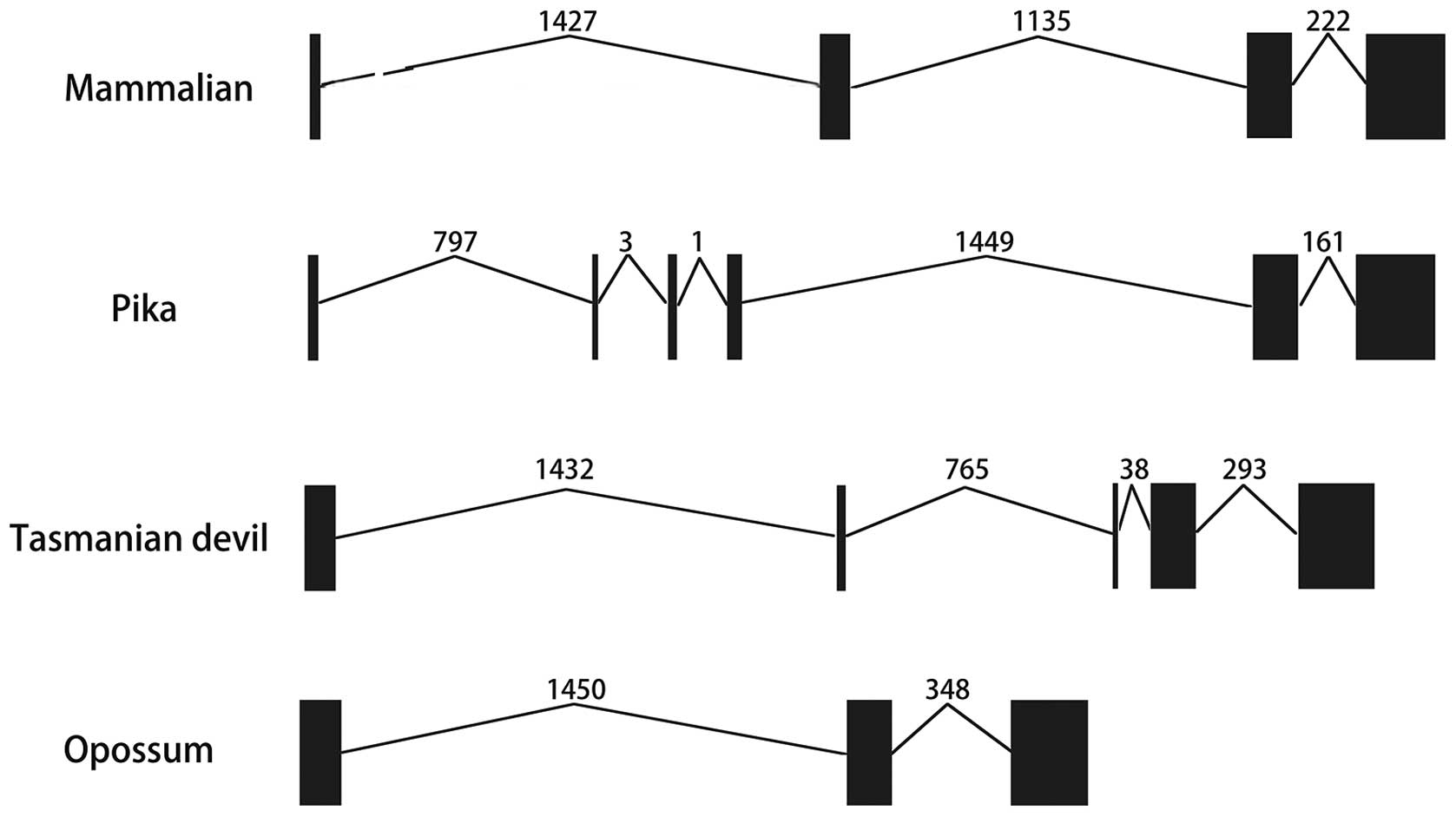
not shown). Furthermore, the exon-intron information was collected
from the Ensembl database and presented in Table I and Fig. 3. In most of the mammalian genomes,
IL-36RN genes had four exons and three introns of similar length.
In the primate lineage, IL-36RN genes showed the same exon lengths
and numbers with similar exon-intron conservations (Table I). However, IL-36RN genes had six
exons and five introns in pikas and only three exons and two
introns in opossums. Furthermore, the tasmanian devil was shown to
have five exons and four introns in its IL-36RN genes (Table I and Fig. 3).
 | Table IExon and intron lengths of
IL-36RN. |
Table I
Exon and intron lengths of
IL-36RN.
| Species |
Length (bp)
|
|---|
| Exon 1 | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | Exon 4 | Intron 4 | Exon 5 | Intron 5 | Exon 6 | Total exons |
|---|
| Armadillo | 32 | 734 | 86 | 820 | 128 | 756 | 225 | – | – | – | – | 471 |
| Bushbaby | 29 | 1343 | 86 | 1280 | 128 | 199 | 225 | – | – | – | – | 468 |
| Cat | 32 | 1334 | 86 | 1036 | 128 | 202 | 225 | – | – | – | – | 471 |
| Chimpanzee | 29 | 1384 | 86 | 1187 | 128 | 201 | 225 | – | – | – | – | 468 |
| Cow | 29 | 1334 | 86 | 1014 | 128 | 171 | 225 | – | – | – | – | 468 |
| Dog | 29 | 1553 | 86 | 1058 | 128 | 194 | 225 | – | – | – | – | 468 |
| Dolphin | 29 | 1518 | 86 | 998 | 128 | 171 | 225 | – | – | – | – | 468 |
| Elephant | 32 | 1510 | 86 | 976 | 128 | 220 | 225 | – | – | – | – | 471 |
| Ferret | 32 | 1539 | 86 | 1032 | 128 | 175 | 225 | – | – | – | – | 471 |
| Gibbon | 29 | 1382 | 86 | 1182 | 128 | 201 | 225 | – | – | – | – | 468 |
| Guinea Pig | 32 | 1650 | 86 | 1282 | 131 | 203 | 225 | – | – | – | – | 474 |
| Horse | 29 | 1369 | 86 | 1050 | 128 | 197 | 225 | – | – | – | – | 468 |
| Human | 29 | 1384 | 86 | 1186 | 128 | 201 | 225 | – | – | – | – | 468 |
| Marmoset | 29 | 1380 | 86 | 1187 | 128 | 201 | 225 | – | – | – | – | 468 |
| Mouse Lemur | 29 | 1353 | 86 | 1264 | 128 | 200 | 225 | – | – | – | – | 468 |
| Olive baboon | 29 | 1385 | 86 | 1197 | 128 | 200 | 222 | – | – | – | – | 465 |
| Opossum | 118 | 1450 | 128 | 348 | 219 | – | – | – | – | – | – | 465 |
| Orangutan | 29 | 1384 | 86 | 1184 | 128 | 201 | 225 | – | – | – | – | 468 |
| Panda | 32 | 1579 | 86 | 1055 | 128 | 201 | 225 | – | – | – | – | 471 |
| Pika | 29 | 797 | 16 | 3 | 25 | 1 | 42 | 1449 | 128 | 161 | 225 | 465 |
| Rabbit | 29 | 1491 | 86 | 1141 | 128 | 198 | 225 | – | – | – | – | 468 |
| Rat | 32 | 2070 | 86 | 1069 | 128 | 205 | 225 | – | – | – | – | 471 |
| Sheep | 32 | 1332 | 86 | 974 | 128 | 174 | 225 | – | – | – | – | 471 |
| Tarsier | 29 | 1348 | 86 | 1774 | 128 | 218 | 225 | – | – | – | – | 468 |
| Tasmanian
devil | 88 | 1432 | 25 | 765 | 5 | 38 | 128 | 293 | 216 | – | – | 462 |
| Tree Shrew | 29 | 1509 | 86 | 1084 | 128 | 237 | 225 | – | – | – | – | 468 |
| Vervet African
green monkey | 29 | 1392 | 86 | 1201 | 128 | 201 | 225 | – | – | – | – | 468 |
Expression profile of the human IL-36RN
gene
A search of the EST sequence database revealed that
the human IL-36RN gene was expressed in the placenta, cervix, lung,
head and neck, eye, fetal heart and testis, and furthermore, that
it was highly expressed in bladder and parathyroid tumors.
Examination of microarray analyses and 'virtual northern blot
analysis' revealed a predominant expression of IL-36RN in cervix,
larynx, lung, mouth, muscle, parathyroid, pharynx, placenta and
testis. A search of the PrognoScan database revealed that human
IL-36RN was also expressed in bladder, blood, brain, breast,
colorectal, esophageal, eye, head and neck, lung, ovarian, skin and
soft tissue cancer.
Comparative genomics analysis of human
IL-36RN
Activator protein 1 (AP-1), c-Fos, c-Jun and nuclear
factor (NF)-κB binding sites were identified within the upstream
regions of the transcriptional start site of human IL-36RN.
Functionally relevant SNP evaluation of
the human IL-36RN gene and identification of somatic mutations in
human cancer
A total of 543 SNPs were identified in the human
IL-36RN gene through searching the NCBI SNP and Ensembl databases.
Among these SNPs, 30 were functionally relevant, causing missense
and nonsense mutations (Table
II). As presented in Table
III, by searching the COSMIC database, 31 somatic mutations of
the IL-36RN gene were identified in cancer.
 | Table IIEvaluation of the functionally
relevant SNP in the human IL-36RN gene. |
Table II
Evaluation of the functionally
relevant SNP in the human IL-36RN gene.
| SNP ID | Chr 2 position
sequence | Sequence | Type | Amino acid
change |
|---|
| rs143724424 | 113820120 |
GCTTC[A/G]AGTCG | Missense | EK |
| rs144478519 | 113820124 |
CGAGT[C/T]GGCTG | Missense | SL |
| rs151325121 | 113819727 |
CCAAT[C/T]GGTGG | Missense | RW |
| rs387906914 | 113818479 |
GCTTC[C/T]AGCTG | Missense | LP |
| rs397514629 | 113820154 |
GTGCA[C/G]GGTGC | Missense | TR |
| rs28938777 | 113819725 |
CCCCA[A/G]TCGGT | Missense | NS |
| rs77864207 | 113819754 |
CCCCC[A/G]TCATC | Missense | VI |
| rs139497891 | 113819812 |
GGAGC[C/T]GACTC | Missense | PL |
| rs141341649 | 113820136 |
CTACC[C/T]GGGCT | Missense | PL |
| rs144182857 | 113820031 |
GCAGC[C/T]AGTGA | Missense | PL |
| rs144420774 | 113820103 |
CATGG[C/G]GCTCA | Missense | GA |
| rs145099228 | 113819721 |
TGGTC[C/T]CCAAT | Missense | PS |
| rs147389610 | 113818487 |
CTGGA[A/G]GGCTG | Missense | GR |
| rs147410197 | 113820087 |
CCTTC[C/T]ACCGG | Missense | YH |
| rs187015338 | 113818503 |
AGGGA[A/G]GGTCA | Missense | KR |
| rs199932303 | 113820090 |
TCTAC[C/T]GGCGG | Missense | RW |
| rs202059991 | 113820222 |
CCCCC[A/G]TCACA | Missense | IV |
| rs369259981 | 113820048 |
TGGAG[C/T]TCTAT | Missense | LF |
| rs371819085 | 113820091 |
CTACC[A/G]GCGGG | Missense | RQ |
| rs372880215 | 113819815 |
GCCGA[C/T]TCTAA | Missense | TI |
| rs374900764 | 113820247 |
GCAGT[A/G]TGACT | Missense | CY |
| rs375207169 | 113820093 |
ACCGG[C/T]GGGAC | Missense | RW |
| rs375718709 | 113819793 |
TGTCA[C/T]GTGGG | Missense | CR |
| rs377330697 | 113820172 |
CGATC[A/G]GCCTG | Missense | QR |
| rs537559199 | 113820044 |
ATCAT[A/G]GAGCT | Missense | MI |
| rs542606182 | 113820094 |
CCGGC[A/G]GGACA | Missense | RQ |
| rs545202535 | 113820237 |
TCTAC[A/T]TCCAG | Missense | FI |
| rs545673991 | 113818451 |
TGAAG[G/T]TGCTT | Missense | VL |
| rs397514630 | 113817043 |
GCTTC[C/T]GGTGA | Nonsense | R-Ter |
| rs368461730 | 113819805 |
TGGGG[C/T]AGGAG | Nonsense | Q-Ter |
 | Table IIISomatic mutations of IL-36RN in tumor
tissues. |
Table III
Somatic mutations of IL-36RN in tumor
tissues.
| Position (AA) | Mutation (CDS) | Mutation (amino
acid) | Mutation ID
(COSM) | Count | Mutation type |
|---|
| 3 | c.9G>C | p.L3L | COSM3836628 | 1 | Substitution-coding
silent |
| 5 | c.15G>A | p.G5G | COSM3894558 | 1 | Substitution-coding
silent |
| 6 | c.17C>T | p.A6V | COSM240220 | 1 |
Substitution-missense |
| 10 | c.28C>T |
p.R10* | COSM126741 | 1 |
Substitution-nonsense |
| 14 | c.41C>T | p.S14L | COSM714706 | 1 |
Substitution-missense |
| 15 | c.44C>A | p.A15E | COSM714705 | 1 |
Substitution-missense |
| 21 | c.63G>T | p.L21L | COSM381474 | 1 | Substitution-coding
silent |
| 29 | c.85G>A | p.G29R | COSM1690946 | 1 |
Substitution-missense |
| 34 | c.102G>A | p.G34G | COSM3894559 | 1 | Substitution-coding
silent |
| 36 | c.108C>A | p.V36V | COSM169172 | 1 | Substitution-coding
silent |
| 37 | c.110T>C | p.I37T | COSM4084297 | 1 |
Substitution-missense |
| 46 | c.137C>T | p.P46L | COSM1690947 | 1 |
Substitution-missense |
| 48 | c.142C>T | p.R48W | COSM441016 | 1 |
Substitution-missense |
| 54 | c.160C>A | p.L54M | COSM300070 | 1 |
Substitution-missense |
| 54 | c.160C>T | p.L54L | COSM3565457 | 1 | Substitution-coding
silent |
| 55 | c.164C>T | p.S55F | COSM1690948 | 1 |
Substitution-missense |
| 56 | c.168C>A | p.P56P | COSM3565458 | 1 | Substitution-coding
silent |
| 71 | c.212G>A | p.G71E | COSM3565459 | 1 |
Substitution-missense |
| 73 | c.218G>C | p.G73A | COSM1527707 | 1 |
Substitution-missense |
| 86 | c.258G>T | p.M86I | COSM3565460 | 1 |
Substitution-missense |
| 92 | c.275C>A | p.A92D | COSM4133012 | 1 |
Substitution-missense |
| 95 | c.284C>T | p.S95F | COSM3565461 | 1 |
Substitution-missense |
| 97 | c.290G>A | p.S97N | COSM3565462 | 1 |
Substitution-missense |
| 106 | c.317G>A | p.G106E | COSM3565463 | 1 |
Substitution-missense |
| 112 | c.334G>A | p.E112K | COSM107437 | 1 |
Substitution-missense |
| 117 | c.350C>A | p.P117Q | COSM3961011 | 1 |
Substitution-missense |
| 126 | c.378A>G | p.E126E | COSM4084298 | 1 | Substitution-coding
silent |
| 136 | c.406C>A | p.L136I | COSM4084299 | 1 |
Substitution-missense |
| 137 | c.411C>T | p.P137P | COSM3565464 | 1 | Substitution-coding
silent |
| 138 | c.412G>A | p.E138K | COSM275559 | 2 |
Substitution-missense |
| 142 | c.425G>T | p.W142L | COSM336664 | 1 |
Substitution-missense |
Meta-analysis of the prognostic value of
IL-36RN gene in cancer
PrognoScan employs the minimum P-value approach for
grouping patients with varied cancer types for survival analysis
and produces a data-set of results, including cancer type, subtype,
endpoint, cohort, contributor, array type, probe ID, number of
patients, optimal cut-off point, Pmin and Pcor. For the IL-36RN
gene, 7 out of the 84 cancer cases showed correlations between
microarray expression in the IL-36RN gene and cancer prognosis
(bladder cancer, 1/2; blood cancer, 0/9; brain cancer, 0/4; breast
cancer, 1/30; colorectal cancer, 1/9; esophageal cancer, 0/1; eye
cancer, 0/1; head and neck cancer, 0/1; lung cancer, 2/15; ovarian
cancer, 2/9; skin cancer, 0/1; soft tissue cancer, 0/1) with a 5%
significance level (Table IV).
Among the two ovarian cancer cases, poor survival in one case was
associated with elevated expression of IL-36RN (DUKE-OC), and the
other one was associated with decreased expression of IL-36RN
(GSE17260). While one case out of nine cases of colorectal cancer
showed poor survival associated with decreased expression of
IL-36RN, elevated expression of IL-36RN in one case of bladder
cancer, one case of breast cancer and two cases of lung cancer was
found to be associated with poor survival.
 | Table IVDataset contents from PrognoScan
showing an association between microarray expression of IL-36RN and
cancer prognosis. |
Table IV
Dataset contents from PrognoScan
showing an association between microarray expression of IL-36RN and
cancer prognosis.
| Database | Cancer type | Patients (n) | Endpoint | Cut-off point | P-value | Prognosis | Reference |
|---|
| GSE13507 | Bladder cancer | 165 | Overall
survival | 0.87 | 0.046 | 2 | (32) |
| GSE12276 | Breast cancer | 204 | Relapse-free
survival | 0.46 | 0.042 | 2 | (33) |
| GSE17536 | Colorectal
cancer | 177 | Overall
survival | 0.21 | 0.033 | 1 | (34) |
| GSE31210 | Lung cancera | 204 | Overall
survival | 0.84 | <0.001 | 2 | (35) |
| GSE31210 | Lung cancera | 204 | Relapse-free
survival | 0.89 | 0.002 | 2 | (35) |
| DUKE-OC | Ovarian cancer | 133 | Overall
survival | 0.44 | 0.031 | 2 | |
| GSE17260 | Ovarian cancer | 110 | Overall
survival | 0.12 | 0.009 | 1 | (36) |
Discussion
The IL-36RN gene encodes the anti-inflammatory
cytokine IL-36Ra, which was previously known as IL-1F5 and later
re-defined as a member of the IL-36 cytokine family.
The present study identified IL-36RN from 27 genomes
and found that IL-36RN exists in all types of mammals, including
primates, rodents and carnivora, as well as elephant, dolphin,
sheep, rabbit, horse and armadillo. In the phylogenetic tree, all
of the primates were clustered. Furthermore, the exon-intron
information indicated that all primates were almost identical with
regard to the IL-36RN gene. According to the alignment and
phylogenetic tree, IL-36RN was evolutionarily conserved among
mammals, indicating a significant biological function of this gene.
It is known that IL-36 cytokines are expressed in various tissue
types and contribute to inflammatory diseases (7), confirming its biological importance
indicated by the present study.
EST sequence analysis revealed that the IL-36RN gene
is expressed in the placenta, cervix, lung, head and neck, eye,
fetal heart and testis; furthermore, high expression had been
detected in bladder and parathyroid tumors. This result implied
that IL-36RN is extensively expressed in a large variety of organ
and tissue types. A total of 30 SNPs, including 28 SNPs causing
missense mutations and 2 SNPs causing nonsense mutations, were
analyzed from 543 available SNPs in human IL-36RN genes. Recently,
several IL-36RN mutations among the 28 SNPs have been reported as
causative genetic defects associated with GPP and related pustular
disorders (18–20,31),
which indicates that changes in IL-36RN SNPs truly contribute to
physiological and pathological functions of IL-36Ra. However,
another reported IL-36RN mutation in the intron region,
rs148755083, which causes GPP (31), was not included in the present
study; therefore, further investigation is required to reveal the
effects of the other SNPs on the links between IL-36RN and
diseases.
In the present study, assessment of the prognostic
value of IL-36RN in cancer using the PrognoScan database revealed
that IL-36RN is expressed in various cancer types including bladder
(32), breast (33), colorectal (34), lung (35) and ovarian cancer (36). In 7 out of 84 cancer cases, IL-36RN
was identified as a promising prognostic factor. Furthermore,
IL-36RN expression varied among different types of cancer and the
prognostic value varied within entries of different databases for
the same cancer type. These results suggested that IL-36RN may have
multiple roles in cancer development. In addition, 31 somatic
mutations of IL-36RN in cancer tissues were identified in the
present study. Thus, additional study is required to confirm the
preliminary findings of the present study, which indicated that
IL-36RN takes part in cancer development, and to assess the
underlying mechanisms.
The IL-36RN gene was identified to bind with the
AP-1, c-Fos, c-Jun, and NF-κB regulatory transcription factors in
the upstream (promoter) region. Transcription factor AP-1 regulates
a broad range of genes involved in cell cycle and inflammation. It
mediates the anti-apoptotic response to hypoxic conditions and
contributes to resistance to chemo- and radiotherapy in colon
cancer cells (37), while it
influences pivotal regulators of cell proliferation, migration and
survival involved in melanoma progression (38) as well as in the carcinogenesis of
the respiratory epithelium (39).
c-Fos has been found to be associated with lipid- and phospholipid
synthesis in several cell types (40) and activates biogenesis in certain
types of tumor cell to support tumor growth (41,42).
c-Jun is a critical transcription factor involved in major
cell-biological activities, including cell proliferation,
apoptosis, angiogenesis and invasiveness by specific regulation of
epidermal growth factor receptor, keratinocyte growth factor,
cyclin D1, p53, proliferin and CD44 (43–46).
NF-κB is known to be the key regulator of apoptosis and controlled
cell suicide by means of controlling pro-apoptotic and
anti-apoptotic genes (47–50). NF-κB exacerbates
inflammation-induced cancer types, while it suppresses chemically
induced skin and liver cancers (51–53),
which suggests that NF-κB has a dual role in cancer. These
transcription factors associated with tumorigenesis may represent a
link between IL-36RN and tumorigenesis or cancer progression.
In conclusion, the present study investigated
IL-36RN in various species and types of cancer at the gene and
protein levels, and the results demonstrated that IL-36RN may have
an important role in cancer progression through tumor-associated
transcription factors and signaling pathways, but this hypothesis
requires further investigation.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (no. 81072400), the Research
Fund for the Doctoral Program of Higher Education of China (no.
20130142110066), the Scientific Research Foundation of Hubei Health
Department (no. JX5B54), the Natural Science Foundation of Hubei
province (no. 2009CDB148), the Wuhan Planning Project of Science
and Technology (no. 201161038340-01) and the Independent Innovation
Research Foundation of Huazhong University of Science and
Technology (no. 2011JC016).
References
|
1
|
Sims JE, Nicklin MJ, Bazan JF, Barton JL,
Busfield SJ, Ford JE, Kastelein RA, Kumar S, Lin H, Mulero JJ, et
al: A new nomenclature for IL-1-family genes. Trends Immunol.
22:536–537. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dinarello C, Arend W, Sims J, Smith D,
Blumberg H, O'Neill L, Goldbach-Mansky R, Pizarro T, Hoffman H,
Bufler P, et al: IL-1 family nomenclature. Nat Immunol. 11:9732010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharaf N, Nicklin MJ and di Giovine FS:
Long-range DNA interactions at the IL-1/IL-36/IL-37 gene cluster
(2q13) are induced by activation of monocytes. Cytokine. 68:16–22.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mulero JJ, Pace AM, Nelken ST, Loeb DB,
Correa TR, Drmanac R and Ford JE: IL1HY1: A novel interleukin-1
receptor antagonist gene. Biochem Biophys Res Commun. 263:702–706.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Towne JE, Renshaw BR, Douangpanya J,
Lipsky BP, Shen M, Gabel CA and Sims JE: Interleukin-36 (IL-36)
ligands require processing for full agonist (IL-36α, IL-36β and
IL-36γ) or antagonist (IL-36Ra) activity. J Biol Chem.
286:42594–42602. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costelloe C, Watson M, Murphy A, McQuillan
K, Loscher C, Armstrong ME, Garlanda C, Mantovani A, O'Neill LA,
Mills KH and Lynch MA: IL-1F5 mediates anti-inflammatory activity
in the brain through induction of IL-4 following interaction with
SIGIRR/TIR8. J Neurochem. 105:1960–1969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gresnigt MS and van de Veerdonk FL:
Biology of IL-36 cytokines and their role in disease. Semin
Immunol. 25:458–465. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen H, Wang Y, Bai C and Wang X:
Alterations of plasma inflammatory biomarkers in the healthy and
chronic obstructive pulmonary disease patients with or without
acute exacerbation. J Proteomics. 75:2835–2843. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ramadas RA, Li X, Shubitowski DM, Samineni
S, Wills-Karp M and Ewart SL: IL-1 Receptor antagonist as a
positional candidate gene in a murine model of allergic asthma.
Immunogenetics. 58:851–855. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Asseldonk EJ, Stienstra R, Koenen TB,
van Tits LJ, Joosten LA, Tack CJ and Netea MG: The effect of the
interleukin-1 cytokine family members IL-1F6 and IL-1F8 on
adipocyte differentiation. Obesity (Silver Spring). 18:2234–2236.
2010. View Article : Google Scholar
|
|
11
|
Kim TJ, Kim TH, Lee HJ, Peddle L, Rahman
P, Hu P, Greenwood CM and Inman RD: Interleukin 1 polymorphisms in
patients with ankylosing spondylitis in Korea. J Rheumatol.
35:1603–1608. 2008.PubMed/NCBI
|
|
12
|
Frey S, Derer A, Messbacher ME, Baeten DL,
Bugatti S, Montecucco C, Schett G and Hueber AJ: The novel cytokine
interleukin-36α is expressed in psoriatic and rheumatoid arthritis
synovium. Ann Rheum Dis. 72:1569–1574. 2013. View Article : Google Scholar
|
|
13
|
Mattii M, Ayala F, Balato N, Filotico R,
Lembo S, Schiattarella M, Patruno C, Marone G and Balato A: The
balance between pro-and anti-inflammatory cytokines is crucial in
human allergic contact dermatitis pathogenesis: The role of IL-1
family members. Exp Dermatol. 22:813–819. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heinemann A, He Y, Zimina E, Boerries M,
Busch H, Chmel N, Kurz T, Bruckner-Tuderman L and Has C: Induction
of phenotype modifying cytokines by FERMT1 mutations. Hum Mutat.
32:397–406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karumbaiah L, Norman SE, Rajan NB, Anand
S, Saxena T, Betancur M, Patkar R and Bellamkonda RV: The
upregulation of specific interleukin (IL) receptor antagonists and
paradoxical enhancement of neuronal apoptosis due to electrode
induced strain and brain micromotion. Biomaterials. 33:5983–5996.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Towne JE and Sims JE: IL-36 in psoriasis.
Curr Opin Pharmacol. 12:486–490. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marrakchi S, Guigue P, Renshaw BR, Puel A,
Pei XY, Fraitag S, Zribi J, Bal E, Cluzeau C, Chrabieh M, et al:
Interleukin-36-receptor antagonist deficiency and generalized
pustular psoriasis. N Engl J Med. 365:620–628. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sugiura K, Takeichi T, Kono M, Ogawa Y,
Shimoyama Y, Muro Y and Akiyama M: A novel IL36RN/IL1F5 homozygous
nonsense mutation, p.Arg10X, in a Japanese patient with adult-onset
generalized pustular psoriasis. Br J Dermatol. 167:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Onoufriadis A, Simpson MA, Pink AE, Di
Meglio P, Smith CH, Pullabhatla V, Knight J, Spain SL, Nestle FO,
Burden AD, et al: Mutations in IL36RN/IL1F5 are associated with the
severe episodic inflammatory skin disease known as generalized
pustular psoriasis. Am J Hum Genet. 89:432–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanazawa N, Nakamura T, Mikita N and
Furukawa F: Novel IL36RN mutation in a Japanese case of early onset
generalized pustular psoriasis. J Dermatol. 40:749–751. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar S, Nei M, Dudley J and Tamura K:
MEGA: A biologist-centric software for evolutionary analysis of DNA
and protein sequences. Brief Bioinform. 9:299–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang Z, Nielsen R, Goldman N and Pedersen
AM: Codon-substitution models for heterogeneous selection pressure
at amino acid sites. Genetics. 155:431–449. 2000.PubMed/NCBI
|
|
23
|
Yang Z: PAML 4: Phylogenetic analysis by
maximum likelihood. Mol Biol Evol. 24:1586–1591. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang B, Chen K, Xu W, Chen D, Tang W and
Xia TS: Integrative genomic analyses of secreted protein acidic and
rich in cysteine and its role in cancer prediction. Mol Med Rep.
10:1461–1468. 2014.PubMed/NCBI
|
|
25
|
Kolker E, Higdon R, Morgan P, Sedensky M,
Welch D, Bauman A, Stewart E, Haynes W, Broomall W and Kolker N:
SPIRE: Systematic protein investigative research environment. J
Proteomics. 75:122–126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kolker E, Higdon R, Haynes W, Welch D,
Broomall W, Lancet D, Stanberry L and Kolker N: MOPED: Model
organism protein expression database. Nucleic Acids Res.
40:D1093–D1099. 2012. View Article : Google Scholar :
|
|
27
|
Wang B, Xu W, Tan M, Xiao Y, Yang H and
Xia TS: Integrative genomic analyses of a novel cytokine,
interleukin-34 and its potential role in cancer prediction. Int J
Mol Med. 35:92–102. 2015.
|
|
28
|
Wang M, Wei X, Shi L, Chen B, Zhao G and
Yang H: Integrative genomic analyses of the histamine H1 receptor
and its role in cancer prediction. Int J Mol Med. 33:1019–1026.
2014.PubMed/NCBI
|
|
29
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the catalogue of somatic
mutations in cancer. Nucleic Acids Res. 39:D945–D950. 2011.
View Article : Google Scholar :
|
|
30
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sugiura K, Takemoto A, Yamaguchi M,
Takahashi H, Shoda Y, Mitsuma T, Tsuda K, Nishida E, Togawa Y,
Nakajima K, et al: The majority of generalized pustular psoriasis
without psoriasis vulgaris is caused by deficiency of
interleukin-36 receptor antagonist. J Invest Dermatol.
133:2514–2521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9:32010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bos PD, Zhang XH, Nadal C, Shu W, Gomis
RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA and
Massagué J: Genes that mediate breast cancer metastasis to the
brain. Nature. 459:1005–1009. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar
|
|
35
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar
|
|
36
|
Yoshihara K, Tajima A, Yahata T, Kodama S,
Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H, et
al: Gene expression profile for predicting survival in
advanced-stage serous ovarian cancer across two independent
datasets. PLoS One. 5:e96152010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kappelmann M, Bosserhoff A and Kuphal S:
AP-1/c-Jun transcription factors: Regulation and function in
malignant melanoma. Eur J Cell Biol. 93:76–81. 2014. View Article : Google Scholar
|
|
39
|
Karamouzis MV, Konstantinopoulos PA and
Papavassiliou AG: The activator protein-1 transcription factor in
respiratory epithelium carcinogenesis. Mol Cancer Res. 5:109–120.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Caputto BL, Cardozo Gizzi AM and Gil GA:
C-Fos: an AP-1 transcription factor with an additional cytoplasmic,
non-genomic lipid synthesis activation capacity. Biochim Biophys
Acta. 1841:1241–1246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Milde-Langosch K: The Fos family of
transcription factors and their role in tumourigenesis. Eur J
Cancer. 41:2449–2461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Motrich RD, Castro GM and Caputto BL: Old
players with a newly defined function: Fra-1 and c-Fos support
growth of human malignant breast tumors by activating membrane
biogenesis at the cytoplasm. PLoS One. 8:e532112013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bakiri L, Lallemand D, Bossy-Wetzel E and
Yaniv M: Cell cycle-dependent variations in c-Jun and JunB
phosphorylation: A role in the control of cyclin D1 expression.
EMBO J. 19:2056–2068. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zenz R, Scheuch H, Martin P, Frank C,
Eferl R, Kenner L, Sibilia M and Wagner EF: C-Jun regulates eyelid
closure and skin tumor development through EGFR signaling. Dev
Cell. 4:879–889. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mils V, Piette J, Barette C, Veyrune J,
Tesnière A, Escot C, Guilhou JJ and Basset-Séguin N: The
proto-oncogene c-fos increases the sensitivity of keratinocytes to
apoptosis. Oncogene. 14:1555–1561. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schreiber M, Kolbus A, Piu F, Szabowski
kA, Möhle-Steinlein U, Tian J, Karin M, Angel P and Wagner EF:
Control of cell cycle progression by c-Jun is p53 dependent. Genes
Dev. 13:607–619. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen F and Castranova V: Nuclear
factor-kappaB, an unappreciated tumor suppressor. Cancer Res.
67:11093–11098. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Horst D, Budczies J, Brabletz T, Kirchner
T and Hlubek F: Invasion associated up-regulation of nuclear factor
kappaB target genes in colorectal cancer. Cancer. 115:4946–4958.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zubair A and Frieri M: Role of nuclear
factor-kB in breast and colorectal cancer. Curr Allergy Asthma Rep.
13:44–49. 2013. View Article : Google Scholar
|
|
50
|
Yu LL, Yu HG, Yu JP, Luo HS, Xu XM and Li
JH: Nuclear factor-kappaB p65 (RelA) transcription factor is
constitutively activated in human colorectal carcinoma tissue.
World J Gastroenterol. 10:3255–3260. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vainer GW, Pikarsky E and Ben-Neriah Y:
Contradictory functions of NF-kappaB in liver physiology and
cancer. Cancer Lett. 267:182–188. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Arsura M and Cavin LG: Nuclear
factor-kappaB and liver carcinogenesis. Cancer Lett. 229:157–169.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Karin M: Nuclear factor-kappaB in cancer
development and progression. Nature. 441:431–436. 2006. View Article : Google Scholar : PubMed/NCBI
|