Introduction
Myocardial infarction, which is predominantly caused
by coronary heart disease, and by coronary thrombosis in
particularly, is a major contributor to mortality rates globally
(1). With developments in research
into myocardial infarction, the biological-psychological-social
medicine nexus has provided a more detailed understanding of the
risks of myocardial infarction. Previous studies have reported that
long non-coding RNAs (lncRNAs) are involved in cardiac development
(2,3). The lncRNAs comprise a group of
non-coding RNAs, in addition to microRNA, PIWI-interacting RNAand
endogenous small interfering RNA, and are defined as transcripts
>200 nt in length with no known protein-coding function
(4). They have a large range of
functions, including in cell proliferation, apoptosis and cell
invasion (5,6). It has been shown that lncRNAs are
associated with cardiac hypertrophy (7), cardiovascular ageing (8) and cardiac tissues following
myocardial infarction (9).
Although there is no direct evidence between lncRNAs and myocardial
infarction, certain lncRNAs are associated with the risks of
myocardial infarction. For example, GAS5 functions as a starvation-
or growth arrest-linked riborepressor (10), and this condition is similar to
myocardial infarction. In addition, lncRNA-p21 and lncRNA PANDA are
induced by DNA damage in a p53-dependent manner (11,12)
which also occurs in the cardiomyocyte death that is associated
with myocardial infarction.
MicroRNAs have been well demonstrated in the
development of cardiovascular diseases (13), however, there are few reports on
lncRNAs in myocardial infarction (14). RNA sequencing (RNA-seq) is a
prevalent technique used to profile lncRNAs, however, the publicly
available RNA-seq data are limited due to relatively high costs of
the RNA-seq technique. In addition, RNA-seq data are lacking in
sample numbers, compared with microarray expression profile data,
which often contained dozens to hundreds of pair-matched samples
(15). Therefore, the present
study adopted a re-annotation method to identify lncRNAs associated
with myocardial infarction. Furthermore, increasing evidence shows
that lncRNAs may be important in regulating gene expression, and
that the functions of lncRNAs are performed predominantly by their
secondary structures, which is difficult to decipher (15). Due to the considerable challenges
in investigating the functions of lncRNA, the present study used a
co-expression-based method, in which lncRNA functions were
predicted, based on the functions of their co-expressed
protein-coding genes (15).
Therefore, the present study aimed to identify the
lncRNAs involved in myocardial infarction. lncRNA functions can be
predicted based on the functions of their co-expressed protein
coding genes, and alterations in the associations between these
genes between the different samples (normal or myocardial
infarction) can be used to identify key lncRNAs in myocardial
infarction. By re-annotating an affymetrix microarray associated
with myocardial infarction, a myocardial infarction-related
differential lncRNA-mRNA co-expression network (MILMN) was
constructed in the current study, then pathway enrichment analysis
was conducted. The present study aimed to identify potential
non-coding RNA biomarkers, in addition to providing further insight
into the understanding of the molecular mechanism of lncRNAs.
Materials and methods
Microarray data
The microarray data set, GSE48060, was downloaded
from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48060).
This dataset applied the methodology on the blood samples from 21
healthy control individuals and 31 patients with myocardial
infarction using an Affymetrix HG-U133 Plus 2.0 Microarray
(16). The whole-genome microarray
profiling was performed on blood samples from control individuals
with normal cardiac function and from patients with first-time
acute myocardial infarction, within 48 h following myocardial
infarction (16).
Functional re-annotation of lncRNAs
To re-annotate the microarray data obtained, a
non-coding RNA function annotation server (ncFANs) was used to
re-annotate the probes on a HG-U133 Plus 2.0 array, following the
steps on its website (17). A
total of 2,495 lncRNAs were re-annotated, and each lncRNA and mRNA
probe was converted into gene Ensembl Gene IDs (http://www.ensembl.org/index.html). If one gene
matched more than one probe, the expression value of this mRNA or
lncRNA was computed by determining the average expression value of
all its corresponding probes.
Construction of the MILMN
Following re-annotation of the microarray data, the
expression values of the lncRNAs and mRNAs were obtained.
Subsequently, Pearson's correlation coefficient (PCC) was
calculated between the expression values of each of the lncRNA-mRNA
pairs across the normal samples and the myocardial infarction
samples, respectively (Fig. 1).
The lncRNA-mRNA pairs with a PCC >0.85 in one sample group, but
<0.5 in the other sample group were selected (Fig. 1), as these parameters indicated
that the lncRNA-mRNA pairs were differentially co-expressed in the
two sample groups. Finally, the MILMN was constructed, in which
nodes were lncRNAs or mRNAs, and were connected if they were
differentially co-expressed (Fig.
2). The top 20 mRNA nodes, which were those with the highest
degree, were determined (Table
I).
 | Table ITop 20 mRNAs with the highest degree
in the myocardial infarction-related differential long non-coding
RNA-mRNA co-expression network. |
Table I
Top 20 mRNAs with the highest degree
in the myocardial infarction-related differential long non-coding
RNA-mRNA co-expression network.
| Symbol | Degree | Gene name |
|---|
| Cilp2 | 49 | Cartilage
intermediate layer protein 2 |
| ANGPTL2 | 46 | Angiopoietin-like
2 |
| Ptgis | 45 | Prostaglandin I2
(prostacyclin) synthase |
| CORO6 | 40 | Coronin 6 |
| CLDN10 | 39 | Claudin 10 |
| piwil2 | 39 | Hydroxy-δ-5-steroid
dehydrogenase, 3 β- and steroid δ-isomerase 7; piwi-like 2
(Drosophila) |
| mfap4 | 37 |
Microfibrillar-associated protein 4 |
| Hus1b | 35 | HUS1 checkpoint
homolog b (S. pombe) |
| PCGF1 | 34 | Polycomb group ring
finger 1 |
| Slc22a9 | 33 | Solute carrier
family 22 (organic anion transporter), member 9 |
| TROAP | 32 | Trophinin
associated protein (tastin) |
| zar1 | 30 | Zygote arrest
1 |
| CLDN19 | 30 | Claudin 19 |
| lmf2 | 30 | Lipase maturation
factor 2 |
| ppyr1 | 28 | Pancreatic
polypeptide receptor 1 |
| ephX1 | 28 | Epoxide hydrolase
1, microsomal (xenobiotic) |
| Fkbp1b | 28 | FK506 binding
protein 1B, 12.6 kDa |
| SLC5A4 | 27 | Solute carrier
family 5 (low affinity glucose cotransporter), member 4 |
| RNF207 | 25 | Ring finger protein
207 |
| HSPB8 | 25 | Heat shock 22 kDa
protein 8 |
Identification of key lncRNAs in
myocardial infarction
To identify the key lncRNAs in myocardial
infarction, the present study initially implemented pathway
enrichment of the mRNAs in the MILMN, using the Database for
Annotation, Visualization and Integrated Discovery 6.7 (18). P<0.05 was considered to indicate
a statistically significant difference. Subsequently an
lncRNA-pathway network was constructed, in which nodes represented
lncRNAs or pathways, and they were connected if the corresponding
co-expressed mRNAs of an lncRNA were enriched in the corresponding
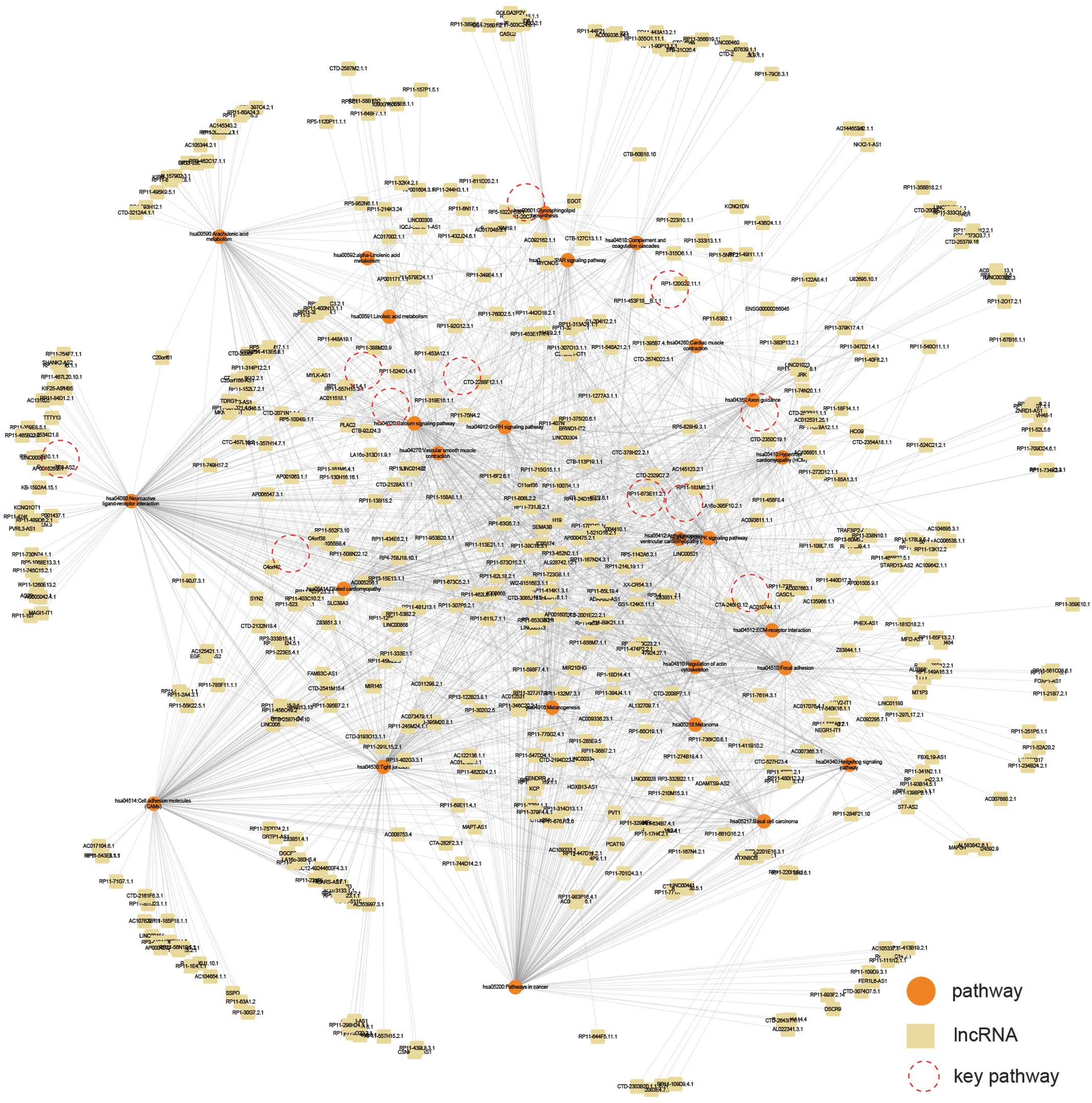
pathway (Fig. 3), suggesting that
these pathways were potentially regulated by the corresponding
lncRNAs. From these pathways, crucial pathways in myocardial
infarction were selected by a literature search using the following
criteria: (i) cardiovascular disease pathway; (ii) important
signaling pathway; (iii) cardiovascular muscle-related.
Finally, the lncRNAs which were linked with at least
six of the 11 crucial pathways in the lncRNA-pathway network
(Fig. 3) were considered to be key
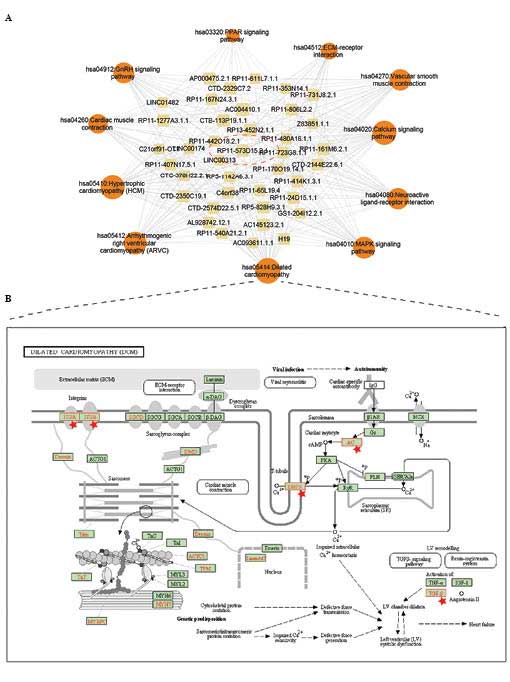
regulating lncRNAs in myocardial infarction. The key lncRNAs and
corresponding crucial pathways are shown in Fig. 4A and in Tables II and III. To better illustrate the
potentially regulated process, the co-expressing mRNAs were
annotated into a dilated cardiomyopathy pathway and a
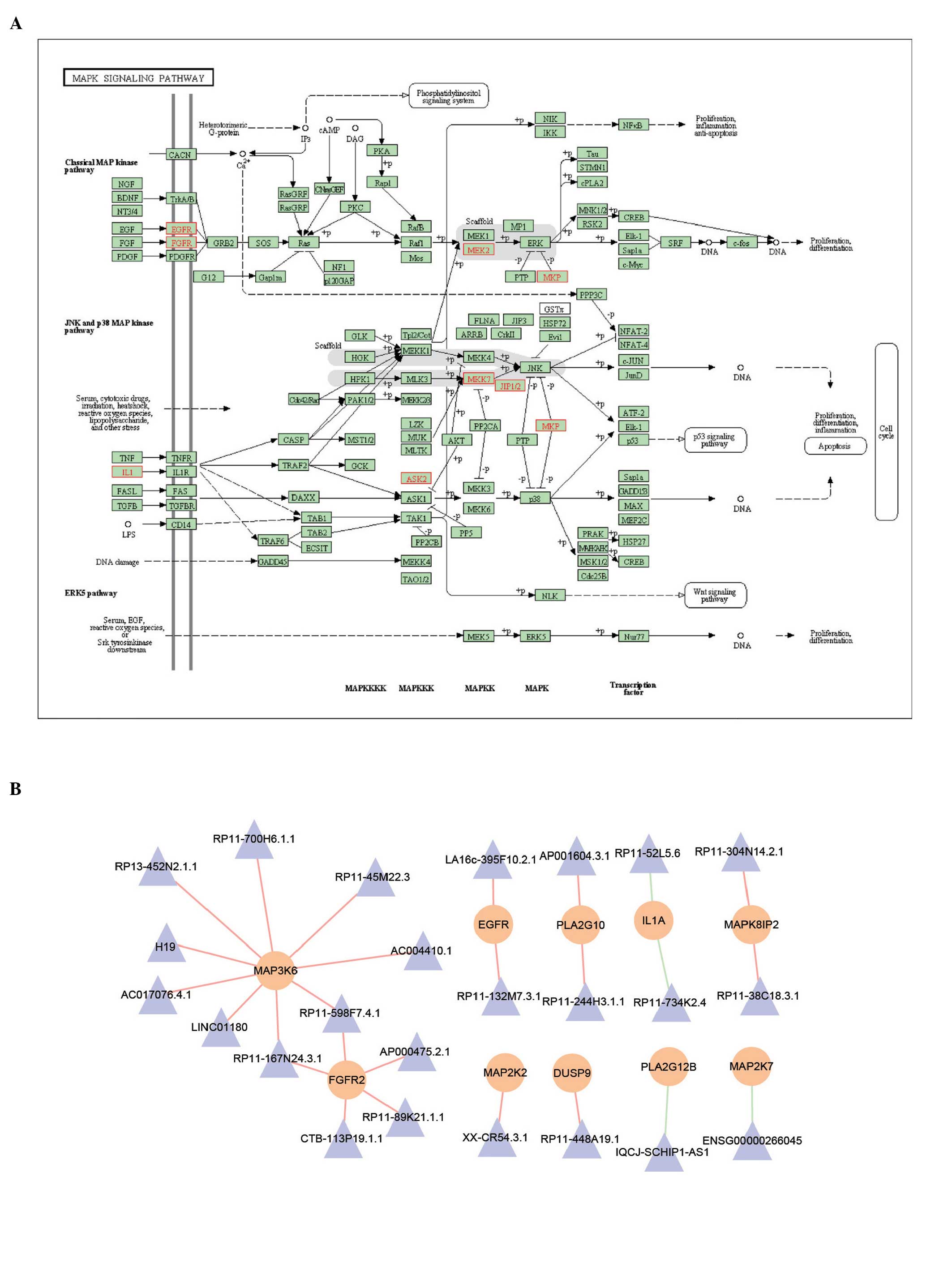
mitogen-activated protein kinase (MAPK) signaling pathway (Figs 4B and 5). Pathway annotation was used performed
using the search and color tools of the Kyoto Encyclopedia of Genes
and Genomes (KEGG) database (http://www.kegg.jp/kegg/tool/map_pathway2.html).
 | Table IIKey pathways in myocardial
infarction. |
Table II
Key pathways in myocardial
infarction.
| Pathway | Regulatory lncRNAs
(n) | PMID |
|---|
| hsa04010: MAPK
signaling pathway | 38 | 25154304 |
| hsa04270: Vascular
smooth muscle contraction | 35 | |
| hsa04260: Cardiac
muscle contraction | 35 | |
| hsa03320: PPAR
signaling pathway | 32 | 10073956 |
| hsa04512:
ECM-receptor interaction | 30 | 19747544 |
| hsa05412:
Arrhythmogenic right ventricular cardiomyopathy | 29 | |
| hsa04080:
Neuroactive ligand-receptor interaction | 29 | 23916832 |
| hsa04020: Calcium
signaling pathway | 28 | 24548334 |
| hsa05410:
Hypertrophic cardiomyopathy | 25 | |
| hsa05414: Dilated
cardiomyopathy | 19 | |
| hsa04912: GnRH
signaling pathway | 12 | 19554077 |
 | Table IIIKey lncRNAs in myocardial
infarction. |
Table III
Key lncRNAs in myocardial
infarction.
| lncRNA | Regulatory pathways
(n) | Function |
|---|
| AC004410.1 | 11 |
Uncharacteristic |
| RP13-452N2.1.1 | 10 |
Uncharacteristic |
| CTB-113P19.1.1 | 10 |
Uncharacteristic |
|
RP11-1277A3.1.1 | 9 |
Uncharacteristic |
| RP11-731J8.2.1 | 9 |
Uncharacteristic |
| RP5-828H9.3.1 | 9 |
Uncharacteristic |
| CTD-2350C19.1 | 9 |
Uncharacteristic |
| RP11-161M6.2.1 | 9 |
Uncharacteristic |
|
CTD-2144E22.6.1 | 9 |
Uncharacteristic |
| RP11-806L2.2 | 9 |
Uncharacteristic |
|
RP11-573D15.2.1 | 9 |
Uncharacteristic |
| RP11-414K1.3.1 | 9 |
Uncharacteristic |
| AP000475.2.1 | 9 |
Uncharacteristic |
| RP11-24D15.1.1 | 9 |
Uncharacteristic |
| C4orf38 | 9 |
Uncharacteristic |
| CTD-2329C7.2 | 9 |
Uncharacteristic |
| AC145123.2.1 | 9 |
Uncharacteristic |
| RP5-1142A6.3.1 | 8 |
Uncharacteristic |
|
RP11-540A21.2.1 | 8 |
Uncharacteristic |
| H19 | 8 | Tumor
suppressor |
|
CTD-2574D22.5.1 | 8 |
Uncharacteristic |
|
RP11-407N17.5.1 | 8 |
Uncharacteristic |
| CTC-378H22.2.1 | 8 |
Uncharacteristic |
| RP11-723G8.1.1 | 8 |
Uncharacteristic |
|
RP11-167N24.3.1 | 8 |
Uncharacteristic |
| AL928742.12.1 | 8 |
Uncharacteristic |
| LINC00174 | 7 |
Uncharacteristic |
| RP11-65L19.4 | 7 |
Uncharacteristic |
|
RP11-442O18.2.1 | 7 |
Uncharacteristic |
|
RP1-170O19.14.1 | 7 |
Uncharacteristic |
|
RP11-480A16.1.1 | 7 |
Uncharacteristic |
| GS1-204I12.2.1 | 6 |
Uncharacteristic |
| RP11-611L7.1.1 | 6 |
Uncharacteristic |
| Z83851.1.1 | 6 |
Uncharacteristic |
| RP11-353N14.1 | 6 |
Uncharacteristic |
| LINC01482 | 6 |
Uncharacteristic |
| AC093611.1.1 | 6 |
Uncharacteristic |
| C21orf91-OT1 | 6 |
Uncharacteristic |
| LINC00313 | 6 |
Uncharacteristic |
Results
Construction of the MILMN
In the present study, the MILMN was constructed,
based on the co-expression associations identified between the
lncRNAs and mRNAs. This network contained a total of 1,476 lncRNAs,
4,444 mRNAs and 12,098 edges (Fig.
2). The top 20 mRNA nodes, which comprised those with the
highest degree, are shown in Table
I.
Detection of key lncRNAs regulating
crucial pathways in myocardial infarction
To investigate the biological functions of lncRNAs
during the development of myocardial infarction, the present study
annotated the mRNAs in the MILMN into KEGG pathways. This resulted
in a total of 26 pathways being detected (Fig. 3), which included certain
cardiovascular disease pathways, including the Dilated
cardiomyopathy and Hypertrophic cardiomyopathy pathways, and
certain important signaling pathways, including the Calcium
signaling pathway and MAPK signaling pathway. Subsequently an
lncRNA-pathway network was constructed based on these pathways, in
which edges indicated lncRNAs, which potentially regulated the
corresponding pathways (Fig. 3).
From these pathways, 11 crucial pathways were selected, following a
literature review. These pathways were either cardiovascular
disease pathways or were important signaling pathways in myocardial
infarction (Table II). The key
lncRNAs, which regulated at least six of the 11 crucial pathways
were selected (Fig. 4A; Table III). A total of 39 key lncRNAs
were identified (Table III), and
three of the lncRNAs (AC004410.1, CTB-113P19.1.1 and
RP13-452N2.1.1) were found to regulate the most crucial pathways
(Table III).
Investigating the regulatory mechanism of
lncRNAs in myocardial infarction
To examine the detailed regulatory mechanism of the
key lncRNAs identified, the co-expressing mRNAs of the key lncRNAS
were mapped into the Dilated cardiomyopathy pathway and MAPK
signaling pathway (Figs. 4B and
5). In the Dilated cardiomyopathy
pathway, certain key proteins were potentially regulated by
lncRNAs, including dihydropteridine reductase (DHPR) and
transforming growth factor (TGF)-β (Fig. 4B). In the MAPK signaling pathway,
lncRNAs were also found, which potentially regulated certain key
proteins, including MAPK kinase 2 (MEK2), MAPK kinase 7 (MKK7) and
epidermal growth factor receptor (EGFR), as shown in Fig. 5A. Subsequently, the sub-network was
extracted from the MILMN, within which were the mRNAs that were in
the MAPK signaling pathway. mRNA-MAP3K6 was found to be
differentially co-expressed with nine lncRNAs, including H19
(Fig. 5B).
Discussion
In previous years, the significant functional
molecular mechanism of lncRNAs has been recognized, particularly in
cardiovascular diseases. Furthermore, increasing evidence shows
that lncRNAs may be important in regulating gene expression
(15). Due to considerable
challenges in examining the functions of lncRNA, a
co-expression-based method was developed, in which lncRNA functions
were predicted based on the functions of their co-expressed
protein-coding genes (15), as
genes that exhibit similar expression patterns under multiple
conditions have a tendency to be involved in the same pathways
(19). Therefore, these
co-expressed protein-coding genes are potentially regulated by the
corresponding lncRNAs (15,20).
Therefore, in the present study, an affymetrix
microarray associated with myocardial infarction was re-annotated,
following which an MILMN was constructed. This network contained a
total of 1,476 lncRNAs and 4,444 mRNAs. In this network, a number
of mRNAs were identified, which were linked with several lncRNAs,
indicating that they are potentially regulated by these lncRNAs in
myocardial infarction. The most connected mRNA was CLIP2, with a
degree of 49 (Table I). Studies
have shown that the minor T allele of the CLIP2 gene has a
protective effect against elevated serum lipid and lipoprotein
levels, thus being associated with the risk of cardiovascular
diseases (21). Another
highly-connected mRNA was PTGIS, which is a catalyst for the
synthesis of PGI2 from prostaglandin H2, is widely distributed and
is predominantly found in vascular endothelial cells and smooth
muscle cells (22).
To examine the key lncRNAs and their potential
functions in myocardial infarction, pathway enrichment of all the
mRNAs in the MILMN was performed, from which an lncRNA-pathway
network was constructed (Fig. 3).
From a total of 26 pathways, 11 crucial pathways, which comprised
cardiovascular disease pathways or important signaling pathways,
were selected (Table II). For
example, the Calcium signaling pathway (23), peroxisome proliferator-activated
receptor signaling pathway (24)
and MAPK signaling pathway (25)
were found to be important in the development of myocardial
infarction.
Subsequently, 39 key lncRNAs were identified that
appeared to regulate the majority (6/11) of the crucial pathways
(Table III). Of these lncRNAs,
AC004410.1, CTB-113P19.1.1 and RP13-452N2.1.1 regulated almost all
the crucial pathways (Table
III). To examine the detailed regulatory mechanism of the key
lncRNAs, the co-expressed mRNAs of the key lncRNAs were mapped into
the Dilated cardiomyopathy pathway. A number of crucial membrane
proteins were annotated (Fig. 4B).
For example, DHPR, a dihydropyridine receptor, is essential in
skeletal muscle excitation-contraction coupling, which leads to an
increase in [Ca2+] via the activation of ryanodine
receptors (26,27), and indicates that lncRNAs may be
involved in inter/intra-cardiac cell communication. Of note,
previous studies have suggested that several classes of RNA
molecules are used for the horizontal transfer of information
between different types of cells in the heart (28). Furthermore, the present study
revealed that another important protein, TGF-β, which is considered
a potential biomarker of myocardial infarction (29), was also regulated by the
lncRNAs.
The present study subsequently focused on the MAPK
signaling pathway, due to its vast implications in signaling and
crosstalk with other signaling networks (Fig. 5A). For example, MAPK is involved in
crosstalk with mitochondria, which are the powerhouses of the cell
that provide >80% of the adenosine triphosphate required for
normal cardiomyocyte function, and have a crucial role in cell
death (30). The results of the
present study showed that certain pivotal proteins in this pathway
were regulated by lncRNAs. These proteins were found to be
important in myocardial infarction, for example MKK cascades
modulate the hypertrophic response of the heart to pressure
overload (31,32) and, in the ras-Raf-MEK-ERK signaling
cascade, overexpression of an activated form of MKK1 has been shown
to lead to profound cardiac hypertrophy without fibrosis (33). In addition, these cascades also
function as molecular switches in response to
spatiotemporal-specific cell-cell communication in myocardial
infarction (30,31). To better illustrate the potential
mechanism of lncRNAs, the present study extracted the sub-network
from the MILMN, which contained key lncRNAs and their regulating
mRNAs in the MAPK signaling pathway. Of these lncRNAs, only H19 had
functional annotation. Although it has been shown to be important
in several types of cancer (34),
its role in myocardial infarction remains to be fully elucidated.
The present study found that H19 potentially regulated mRNA-MAP3K6,
coding the ASK2 protein, which promotes pathological cardiac
remodeling following myocardial infarction (35), indicating the potential roles of
H19. Although the results of the present study require further
experimental verification, the results provide further insight into
understanding the roles of lncRNAs in myocardial infarction.
Acknowledgments
The present study was supported by grants from the
National Science Foundation of China (grant no. 30873131) and the
Jilin Province Development and Reform Commission (grant no.
3J113AX33426).
References
|
1
|
Wang Y, Zhang H, Chai F, Liu X and Berk M:
The effects of escitalopram on myocardial apoptosis and the
expression of Bax and Bcl-2 during myocardial ischemia/reperfusion
in a model of rats with depression. BMC Psychiatry. 14:3492014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grote P, Wittler L, Hendrix D, Koch F,
Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M and
Herrmann BG: The tissue-specific lncRNA Fendrr is an essential
regulator of heart and body wall development in the mouse. Dev
Cell. 24:206–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang KC, Yamada KA, Patel AY, Topkara VK,
George I, Cheema FH, Ewald GA, Mann DL and Nerbonne JM: Deep RNA
sequencing reveals dynamic regulation of myocardial noncoding RNAs
in failing human heart and remodeling with mechanical circulatory
support. Circulation. 129:1009–1021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mitra SA, Mitra AP and Triche TJ: A
central role for long non-coding RNA in cancer. Front Genet.
3:172012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang K, Liu F, Zhou LY, Long B, Yuan SM,
Wang Y, Liu CY, Sun T, Zhang XJ and Li PF: The long noncoding RNA
CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res.
114:1377–1388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gupta SK, Piccoli MT and Thum T:
Non-coding RNAs in cardiovascular ageing. Ageing Res Rev. 17:79–85.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ounzain S, Micheletti R, Beckmann T,
Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A,
Johnson R, et al: Genome-wide profiling of the cardiac
transcriptome after myocardial infarction identifies novel
heart-specific long non-coding RNAs. Eur Heart J. 36:353–368a.
2015. View Article : Google Scholar :
|
|
10
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee S, Choi E, Cha MJ, Park AJ, Yoon C and
Hwang KC: Impact of miRNAs on cardiovascular aging. J Geriatr
Cardiol. 12:569–574. 2015.PubMed/NCBI
|
|
14
|
Calore M, De Windt LJ and Rampazzo A:
Genetics meets epigenetics: Genetic variants that modulate
noncoding RNA in cardiovascular diseases. J Mol Cell Cardiol. Nov
3–2015.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suresh R, Li X, Chiriac A, Goel K, Terzic
A, Perez-Terzic C and Nelson TJ: Transcriptome from circulating
cells suggests dysregulated pathways associated with long-term
recurrent events following first-time myocardial infarction. J Mol
Cell Cardiol. 74:13–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liao Q, Xiao H, Bu D, Xie C, Miao R, Luo
H, Zhao G, Yu K, Zhao H, Skogerbø G, et al: ncFANs: A web server
for functional annotation of long non-coding RNAs. Nucleic Acids
Res. 39:W118–W124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Da Sacco L, Baldassarre A and Masotti A:
Bioinformatics tools and novel challenges in long non-coding RNAs
(lncRNAs) functional analysis. Int J Mol Sci. 13:97–114. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luptáková L, Benčová D, Siváková D and
Cvíčelová M: Association of CILP2 and ACE gene polymorphisms with
cardiovascular risk factors in Slovak midlife women. Biomed Res
Int. 2013:6342072013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakayama T: Genetic polymorphisms of
prostacyclin synthase gene and cardiovascular disease. Int Angiol.
29(Suppl 2): S33–S42. 2010.
|
|
23
|
Zhao Y, Hu HY, Sun DR, Feng R, Sun XF, Guo
F and Hao LY: Dynamic alterations in the CaV1.2/CaM/CaMKII
signaling pathway in the left ventricular myocardium of ischemic
rat hearts. DNA Cell Biol. 33:282–290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marx N, Bourcier T, Sukhova GK, Libby P
and Plutzky J: PPARgamma activation in human endothelial cells
increases plasminogen activator inhibitor type-1 expression:
PPARgamma as a potential mediator in vascular disease. Arterioscler
Thromb Vasc Biol. 19:546–551. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei N, Zhang C, He H, Wang T, Liu Z, Liu
G, Sun Z, Zhou Z, Bai C and Yuan D: Protective effect of saponins
extract from Panax japonicus on myocardial infarction: Involvement
of NF-kB, Sirt1 and mitogen-activated protein kinase signalling
pathways and inhibition of inflammation. J Pharm Pharmacol.
66:1641–1651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eltit JM, Franzini-Armstrong C and Perez
CF: Amino acid residues 489–503 of dihydropyridine receptor (DHPR)
beta1a subunit are critical for structural communication between
the skeletal muscle DHPR complex and Type-1 ryanodine receptor. J
Biol Chem. 289:36116–36124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Viola HM, Adams AM, Davies SM, Fletcher S,
Filipovska A and Hool LC: Impaired functional communication between
the L-type calcium channel and mitochondria contributes to
metabolic inhibition in the mdx heart. Proc Natl Acad Sci USA.
111:E2905–E2914. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sluijter JP, Verhage V, Deddens JC, van
den Akker F and Doevendans PA: Microvesicles and exosomes for
intra-cardiac communication. Cardiovasc Res. 102:302–311. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lax A, Sanchez-Mas J, Asensio-Lopez MC,
Fernandez-Del Palacio MJ, Caballero L, Garrido IP, Pastor-Perez FJ,
Januzzi JL and Pascual-Figal DA: Mineralocorticoid receptor
antagonists modulate galectin-3 and interleukin-33/ST2 signaling in
left ventricular systolic dysfunction after acute myocardial
infarction. JACC Heart Fail. 3:50–58. 2015. View Article : Google Scholar
|
|
30
|
Javadov S, Jang S and Agostini B:
Crosstalk between mitogen-activated protein kinases and
mitochondria in cardiac diseases: Therapeutic perspectives.
Pharmacol Ther. 144:202–225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Muslin AJ: MAPK signalling in
cardiovascular health and disease: Molecular mechanisms and
therapeutic targets. Clin Sci (Lond). 115:203–218. 2008. View Article : Google Scholar
|
|
32
|
Foltz IN, Gerl RE, Wieler JS, Luckach M,
Salmon RA and Schrader JW: Human mitogen-activated protein kinase
kinase 7 (MKK7) is a highly conserved c-Jun N-terminal
kinase/stress-activated protein kinase (JNK/SAPK) activated by
environmental stresses and physiological stimuli. J Biol Chem.
273:9344–9351. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bueno OF, De Windt LJ, Tymitz KM, Witt SA,
Kimball TR, Klevitsky R, Hewett TE, Jones SP, Lefer DJ, Peng CF, et
al: The MEK1-ERK1/2 signaling pathway promotes compensated cardiac
hypertrophy in transgenic mice. EMBO J. 19:6341–6350. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamaguchi O, Higuchi Y, Hirotani S,
Kashiwase K, Nakayama H, Hikoso S, Takeda T, Watanabe T, Asahi M,
Taniike M, et al: Targeted deletion of apoptosis signal-regulating
kinase 1 attenuates left ventricular remodeling. Proc Natl Acad Sci
USA. 100:15883–15888. 2003. View Article : Google Scholar : PubMed/NCBI
|