Introduction
Neuropathic pain, which is characterized by
spontaneous pain, hyperalgesia and allodynia, is a major clinical
problem that remains difficult to treat (1). At present, pain relief can only be
achieved in 40–60% of patients; therefore, it is important to study
the underlying mechanisms of pain, with the aim of eventually
developing novel therapeutic drugs (2). Previous studies have suggested that
glial cell-mediated neuroinflammation has an important role in
neuropathic pain; however, exactly how glial cells are involved
remains unclear (3–5).
Adenosine cyclic 3,5-monophosphate (cAMP) is an
ubiquitous regulator of inflammation and is a key second messenger
that influences glial activity (6,7). It
has previously been reported that increasing levels of cAMP via
various methods may suppress the activation of glial cells (both
microglia and astrocytes), decrease the production of
proinflammatory mediators, including tumor necrosis factor (TNF)-α,
interleukin (IL)-1β, IL-6, IL-12 and nitric oxide, and increase the
expression of anti-inflammatory factor IL-10 (8–10).
Phosphodiesterases (PDEs), which are responsible for
the degradation of cAMP, have an important role regulating the
intracellular levels of cAMP (11,12).
PDEs comprise 11 subfamilies and numerous spliced transcripts.
Various non-specific PDE inhibitors, including propentofylline,
pentoxifylline and inbudilast, have been reported to attenuate
hypersensitivity in numerous animal models of neuropathic pain
(13–15). Furthermore, it has been suggested
that the PDE4 family may be the prevailing target of these PDE
inhibitors. PDE4 comprises four subtypes: A, B, C and D; however,
it remains to be elucidated which subtypes are the major targets
until now.
One method that may be used to determine which
subtype is associated with neuropathic pain is RNA interference
(RNAi), which uses double-stranded RNA to effectively and
specifically suppress gene expression and induce the degradation of
target mRNA (16). Since the
effects of this technique are transient, silencing a particular
gene for 48 h in rat tissues offers the potential to map out the
dynamic time course of the expression of various subtypes (17).
In the present study, RNAi was used to specifically
silence various subtypes of the PDE4 gene, and to observe its
effects on mechanical and thermal hyperalgesia, and glia-mediated
spinal inflammation in rats with L5 spinal nerve ligation (SNL).
The results of the present study may contribute to understanding
regarding which subtype of PDE4 is associated with glial
activation, and shed light on the development of novel therapeutic
strategies for the treatment of neuropathic pain.
Materials and methods
Experimental animals
A total of 286 adult male Sprague-Dawley (SD) rats
(weight, 180–200 g; age, 8–12 weeks) were obtained from the
Experimental Animal Center of Jinling Hospital (Nanjing, China).
The rats were maintained at 24±1°C, under a 12 h light-dark cycle,
with ad libitum access to food and water. The present study
followed the International Association for the Study of Pain
guidelines for pain research in animals (18), and all animal studies were approved
by the Animal Care and Use Committee of Jinling Hospital.
Detection of PDE4A, B, C and D protein
expression using western blotting
A total of 16 SD rats were randomly divided into
four groups (n=4/group), to determine the levels of PDE4A, B, C and
D in the lumbar spinal cord. The rats were sacrificed under
anesthesia with sodium pentobarbital (40 mg/kg, i.p.; Suolaibao
Biotechnology Co,. Ltd., Beijing, China) and lumbar spinal cords
(L4 and L5) were harvested and stored at −80°C. Frozen tissues were
homogenized in radioimmunoprecipitation assay lysis buffer
containing 1 mM phenylmethylsulfonyl fluoride (Beyotime Institute
of Biotechnology, Haimen, China) and centrifuged at 4°C for 10 min
at 10,000 × g. The supernatants were harvested and protein
concentrations were determined using the Bradford method (Leagene
Biotechnology Company, Beijing, China). Protein samples (100
µg) were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and were transferred to
0.45-µm polyvinylidene difluoride membranes (EMD Millipore,
Billerica, MA, USA). The membranes were blocked with 5% nonfat dry
milk, and were then incubated with the following rabbit anti-rat
polyclonal antibodies: Anti-PDE4A (1:1,000; cat. no. ab14607),
anti-PDE4B (1:1,000; cat. no. ab14611), anti-PDE4D (1:500; cat. no.
ab14613) (Abcam, Cambridge, MA, USA) and anti-PDE4C (1:300; cat.
no. PD4C-301AP: Fabgennix International Inc., Frisco, TX, USA)
overnight at 4°C. Subsequently, the membranes were incubated with a
horseradish peroxidase (HRP)-labeled goat anti-rabbit
immunoglobulin (Ig)G antibody (1:1,000; cat. no. 7074; Cell
Signaling Technology, Inc., Danvers, MA, USA) for 1 h at room
temperature. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
used as a loading control (1:1,000; cat. no. 3683; Cell Signaling
Technology, Inc.). The proteins were visualized using
chemiluminescence reagents provided within an enhanced
chemiluminescence kit (GE Healthcare Life Sciences, Chalfont, UK)
and were exposed to film. Scanning densitometry (ImageJ2x; Rawak
Software, Inc.; National Institutes of Health, Bethesda, MD, USA)
was used for the semi-quantitative analysis of the data.
Small interfering (si)RNA
preparation
siRNA were designed to target the sequences of rat
PDE4A (GenBank accession NM_013101), PDE4B (GenBank accession
NM_017031) and PDE4D (GenBank accession NM_017032). The BLOCK-iT™
Alexa Fluor Red Oligo (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used as a mismatch control. All of the
oligonucleotide sequences were examined against the GenBank
database using the Basic Local Alignment Search Tool algorithm
(http://blast.ncbi.nlm.nih.gov/Blast.cgi), in order to
exclude non-specific matches with any unintended nucleotide
sequences. The oligonucleotides, which had been synthesized,
individually deprotected, and purified using RNase-free
high-performance liquid chromatography, were purchased from
Invitrogen; Thermo Fisher Scientific, Inc. The siRNA stocks were
aliquoted and stored at −20°C, at a concentration of 200 µM
in annealing buffer (19). siRNA
sequences used in the present study are presented in Table I.
 | Table IsiRNA sequences. |
Table I
siRNA sequences.
| siRNA | Sequence |
|---|
| siR-A | S:
5′-AAGAGUGAGAAGUUGCUUCGAACGC-3′
A: 5′-UUCUCACUCUUCAACGAAGCUUGCG-3′ |
| siR-B | S:
5′-UUCACCAUCCACAACAACAGUCUUG-3′
A: 5′-AAGUGGUAGGUGUUGUUGUCAGAA-3′ |
| siR-D | S:
5′-AUGGAUGGUUGGUUGCACAUGGGUG-3′
A: 5′-CACCCAUGUGCAACCAACCAUCCAU-3′ |
Intrathecal catheter implantation
Under sodium pentobarbital (40 mg/kg, i.p.)
anesthesia, an intrathecal catheter (PE-10 tubing, 18 cm) was
inserted 1–2 cm cephalad into the rat lumbar subarachnoid space at
the L4–L5 intervertebral discs. The catheter implantation procedure
was successfully performed in 224 of the 270 remaining rats. The
tip of the catheter was placed near the lumbar enlargement of the
spinal cord (20). The catheter,
with ~20 µl dead space, was subcutaneously tunneled and
externalized through the skin of the neck region. After 5 days, 2%
lidocaine (10 µl; Jinling Pharmaceutical Co., Ltd., Nanjing,
China) was injected intrathecally into the rats with no impaired
movement. Rats that exhibited lower limb paralysis within 1 min
indicated successful catheterization, and those that exhibited
neurological deficits resulting from the surgical procedure were
excluded from further experiments.
Surgery and siRNA administration
The L5 spinal nerve ligation model was generated
according to methods described by Chung et al (21). Briefly, following anesthetization
with sodium pentobarbital (40 mg/kg, i.p.), the rats with
intrathecal catheters were placed in the prone position. The left
paraspinal muscles were separated from the spinous processes at the
L4–S2 level under aseptic conditions. The L5 transverse process was
carefully removed, in order to identify the spinal nerves, and the
L5 spinal nerve was ligated with 7-0 silk thread. In the sham
group, the surgical procedure was identical, except that the left
L5 spinal nerve was not ligated. The rats were returned to their
cages and observed for any signs of motor deficits. None of the
rats exhibited motor dysfunction after surgery.
The rats were divided into seven groups
(n=32/group): The sham group (sham surgery + saline), the saline
group (SNL + saline), the vehicle group (SNL +
Lipofectamine® RNAiMAX; Invitrogen; Thermo Fisher
Scientific, Inc.), the mismatch siRNA group (SNL + mismatch siRNA),
the PDE4A-siRNA group (SNL + PDE4A-siRNA), the PDE4B-siRNA group
(SNL + PDE4B-siRNA) and the PDE4D-siRNA group (SNL + PDE4D-siRNA).
siRNA or mismatch RNA complexes were prepared immediately prior to
administration by mixing the RNA solution with
Lipofectamine® RNAiMAX transfection reagent, at a ratio
of 1:4 (w:v). The final concentration of siRNA was 2 µg in
10 µl. siRNAs, mismatch RNA, saline or
Lipofectamine® RNAiMAX were administered intrathecally
just after ligation, and at 1, 3, 5 and 7 days after surgery.
Evaluation of thermal and mechanical
hyperalgesia
Thermal and mechanical hyperalgesia were measured 1
day prior to and 2, 4, 6 and 8 days after surgery. Behavioral
studies were carried out in a quiet, temperature-controlled (24°C)
room between the hours of 8:00 AM and 10:00 AM.
Mechanical withdrawal threshold (MWT) was measured
using an Electro Von Frey anesthesiometer (Model 2390CE; IITC,
Inc., Woodland Hills, CA, USA). Briefly, the rats were individually
placed beneath an inverted ventilated Plexiglas cage with a
metal-mesh floor, allowing access to the plantar surface of the
hind paw. After 30 min of acclimation, gentle incremental pressure
(maximum 200 g) was applied using a rigid von Frey hair to the
plantar surface of the ipsilateral hind paw, until the paw was
withdrawn. Five tests were conducted at intervals of 5 min and the
force (g) applied was recorded.
Thermal withdrawal latency (TWL) was determined
using radiant heat (Model 390; IITC, Inc.). Following acclimation
to the Plexiglas cage (23×18×13 cm; 3-mm-thick glass floor), the
radiant heat source beneath the glass floor was focused on the
plantar surface of the ipsilateral hind paw when in contact with
the floor. The paw TWL was obtained five times per animal with
intervals of 5 min. Light intensity was preset, in order to obtain
a baseline latency of ~10 sec and the cutoff time was set at 20 sec
to avoid tissue damage.
Detection of PDE4A, B and D mRNA
expression
Following behavioral testing at 2, 4, 6 and 8 days
after the operation, all of the rats were anesthetized with sodium
pentobarbital (80 mg/kg, i.p.) and were sacrificed by decapitation,
and all appliances were treated with diethylpyrocarbonate to
prevent the degradation of mRNA. The lumbar spinal cords were
harvested and the mRNA expression levels of PDE4A, B and D were
assessed using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) (n=6 at each time point for each group). The
protein expression levels of PDE4A, B and D were assessed by
western blotting, as previously described.
Following homogenization of the tissue samples,
total RNA was extracted from the ipsilateral lumbar spinal cord
using an RNA Isolation kit (Invitrogen; Thermo Fisher Scientific,
Inc.) and RNA was detected using a spectrophotometer (Epoch 2;
BioTek Instruments, Inc., Winooski, VT, USA). cDNA was synthesized
by reverse transcription (RT). The RT reaction was carried out in a
20 µl total reaction volume, containing 4 µl 5X RT
buffer, 4 µl 2.5 mM dNTPs, 1 µl Multiscribe reverse
transcriptase (50 U/µl) (Promega Corporation, Madison, WI,
USA), 1 µl RNase inhibitor, 5 µl RNase-free water,
and 3 µg DNase-treated total RNA in a 5 µl volume.
The RT reaction was carried out at 25°C for 10 min, 37°C for 120
min, and 95°C for 5 min. qPCR was conducted using a Rotor-Gene 3000
Real Time PCR system (Qiagen, Inc., Valencia, CA, USA). The
sequences of the primers (Invitrogen; Thermo Fisher Scientific,
Inc.) used in the present study are presented in Table II. qPCR was performed using SYBR
Green I (1:20,000; Qiagen, Inc., Valencia, CA, USA), with the
following cycling conditions: 1 cycle at 95°C for 3 min, followed
by 40 cycles at 95°C for 45 sec, 61°C for 45 sec, 72°C for 40 sec
and 80°C for 5 sec. The PCR reaction volume (25 µl)
consisted of 3 units Platinum Taq DNA polymerase, 1.5 mM
MgCl2, 205 µM dGTP, dCTP, dATP and dTTP, 400 nM
forward and reverse primers, 2.5 µl 10X PCR buffer (all
Promega Corporation), 10 µl SYBR Green I and 1 µl
cDNA. The mRNA expression levels were calculated according to
relative standard curves. The curves were generated by plotting the
quantification cycle (Cq) against the log amount of total cDNA
added to the reaction. The relative target gene expression levels
were determined using the 2−ΔΔCq method (22). Results were normalized to
GAPDH.
 | Table IIPrimer sequence used in RT-PCR. |
Table II
Primer sequence used in RT-PCR.
| Gene | Sequence |
|---|
| PDE4A | F:
5′-GAAGACAACCGGGACTCCT-3′
R: 5′-CCTCAGTGGTAGGCAATCC-3′ |
| PDE4B | F:
5′-CCTCCGACACCTTCGTAAC-3′
R: 5′-CCAGGTCTGTGAAGACAGC-3′ |
| PDE4D | F:
5′-CCCTCTTGACTGTTATCATGCACACC-3′
R: 5′-GATCCTACATCATGTATTGCACTGGC-3′ |
| GAPDH | F:
5′-CCATGTTCGTCATGGGTGTGAACCA-3′
R: 5′-GCCAGTAGAGGCAGGGATGATGTTC-5′ |
Detection of extracellular
signal-regulated kinases (ERK), phosphorylated (p)-ERK, CD11 and
glial fibrillary acidic protein (GFAP)
A total of 8 days after surgery, the protein
expression levels of ERK, p-ERK, CD11 and GFAP were detected by
western blotting (n=4), as described previously in the present
study. For ERK and p-ERK, 50 µg samples were separated in
each lane. The following antibodies were used: Anti-ERK1/2
(1:1,000; cat. no. 9102), anti-p-ERK1/2 (Thr202/Tyr204) (1:1,000;
cat. no. 9101) (Cell Signaling Technology, Inc.), anti-GFAP (1:500;
cat. no. sc-9065; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
anti-CD11b (1:1,000; cat. no. ab75476) and HRP-conjugated goat
anti-mouse (cat. no. ab47827)/anti-rabbit (cat. no. ab6721) Ig
(1:1,000) (Abcam).
Detection of TNF-α, IL-1β, IL-6 and IL-10
expression
The expression of cytokines, including TNF-α, IL-1β,
IL-6 and IL-10, was determined using enzyme-linked immunosorbent
assay (ELISA) kits (R&D Systems, Minneapolis, MN, USA) 8 days
after surgery (n=4/group), according to the manufacturer's
protocol. Ipsilateral lumbar spinal cord samples (40–50 mg) were
dissected and homogenized in a buffer containing a protease
inhibitor (Roche Diagnostics, Mannheim, Germany) using a Power Gen
124 tissue tearer (Thermo Fisher Scientific, Inc.). The samples
were then centrifuged at 20,000 × g for 30 min at 4°C. The
supernatants were aliquoted and stored at −80°C for further protein
quantification.
Statistical analysis
Data are presented as the mean ± standard deviation.
Data from the western blotting and RT-qPCR studies were analyzed
using one-way analysis of variance (ANOVA). Data from the thermal
and mechanical hyperalgesia tests were analyzed using two-way
ANOVA, in order to determine the differences between the groups at
various time points. All statistical analyses were performed using
SPSS 18.0 statistical software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Protein expression levels of PDE4A, B and
D in the spinal cord of naïve rats
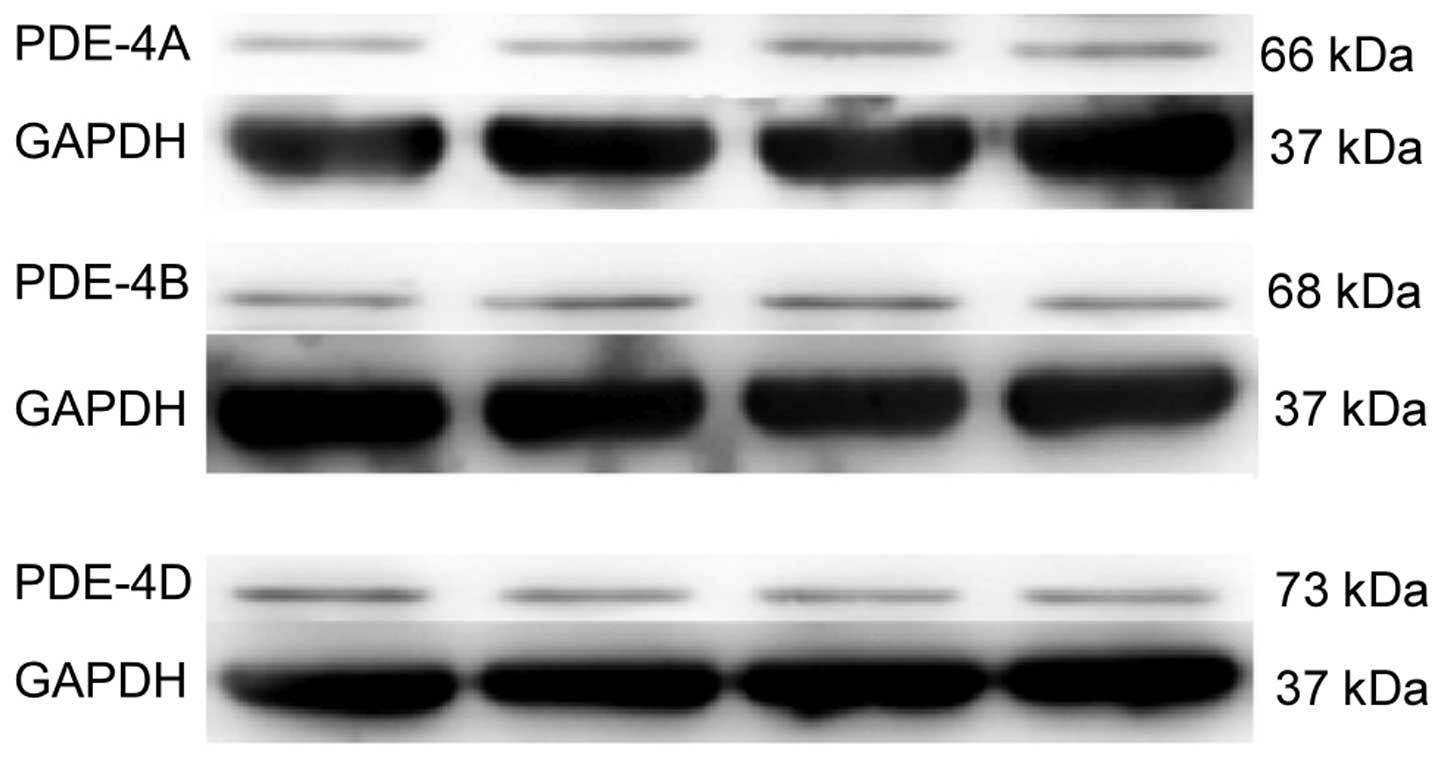
PDE4A, B and D protein expression was detected in
the spinal cord of naïve rats (Fig.
1); however, PDE4C expression was not detected (data not
shown).
siRNA attenuates the protein and mRNA
expression levels of PDE4A, B and D
PDE4A, B and D mRNA expression levels were
significantly upregulated 2, 4, 6 and 8 days after L5 SNL, as
compared with the sham group (Fig.
2). Treatment with saline, vehicle or mismatched RNA did not
reduce the upregulated mRNA expression levels (P<0.05; Fig. 2). However, treatment with 2
µg siRNA-PDE4A, B or D significantly decreased the mRNA
expression levels ofPDE4A, B or D, respectively (P<0.05;
Fig. 2), as compared with the
saline group. This effect, first observed at 2 days post-L5 SNL,
continued to 8 days post-ligation without significant
alterations.
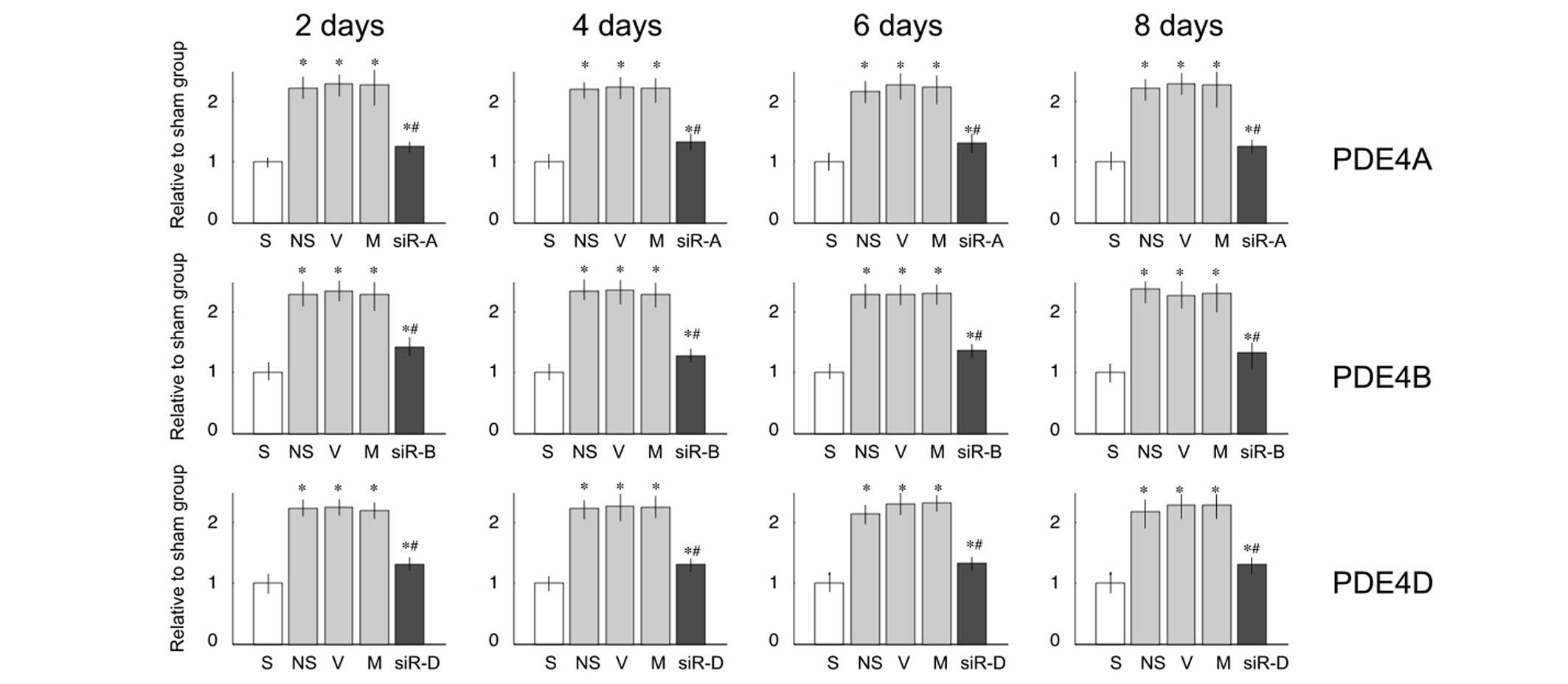 | Figure 2mRNA expression levels of
phosphodiesterase (PDE)4A, B and D in the lumbar spinal cord of
rats. Relative mRNA expression levels of PDE4A, B and D were
significantly increased on days 2, 4, 6 and 8 after L5 ligation, as
compared with the sham (S) group. Compared with the saline (NS)
group, treatment with small interfering (si)RNA decreased the mRNA
expression levels. Data are presented as the mean ± standard
deviation. *P<0.05 vs. the S group;
#P<0.05 vs. the NS group. V, vehicle group; M,
mismatch siRNA group; siR-A, PDE4A-siRNA group; siR-B, PDE4B-siRNA
group; siR-D, PDE4D-siRNA group. |
siRNA exhibited similar effects on the protein
expression levels of PDE4A, B and D (Fig. 3). L5 ligation resulted in
overexpression of PDE proteins, and intrathecal injection with
siRNA-PDE4A, B or D significantly lowered the expression of PDE4A,
B or D, respectively.
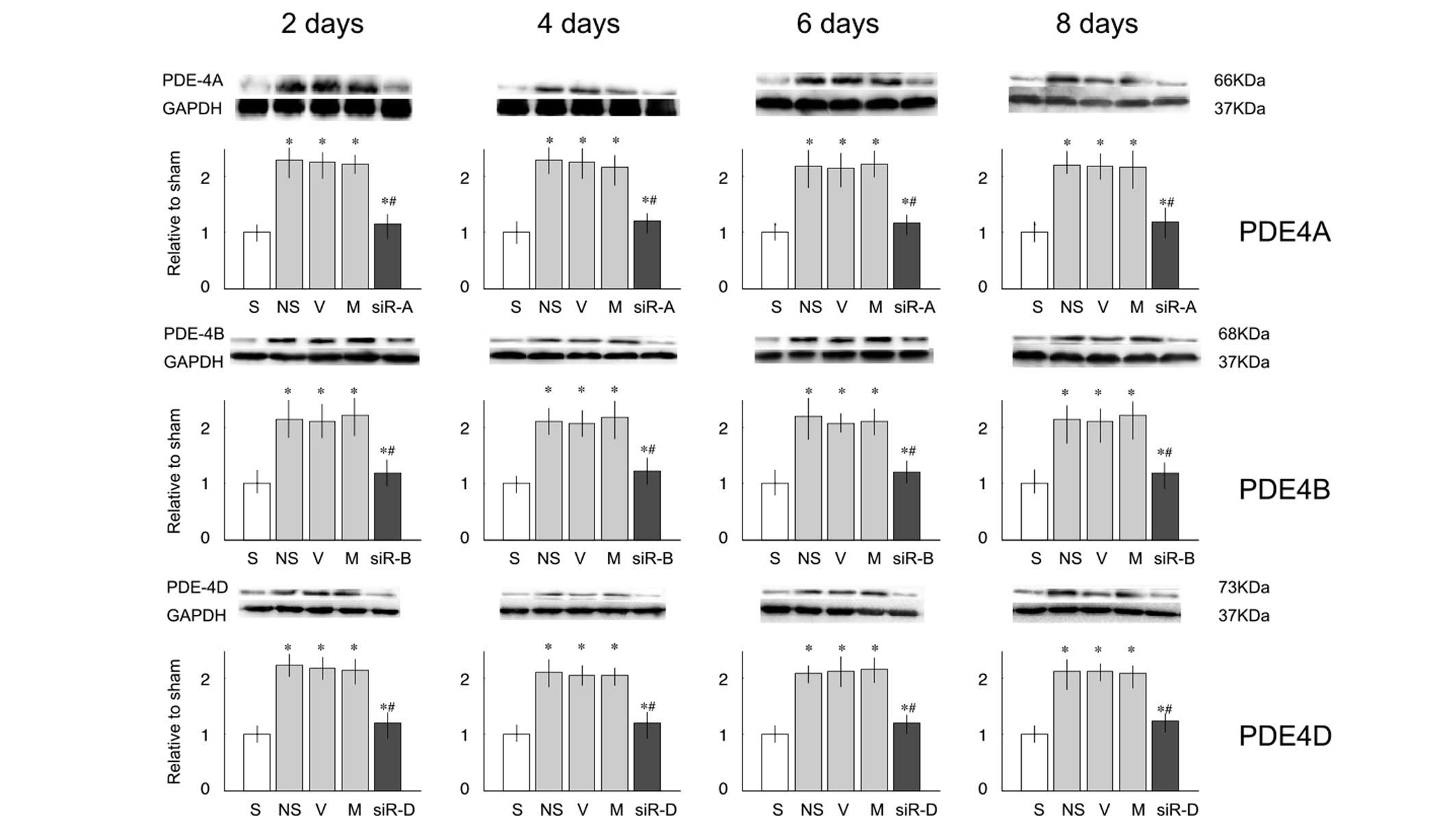 | Figure 3Protein expression levels of
phosphodiesterase (PDE)4A, B and D in the lumbar spinal cord of
rats. Relative protein expression levels of PDE4A, B and C were
significantly increased on days 2, 4, 6 and 8 after nerve injury,
as compared with the sham (S) group. Compared with the saline (NS)
group, treatment with small interfering (si)RNA decreased the
protein expression levels. Data are presented as the mean ±
standard deviation. *P<0.05 vs. the S group;
#P<0.05 vs. the NS group. V, vehicle group; M,
mismatch siRNA group; siR-A, PDE4A-siRNA group; siR-B, PDE4B-siRNA
group; siR-D, PDE4D-siRNA group; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase. |
Intrathecal injection with PDE4B siRNA
improves MWT and TWL in SNL rats
Prior to L5 SNL, all groups exhibited comparable
baseline thresholds for mechanical and thermal stimuli (Fig. 4A and B; -1 day). The values of the
sham group rats were steady during the experimental period
(Fig. 4A and B). L5 SNL led to
significantly reduced MWT and TWL (P<0.05; Fig. 4). No significant differences were
observed between the saline, vehicle and mismatched RNA groups
(P>0.05; Fig. 4). In addition,
intrathecal injection with PDE4A-siRNA and PDE4D-siRNA did not
improve the mechanical allodynia and thermal hyperalgesia, as
compared with the saline group (P>0.05; Fig. 4). However, injection with
PDE4B-siRNA significantly attenuated the development of mechanical
allodynia and thermal hyperalgesia, as compared with the saline
group (P<0.05; Fig. 4).
 | Figure 4Line plots illustrating alterations
in (A) mechanical withdrawal threshold (MWT) and (B) thermal
withdrawal latency (TWL). Tests were conducted 1 day prior to
surgery, and 2, 4, 6 and 8 days after the operation. L5 spinal
nerve ligation (SNL) resulted in an overall significant decrease in
mechanical and thermal threshold, as compared with the sham group
(P<0.05). Treatment with PDE4-A or D-specific small interfering
(si)RNA, had no effect on MWT or TWL (P>0.05), whereas
PDE4B-siRNA significantly attenuated the development of mechanical
and thermal hyperalgesia in L5 SNL rats, as compared with the SNL +
saline (NS) group. Data are presented as the mean ± standard
deviation. *P<0.05 vs. the sham group;
#P<0.05 vs. the SNL + NS group. V, vehicle group; M,
mismatch siRNA group; siR-A, PDE4A-siRNA group; siR-B, PDE4B-siRNA
group; siR-D, PDE4D-siRNA group. |
ERK activation in the spinal cord
L5 SNL resulted in a significant increase in the
expression levels of p-ERK, as compared with the sham group
(Fig. 5A; P<0.05). Compared
with the saline group, only the PDE4B-siRNA group exhibited a
significant decrease in the expression levels of p-ERK (Fig. 5A; P<0.05). However, there was no
difference in the expression of total ERK between the various
groups (Fig. 5B; P>0.05).
Protein expression of CD11b and GFAP in
the spinal cord of rats
CD11b, a microglial marker (Fig. 6A) and GFAP, an astrocyte marker
(Fig. 6B), were measured 8 days
after L5 SNL. CD11b and GFAP protein expression levels were higher
following SNL, as compared with the sham group (P<0.05; Fig. 6). However, intrathecal injection of
PDE4B-siRNA attenuated the upregulated expression of CD11b and
GFAP, as compared with the saline group (P<0.05; Fig. 6).
Effects of PDE4B-siRNA on the expression
of TNF-α, IL-6, IL-1β and IL-10 in the spinal cord
To investigate the effects of PDE4B-siRNA, the
expression levels of numerous inflammatory cytokines, including
TNF-α, IL-6, IL-1β and IL-10, were measured. Compared with the sham
group, the expression levels of TNF-α, IL-6 and IL-1β were
significantly increased in the SNL groups (P<0.05). However, the
PDE4B-siRNA group exhibited significantly reduced levels of the
proinflammatory cytokines, as compared with the saline group
(P<0.05). In addition, PDE4B-siRNA significantly enhanced IL-10
production, as compared with the other SNL groups (P<0.05;
Table III).
 | Table IIIExpression of cytokines 8 days
post-operation (pg/mg, mean ± standard deviation). |
Table III
Expression of cytokines 8 days
post-operation (pg/mg, mean ± standard deviation).
| Cytokine | S | NS | V | siR-M | siR-B |
|---|
| TNF-α | 47.6±4.3 | 129.2±3.8a | 126.5±4.8a | 130.1±3.6a | 96.1±3.5a,b |
| IL-1β | 43.7±6.6 | 144.1±15.9a | 147.6±17.6a | 154.0±11.9a | 84.2±13.3a,b |
| IL-6 | 77.9±12.3 | 259.1±15.8a | 269.4±10.3a | 260.7±17.1a | 162.4±12.7a,b |
| IL-10 | 148.5±12.3 | 156.1±9.6 | 158.9±11.5 | 159.8±10.9 | 241.9±17.4b |
Discussion
In the present study, a model of neuropathic pain
was established using single-sided L5 SNL (21). The model was confirmed, since MWT
and TWL were reduced following SNL. In addition, the mRNA and
protein expression levels of PDE4A, B and D were significantly
upregulated following SNL. Intrathecal injection of the SNL rats
with PDE4A, B or D-specific siRNA markedly reduced the elevated
expression levels of PDE4A, B and D, respectively. However, only in
the group treated with PDE4B-specific siRNA were MWT and TWL
improved. Furthermore, only in rats treated with PDE4B-specific
siRNA was p-ERK activity significantly decreased, the expression
levels of proinflammatory cytokines TNF-α, IL-1β and IL-6
suppressed, and the expression of IL-10 increased 8 days after L5
SNL. These findings suggested that, among the PD4E family, PDE4B
may have an important role in ameliorating neuropathic pain,
potentially via inhibition of ERK activity.
In the past decade, it has been reported that
microglia and astrocytes are activated in the spinal cord in
various animal models of neuropathic pain, and have an important
role in the development and maintenance of hypernociception
(23,24). In our previous study, a
non-specific glial inhibitor, pentoxifylline, was shown to exert a
dose-dependent antihyperalgesic effect when systemically injected
prior to nerve injury; however, it had no effect following the
establishment of hypersensitivity (14). In addition, other non-specific PDE
inhibitors, such as propentofylline and inbudilast, may attenuate
hyperalgesia in both preventive and therapeutic schemes; the
differences may be due to the various inhibitory potencies of these
drugs on PDE4 (13,15). To further elucidate the role of
PDE4, the present study used specific siRNA to suppress the
respective PDE4 subtypes, and observed the behavioral and
neuroinflammatory response in the spinal cord.
The expression levels of PDE4A, B, C and D were
initially detected by western blotting in the spinal cord of naïve
rats. The expression levels of PDE4A, B and D were similar and
quite low, as compared with GAPDH; however, PDE4C protein was not
detected. These results were consistent with those of previous
reports (25,26), which detected the expression of
PDE4A, B and D in oligodendrocytes, and PDE4B in microglia using
immunohistochemistry.
The present study used three siRNAs targeting PDE4A,
B and D to study their potential roles in neuropathic pain. In
order to determine the effects and specificity of the siRNA, we
observed both the mRNA and protein expression levels of the
respective targets 1 day after siRNA injection. After 2, 4, 6 and 8
days, SNL significantly increased the mRNA and protein expression
levels of PDE4A, B and D by ~200%, and only specific siRNA could
significantly inhibit this increase by ~50%. The possible
mechanisms underlying increased PDE4 expression may be via nuclear
factor-κB activation (27) and
enhanced N-methyl-D-aspartate receptor activity (28) after nerve injury. siRNA specificity
was confirmed by the markedly suppressive effects of the siRNA
molecules for up to 8 days after SNL.
All three siRNA were able to suppress the expression
of their respective targets; however, only PDE4B siRNA exhibited
significant attenuation of mechanical and thermal hyperalgesia in
the SNL rats after 8 days. Previous studies reported that PDE4B is
mainly expressed in microglia (29,30);
therefore, the expression levels of CD11b, a microglia marker and
GFAP, an astrocyte marker were detected 8 days after SNL. The
present study demonstrated that PDE4B siRNA could significantly
inhibit the increases in CD11b and GFAP after nerve injury. These
findings indicated that only inhibition of PDE4B could suppress the
SNL-induced activation of microglia and astrocytes.
To further understand the inhibitory effects of
PDE4B siRNA, alterations in ERK activity were observed in the
lumbar spinal cord 8 days after SNL. Previous studies have
demonstrated that phosphorylation of ERK contributes to the
neuroinflammation mediated by microglia and astrocytes in various
models of neuropathic pain (31–33).
The present study demonstrated that PDE4B siRNA was able to
significantly inhibit the activation of ERK. Although the present
study did not measure the levels of cAMP in the spinal cord,
previous studies have shown that an elevated cAMP signal may
inhibit the activation of ERK via protein kinase A (PKA)-dependent
and -independent pathways (34,35).
Cytokines are important factors released by
activated microglia and astrocytes, which influence neuronal
excitivity. Previous studies have reported that enhanced
intracellular cAMP activity in microglia and astrocytes may inhibit
the production of proinflammatory cytokines and enhance IL-10
secretion (36,37). Using primary cultured microglia, we
further demonstrated that PKA was a main downstream effector
(38). In the present study, PDE4B
siRNA exerted inhibitory effects on the expression of TNF-α, IL-1β
and IL-6, and enhanced the expression of IL-10.
Considering the low expressions of PDE, 100
µg protein was loaded for western blotting in the present
study; however, PDE4C expression could not be detected in naïve
rats. Fortunately, previous studies have demonstrated that PDE4A, B
and D are expressed in oligodendrocytes, whereas only PDE4B has
been detected in microglia, as determined using
immunohistochemistry (25,26). Secondly, the levels of cAMP were
not detected in the spinal cord after intrathecal siRNA injection.
Since the present study was focused on the effects of PDE on the
modulation of glial-mediated neuroinflammation, it was hypothesized
that the level of cAMP in the mixed spinal tissue could not reflect
the levels in glia. In our ongoing study using cultured microglia,
cAMP concentration was measured following treatment with PDE4 siRNA
(unpublished data). Only PDE4B siRNA was able to significantly
increase cAMP levels in the microglia 30 min after a
lipopolysaccharide challenge.
The results of the present study indicated that
PDE4B may be considered the main effector for the degradation of
cAMP in glia, and inhibition of PDE4B may inhibit glial activation,
reduce proinflammatory cytokine expression, and subsequently
attenuate hypernociception following nerve injury.
In conclusion, the present study provided evidence
suggesting that inhibition of PDE4B may attenuate neuroinflammation
in the spinal cord, and partly relieve neuropathic pain. PDE4B may
be considered a promising target for the development of novel
therapeutic strategies for the treatment of neuropathic pain.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81102514).
References
|
1
|
Dworkin RH, O'Connor AB, Backonja M,
Farrar JT, Finnerup NB, Jensen TS, Kalso EA, Loeser JD, Miaskowski
C, Nurmikko TJ, et al: Pharmacologic management of neuropathic
pain: Evidence-based recommendations. Pain. 132:237–251. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Attal N, Cruccu G, Baron R, Haanpää M,
Hansson P, Jensen TS and Nurmikko T; European Federation of
Neurological Societies: EFNS guidelines on the pharmacological
treatment of neuropathic pain: 2010 revision. Eur J Neurol.
17:e1113–e1188. 2010. View Article : Google Scholar
|
|
3
|
Gao YJ and Ji RR: Targeting astrocyte
signaling for chronic pain. Neurotherapeutics. 7:482–493. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsuda M, Inoue K and Salter MW:
Neuropathic pain and spinal microglia: A big problem from molecules
in 'small' glia. Trends Neurosci. 28:101–107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Watkins LR and Maier SF: Glia: A novel
drug discovery target for clinical pain. Nat Rev Drug Discov.
2:973–985. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liou JT, Liu FC, Hsin ST, Yang CY and Lui
PW: Inhibition of the cyclic adenosine monophosphate pathway
attenuates neuropathic pain and reduces phosphorylation of cyclic
adenosine monophosphate response element-binding in the spinal cord
after partial sciatic nerve ligation in rats. Anesth Analg.
105:1830–1837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taskén K and Aandahl EM: Localized effects
of cAMP mediated by distinct routes of protein kinase A. Physiol
Rev. 84:137–167. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ottonello L, Morone MP, Dapino P and
Dallegri F: Cyclic AMP-elevating agents down-regulate the oxidative
burst induced by granulocyte-macrophage colony-stimulating factor
(GM-CSF) in adherent neutrophils. Clin Exp Immunol. 101:502–506.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pearse DD, Pereira FC, Marcillo AE, Bates
ML, Berrocal YA, Filbin MT and Bunge MB: cAMP and Schwann cells
promote axonal growth and functional recovery after spinal cord
injury. Nat Med. 10:610–616. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pryzwansky KB and Madden VJ: Type 4A
cAMP-specific phosphodiesterase is stored in granules of human
neutrophils and eosinophils. Cell Tissue Res. 312:301–311. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lugnier C: Cyclic nucleotide
phosphodiesterase (PDE) super-family: A new target for the
development of specific therapeutic agents. Pharmacol Ther.
109:366–398. 2006. View Article : Google Scholar
|
|
12
|
Lynch MJ, Hill EV and Houslay MD:
Intracellular targeting of phosphodiesterase-4 underpins
compartmentalized cAMP signaling. Curr Top Dev Biol. 75:225–259.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gwak YS and Hulsebosch CE: Remote
astrocytic and microglial activation modulates neuronal
hyperexcitability and below-level neuropathic pain after spinal
injury in rat. Neuroscience. 161:895–903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu J, Feng X, Yu M, Xie W, Zhao X, Li W,
Guan R and Xu J: Pentoxifylline attenuates the development of
hyperalgesia in a rat model of neuropathic pain. Neurosci Lett.
412:268–272. 2007. View Article : Google Scholar
|
|
15
|
Hama A, Broadhead A, Lorrain DS and Sagen
J: The antinociceptive effect of the asthma drug ibudilast in rat
models of peripheral and central neuropathic pain. J Neurotrauma.
29:600–610. 2012. View Article : Google Scholar
|
|
16
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Doré-Savard L, Roussy G, Dansereau MA,
Collingwood MA, Lennox KA, Rose SD, Beaudet N, Behlke MA and Sarret
P: Central delivery of Dicer-substrate siRNA: A direct application
for pain research. Mol Ther. 16:1331–1339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zimmerman M: Ethical guidelines for
investigations of experimental pain in conscious animals. Pain.
16:109–110. 1983. View Article : Google Scholar
|
|
19
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kawamata T, Omote K, Kawamata M, Iwasaki H
and Namiki A: Antinociceptive interaction of intrathecal
alpha2-adrenergic agonists, tizanidine and clonidine, with
lidocaine in rats. Anesthesiology. 87:436–448. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chung JM, Kim HK and Chung K: Segmental
spinal nerve ligation model of neuropathic pain. Methods Mol Med.
99:35–45. 2004.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Zhang GH, Lv MM, Wang S, Chen L, Qian NS,
Tang Y, Zhang XD, Ren PC, Gao CJ, Sun XD and Lu LX: Spinal
astrocytic activation is involved in a virally-induced rat model of
neuropathic pain. PLoS One. 6:e230592011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu PY, Lu CL, Wang CC, Lee IH, Hsieh JC,
Chen CC, Lee HF, Lin HC, Chang FY and Lee SD: Spinal microglia
initiate and maintain hyperalgesia in a rat model of chronic
pancreatitis. Gastroenterology. 142:165–173. 2012. View Article : Google Scholar
|
|
25
|
Whitaker CM, Beaumont E, Wells MJ,
Magnuson DS, Hetman M and Onifer SM: Rolipram attenuates acute
oligodendrocyte death in the adult rat ventrolateral funiculus
following contusive cervical spinal cord injury. Neurosci Lett.
438:200–204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schaal SM, Garg MS, Ghosh M, Lovera L,
Lopez M, Patel M, Louro J, Patel S, Tuesta L, Chan WM and Pearse
DD: The therapeutic profile of rolipram, PDE target and mechanism
of action as a neuroprotectant following spinal cord injury. PLoS
One. 7:e436342012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vicini E and Conti M: Characterization of
an intronic promoter of a cyclic adenosine 3′,5′-monophosphate
(cAMP)-specific phosphodiesterase gene that confers hormone and
cAMP inducibility. Mol Endocrinol. 11:839–850. 1997.PubMed/NCBI
|
|
28
|
Hajjhussein H, Suvarna NU, Gremillion C,
Chandler LJ and O'Donnell JM: Changes in NMDA receptor-induced
cyclic nucleotide synthesis regulate the age-dependent increase in
PDE4A expression in primary cortical cultures. Brain Res.
1149:58–68. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sebastiani G, Morissette C, Lagacé C,
Boulé M, Ouellette MJ, McLaughlin RW, Lacombe D, Gervais F and
Tremblay P: The cAMP-specific phosphodiesterase 4B mediates
Abeta-induced microglial activation. Neurobiol Aging. 7:691–701.
2006. View Article : Google Scholar
|
|
30
|
Reyes-Irisarri E, Sánchez AJ,
García-Merino JA and Mengod G: Selective induction of cAMP
phosphodiesterase PDE4B2 expression in experimental autoimmune
encephalomyelitis. J Neuropathol Exp Neurol. 66:923–931. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhuang ZY, Gerner P, Woolf CJ and Ji RR:
ERK is sequentially activated in neurons, microglia, and astrocytes
by spinal nerve ligation and contributes to mechanical allodynia in
this neuropathic pain model. Pain. 114:149–159. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choi DC, Lee JY, Lim EJ, Baik HH, Oh TH
and Yune TY: Inhibition of ROS-induced p38MAPK and ERK activation
in microglia by acupuncture relieves neuropathic pain after spinal
cord injury in rats. Exp Neurol. 236:268–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li W, Li Y, Zhu S, Ji Q, Shu Y, Zhang L
and Liu J: Rosuvastatin attenuated the existing morphine tolerance
in rats with L5 spinal nerve transection through inhibiting
activation of astrocytes and phosphorylation of ERK42/44. Neurosci
Lett. 584:314–319. 2015. View Article : Google Scholar
|
|
34
|
Aronoff DM, Canetti C, Serezani CH, Luo M
and Peters-Golden M: Cutting edge: Macrophage inhibition by cyclic
AMP (cAMP): Differential roles of protein kinase A and exhange
protein directly activated by cAMP-1. J Immunol. 174:595–599. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Emery AC and Eiden LE: Signaling through
the neuropeptide GPCR PAC1, induces neuritogenesis via a single
linear cAMP- and ERK-dependent pathway using a novel cAMP sensor.
FASEB J. 26:3199–3211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Woo MS, Jang PG, Park JS, Kim WK, Joh TH
and Kim HS: Selective modulation of lipopolysaccharide-stimulated
cytokine expression and mitogen-activated protein kinase pathways
by dibutyryl-cAMP in BV2 microglial cells. Brain Res Mol Brain Res.
113:86–96. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao L and Brinton RD: Suppression of
proinflammatory cytokines interleukin-1beta and tumor necrosis
factor-alpha in astrocytes by a V1 vasopressin receptor agonist: A
cAMP response element-binding protein-dependent mechanism. J
Neurosci. 24:2226–2235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu J, Zhao X, Cao J, Xue Q, Feng X, Liu
X, Zhang F and Yu B: Differential roles of PKA and Epac on the
production of cytokines in the endotoxin-stimulated primary
cultured microglia. J Mol Neurosci. 45:186–193. 2011. View Article : Google Scholar
|