Introduction
Esophageal cancer is a prevalent type of cancer
worldwide and is ranked sixth among cancer-associated mortalities
(1). According to a recent study,
in 2008 ~482,000 new esophageal cancer cases were diagnosed and
407,000 cancer-associated mortalities occurred globally (1). Almost half of newly diagnosed
esophageal cancer cases occurred in China (2,3).
Northern China is a high-incidence area for esophageal cancer and
has been termed the Asian esophageal cancer belt (4). The highest-incidence areas of China
include Linxian (Henan), Cixian (Hebei), Huai'an (Jiangsu)
(5–7), a high incidence is also observed in
Xinjiang, which has a population of various ethnic groups (3).
Esophageal squamous cell carcinoma (ESCC) is the
predominant histological type of esophageal cancer in China, which
contributes to >90% of all esophageal cancer incidences. Less
common types include esophageal adenocarcinomas, melanoma,
leiomyosarcoma and small-cell carcinoma (8–14).
Currently, esophageal cancer is treated using
surgery, chemotherapy, radiotherapy, biotherapy, or a combination
of these modalities (15). Despite
improvements in surgical techniques and adjuvant chemoradiation,
the overall 5-year survival rate of esophageal cancer has remained
<10% in the USA (16). As the
long-term survival rate is correlated with the clinical stage of
esophageal cancer (17), early
diagnosis and treatment would contribute to improving survival
rates and the quality of life of esophageal cancer patients.
However, early diagnosis is difficult as the majority of
early-stage cases of esophageal cancer are asymptomatic (18).
Suppression of apoptosis is a notable biological
behavior of cancer, and it is crucial in the oncogenesis and
progression of ESCC (19–21). Elucidation of the regulatory
mechanisms underlying gene expression in the process of apoptosis
may contribute to early diagnosis and personalized therapy for ESCC
patients.
Survivin is the smallest member of the inhibitor of
apoptosis protein (IAP) family, and is important in the regulation
of apoptosis (22). Survivin
expression levels are correlated with the clinicopathological
parameters and prognosis of esophageal cancer patients (23–27).
Furthermore, overexpression of survivin has been associated with an
increased likelihood of tumor relapse and poor overall survival
(28,29). Thus, survivin detection has been
used as a biomarker for monitoring tumor recurrence, and survivin
has been targeted as a personalized therapeutic strategy in
clinical trials (30–35).
The molecular mechanisms of high survivin expression
in tumor tissues include amplification of the survivin locus
(36), demethylation of the
survivin promoter (37) and
increased promoter activity (38).
Furthermore, previous studies have observed that transcription
factors, such as nuclear factor κ-light-chain-enhancer of activated
B cells (NF-κB) (39), are
important for increased survivin transcription activity. Activation
of the NF-κB signaling pathway contributes to tumor progression by
blocking apoptosis via upregulation of survivin (40,41).
NF-κB is a nuclear transcription factor that
regulates immunoglobulin (Ig) κ light chain expression in B
lymphocytes (42,43). The mammalian genome encodes five
NF-κB subunits: RelA (p65), RelB, c-Rel and NF-κB1 (p50 and its
precursor, p105), and NF-κB2 (p52 and its precursor, p100)
(42).
Activation of the NF-κB signaling pathway is
significantly associated with reduced overall survival in patients
with ESCC (44,45). Inhibition of the NF-κB signaling
pathway thus represents a promising approach for treatment of ESCC
(46). However, the mechanism
underlying activation of transcription factor p65 (NF-κB p65),
which participates in the NF-κB canonical signaling pathway in
ESCC, remains to be elucidated.
As NF-κB p65 and survivin contribute to regulating
apoptosis, and are highly expressed or persistently activated in
tumorigenesis and progression of ESCC (32,47),
the present study hypothesized that survivin activates NF-κB p65 by
regulating the expression levels of inhibitor of nuclear factor κB
kinase subunit β (IKKβ) or inhibitor of nuclear factor κB kinase
subunit α (IKKα) in ESCC. Thus, the aim of the current study was to
investigate this hypothesis and establish the role of survivin in
the activation of NF-κB p65 in ESCC.
Materials and methods
Tumor tissue specimens
Forty pairs of ESCC and healthy adjacent esophageal
tissue samples were obtained from surgically excised specimens of
ESCC from patients at the Affiliated Cancer Hospital of Xinjiang
Medical University (Ürümqi, China) between July and December 2013.
The tumor and adjacent healthy tissues were frozen in liquid
nitrogen immediately following resection. The patients in the
current study had not received chemotherapy or radiation therapy
prior to surgery. The present study was approved by the Ethics
Committee of the Affiliated Cancer Hospital of Xinjiang Medical
University. Written informed consent was provided by the families
of all of the patients.
Cell culture
The Eca109 cell line was purchased from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China). The
cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10%
heat-inactivated fetal bovine serum (FBS; Zhejiang Tianhang
Biological Technology Co., Ltd., Zhejiang, China), penicillin (100
U/ml) and streptomycin (100 mg/ml) (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified 5% CO2
atmosphere.
The KYSE150 cell line was purchased from the Beijing
Institute of Cancer (Beijing, China). The cells were cultured in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% heat-inactivated FBS,
penicillin (100 U/ml) and streptomycin (100 mg/ml) at 37°C in a
humidified 5% CO2 atmosphere.
LV3-survivin shRNA interference plasmid
construction
The LV3 vector was purchased from Shanghai
GenePharma Co., Ltd., Shanghai, China). The small hairpin RNA
(shRNA) targeting sequences for survivin and the control are
presented in Table I. The
sequences were inserted between the HindIII and XhoI
sites of the LV3 vector, which yielded LV3-survivin shRNA and
LV3-control shRNA plasmids (synthesized by Shanghai GenePharma Co.
Ltd.). The constructs were verified by DNA sequence analysis.
 | Table IshRNA targeting sequences for
survivin and the control. |
Table I
shRNA targeting sequences for
survivin and the control.
| Target gene | shRNA targeting
sequence, 5′→3′ |
|---|
| Survivin |
GAAAGTGCGCCGTGCCATCTTCAAGAGAGATGGCACGGCGCACTTTCTT |
| Control |
GCGCGCACAATCTACGCTAGTTTCAAGAGAACTAGCGTAGATTGTGCGCGCTT |
GV142-survivin overexpression plasmid
construction
The GV142 plasmid was purchased from GeneChem Co.,
Ltd. (Shanghai, China). For the GV142-survivin overexpression and
GV142-control plasmid construction, GV227 (GeneChem Co., Ltd.) was
used as the template, and the survivin and control polymerase chain
reaction (PCR) primers used are presented in Table II. The resulting PCR products were
inserted into the GV142 vector between HindIII and
XhoI sites, yielding GV142-survivin overexpression and
GV142-control plasmids.
 | Table IIPrimers for GV142-survivin
overexpression plasmid and the GV142-control plasmid
construction. |
Table II
Primers for GV142-survivin
overexpression plasmid and the GV142-control plasmid
construction.
| Target gene | Primers, 5′→3′
|
|---|
| Forward | Reverse |
|---|
| Survivin |
TGCCAAGCTTATGGGTGCCCCGACGTTGC |
TCCGCTCGAGTATCCATGGCAGCCAGCTGCTC |
| Control |
TTATGGGTGCCCCGACGTTGC |
TCCGCTCGAGTATCCTGCCAAGCATGGCAGCCAGCTGCTC |
Plasmid transient transfection
Prior to transfection, 2×105 cells/well
were placed into 6-well plates cultivated in serum-free culture
medium and antibiotics, and grown overnight until they reached
70–80% confluence. Plasmid transfection was performed using
Lipofectamine™ 2000 transfection reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. The plasmids and
Lipofectamine™ 2000 were diluted in Opti-MEM I Reduced Serum Medium
(Thermo Fisher Scientific, Inc.) separately and incubated for 10
min at room temperature. The diluted solutions were mixed and
incubated for 20 min at room temperature. Subsequently, the
mixtures were added to each well containing cells and medium. The
cells with only the transfection reagent served as a blank control.
Cell culture plates were incubated for 6 h at 37°C in a
CO2 incubator. Culture medium containing 10% FBS was
added and cells were incubated under the above-mentioned
conditions.
YM155 treatment and cell viability
assay
Cells were seeded in 96-well plates at a density of
5×103 cells/well in a volume of 100 μl culture
medium per well. After 24 h, cells were exposed to survivin
inhibitor, YM155 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
at various concentrations (0, 0.005, 0.05, 0.5, 5 and 50 μM)
for 48 h. Following incubation at 37°C, 0.4% Trypan Blue solution
(Sigma-Aldrich, St. Louis, MO, USA) was added to each well.
Viability was assessed by counting the relative number of live,
unstained cells to total cells, and the half-inhibitory
concentration (IC50) for YM155 was derived from a
logarithmic plot.
Cells treated with YM155 (concentrations at, above
and below the IC50) were harvested at 48 h, and the
total RNA and protein was extracted to determine the expression
levels of survivin, IKKα, IKKβ and NF-κB p65 by reverse
transcription quantitative (RT-q) PCR and western blotting.
RT-qPCR
At 48 h post-transfection, total cellular RNA was
extracted using TRIzol (Thermo Fisher Scientific, Inc.) following
the manufacturer's protocol. Complementary DNA (cDNA) was
synthesized from 1 μg total RNA using Maloney Murine
Leukemia Virus Reverse Transcriptase (Promega Corporation, Madison,
WI, USA) and an oligo-deoxy-thymine nucleotide primer (Promega
Corporation) according to the manufacturer's protocols. DNA content
was measured using a UV/visible spectrophotometer (Ultrospec 2000;
GE Healthcare Life Sciences, Chalfont, UK). Primer sequences are
presented in Table III. Primers
used in the current study were synthesized by Shanghai Shenggong
Biology Engineering Technology Service, Ltd. (Shanghai, China).
RT-qPCR assays used the TaqMan® Fast Virus 1-Step Master
mix kit (Thermo Fisher Scientific, Inc.). The reaction system was:
12 μl SYBR® green reagent (Thermo Fisher
Scientific, Inc.), 0.2 μM each primer, 1 μl cDNA
template, and 6 μl nuclease-free distilled water (Thermo
Fisher Scientific, Inc.). GAPDH served as an internal standard to
evaluate the relative expression levels of the target genes. qPCR
analysis was performed on an Applied Biosystems® 7500
Fast Real Time PCR instrument (Thermo Fisher Scientific, Inc.). The
PCR conditions, performed for 40 cycles, were as follows: 2 min at
50°C, 2 min at 95°C, 15 sec at 95°C, 15 sec at 55–60°C, 1 min at
72°C. The relative quantification transcript levels were determined
using the 2−ΔΔCq method. Specificities of all PCR
amplifications were confirmed by melting curve analysis. All
experiments were performed in triplicate and results are presented
as the mean ± standard deviation.
 | Table IIIPrimers for survivin, NF-κB p65, IKKα
and IKKβ for quantitative reverse transcription-polymerase chain
reaction. |
Table III
Primers for survivin, NF-κB p65, IKKα
and IKKβ for quantitative reverse transcription-polymerase chain
reaction.
| Target gene | Primers, 5′→3′
| Product size,
bp |
|---|
| Forward | Reverse |
|---|
| Survivin |
ACCGAGACCTAAAGTCCAAC |
AGACAGATAGCCACGACCC | 305 |
| NF-κB |
GTCTTCGTGCTCGGTGATG |
AGGACCTCTGACCCAAATG | 147 |
| IKKβ |
TTTACAGTATGCCTCCACC |
GTTTACTCCCGTCTGCTTC | 365 |
| IKKα |
GCCCTTGTCCCTGTCCCTA |
GCAGAGTATTTCCCTTTGGTTTGA | 374 |
| GAPDH |
TCTTAGGGAAGGTCAGCATT |
GCCTCTAGTTCCYYGGCATCA | 325 |
Western blotting assays
At 48 h post-transfection, total protein was
extracted, by washing twice with ice-cold phosphate-buffered saline
(PBS; Thermo Fisher Scientific, Inc.), and resuspended in lysis
buffer [1 M Tris HCl (pH 7.4), 5M NaCl, 0.5 M ethylene glycol
tetraacetic acid, 0.5 M EDTA, NP-40, 10% SDS (Wuhan Boster
Biological Technology, Ltd., Wuhan, China), glycerine, 10
μg/μl aprotinin, 10 μg/μl leupeptin, 10
μg/μl Pepstatin A, 10 mM phenylmethylsulfonyl
fluoride, double-distilled H2O] for 30 min on ice.
Suspensions were centrifuged at 18,407 × g for 15 min at 4°C. The
supernatant containing the protein was collected and the protein
concentration of each lysate was determined by Pierce BCA protein
assay (Thermo Fisher Scientific, Inc.). Protein (20 μg) was
loaded for each sample. Proteins were denatured, subjected to
SDS-PAGE using 10–15% polyacrylamide gels (Wuhan Boster Biological
Technology, Ltd.), electrophoresed (stacking gel, 60 V for 45 min;
separating gel, 100 V for 90 min) and electrophoretically
transferred onto nitrocellulose membranes (Whatman; GE Healthcare
Life Sciences). The membranes were blocked with 5% non-fat dried
milk (Sigma-Aldrich) in Tris-buffered saline and Tween-20 (TBST;
Beijing Solarbio Science & Technology Co., Ltd.,Beijing,China)
at room temperature for 2 h. The membranes were incubated overnight
at 4°C with the following primary antibodies: Rabbit polyclonal
anti-GAPDH (1:400; cat. no. BA2913; Wuhan Boster Biological
Technology, Ltd.), which served as a loading control; rabbit
polyclonal anti-survivin (1:1,000; cat. no. sc-10811; Santa Cruz
Biotechnology Inc.); rabbit polyclonal anti-phosphorylated
(p)-NF-κB p65 (pSer536) (1:1,000; cat. no. AF2006;
Affinity Biosciences, Cell Signal Transduction, Cincinnati, OH,
USA); rabbit polyclonal anti-NF-κB p65 (1:400; cat. no. BA0610;
Wuhan Boster Biological Technology, Ltd.); rabbit polyclonal
anti-IKKα (1:400; cat. no. BA1594-2; Wuhan Boster Biological
Technology, Ltd.); and rabbit polyclonal anti-IKKβ (1:400; cat. no.
BA4458-2; Wuhan Boster Biological Technology, Ltd.) were blocked in
TBST. Membranes were washed three times (10 min per wash) with TBST
at room temperature. Subsequently, the membranes were incubated
with appropriate horseradish peroxidase-linked goat anti-rabbit
secondary antibodies at a dilution of 1:1,000 (cat. no. BA1054;
Wuhan Boster Biological Technology, Ltd.) diluted in TBST for 1 h
at room temperature. Protein bands were visualized using an
Enhanced Chemiluminescence Detection kit (Thermo Fisher Scientific,
Inc.).
Luciferase reporter gene assay
The human IKKβ promoter sequences (3,000 bp
upstream) and random control sequence (size, 3,000 bp) were
obtained by PCR amplification and inserted between the
HindIII and XhoI sites of the pGL3 vector (Promega
Corporation) yielding pGL3-IKKβ and pGL3-random control plasmids
(synthesized by GeneChem Co., Ltd.). Eca109 and KYSE150 cells were
seeded at 5×105 cells per well in 6-well dishes one day
prior to transfection. The cells were co-transfected with 0.1
μg pGL3-random control, pGL3-IKKβ or pGL3-basic firefly
luciferase reporter construct, 0.01 μg pRL-TK Renilla
luciferase reporter plasmid and the GV142-survivin overexpression
plasmid, using Lipofectamine™ 2000. pRL-TK Renilla
luciferase reporter plasmid was co-transfected to assess the
transfection efficiency. Post-transfection (48 h), cells were
harvested and lysed with 1X lysis buffer (Promega Corporation).
Cell extracts (20 μl) were assayed for luciferase activity
using the Dual-luciferase Reporter assay system kit (Promega
Corporation) according to the manufacturer's protocols. Relative
levels of reporter gene expression were expressed as ratios of
firefly luciferase activity to Renilla luciferase (LU/RL).
All experiments were performed in triplicate.
Chromatin immunoprecipitation assay
(ChIP)
ChIP assays were performed using the EZ ChIP™ kit
(EMD Millipore, Billerica, MA, USA) according to the manufacturer's
protocols. Eca109 and KYSE150 cells were transfected with the
GV142-survivin overexpression plasmid and fixed with 1%
formaldehyde (Sigma-Aldrich) for 10 min. The Eca109 and KYSE150
cells were washed twice with 1X PBS, lysed, and sonicated to reduce
DNA lengths to within the range of 200–1,000 bp. The survivin/DNA
complexes were incubated with 4 μg rabbit antibody against
survivin, 1 μl Normal Mouse IgG (dilution, 1:1,000), which
served as the negative control, and 1 μl Anti-RNA Polymerase
II (dilution, 1:1,000; both included in the EZ-ChIP™ kit) served as
the positive control. The mixes were incubated at room temperature
for 60–90 min. The immune complexes were precipitated, eluted,
reverse-crosslinked and treated with proteinase K [Tiangen Biotech
(Beijing) Co.,Ltd., Beijing, China]. The resulting DNA samples were
amplified using primers for the putative survivin site in the human
IKKα and IKKβ promoter region. The primer sequences are presented
in Table IV. PCR fragments were
separated and visualized on 1.8% agarose gels stained with ethidium
bromide (Shanghai Bioleaf Biotech Co., Ltd., Shanghai, China). The
ratios of the PCR products of survivin (IKKα, IKKβ and NF-κB p65)
to GAPDH were used to determine the expression levels of target
genes.
 | Table IVPromoter-specific primers for IKKα
and IKKβ used in the chromatin immunoprecipitation assay. |
Table IV
Promoter-specific primers for IKKα
and IKKβ used in the chromatin immunoprecipitation assay.
| Target gene | Primer, 5′→3′ | Product site,
bp |
|---|
| IKKα
promoter-1 | F:
TGTGGATGGAGGCGTAGAG
R: AGCCAGAAGGGAAGAATGAG | Upstream,
2596–2576 |
| IKKα
promoter-2 | F:
GAATCCTCCAGGGAGACCAAAGTAA
R: TTACTTTGGTCTCCCTGGAGGATTC | Upstream,
1758–1737 |
| IKKα
promoter-3 | F:
CCCTGACATAACCCCAGCCACA
R: ACAGCCCCACCATCCCCATT | Upstream,
267–247 |
| IKKα
promoter-4 | F:
GCCCTTCAGGAGCAACTAA
R: TGACGCCTACCATAGCACTC | Upstream,
1695–1675 |
| IKKα
promoter-5 | F:
TCAAGGCGATAATGCTCACT
R: TCTCCACTTTCAGCCGTTT | Upstream,
801–781 |
| IKKα
promoter-6 | F:
CAAGGTGGACTAGGGTTGTAAA
R: TGTGAGAATCTAATGCCTGATG | Upstream,
1402–1424 |
| IKKα
promoter-7 | F:
AAATAACTTGCTCCATACCCTG
R: GGGGAATGGCAGTTGTGA | Upstream,
788–808 |
| IKKα
promoter-8 | F:
TCTTGGGTAGGGAAGTATGGG
R: GGTCTGGGAAGTCTTGCTTTA | Upstream,
1197–1006 |
| IKKα
promoter-9 | F:
AGTTTATAGGGCAAGAATCGAG
R: GGTAAAGGTAGTATTGGGCAAC | Upstream,
176–156 |
| IKKβ
promoter-1 | F:
AAAGAAAGAAACCAAGTAGCCG
R: TGAGGTATTGATAGCAGCAGTG | Upstream,
270–248 |
| IKKβ
promoter-2 | F:
TCTTCAGGTTCCTTTGGTAGTT
R: TGAGTTTCTCCGTTTTATGGG | Upstream,
860–849 |
| IKKβ
promoter-3 | F:
GGTAGGCAAGGGCAGTTCT
R: GACCGTGCTCACCGATTT | Upstream,
1520–1502 |
Flow cytometric analysis
For cell cycle analysis, DNA labeling was conducted
using the Cycle Test Plus DNA reagent kit (BestBio Co., Ltd.,
Shanghai, China). Labeling with propidium iodide (PI) and Annexin V
was performed using an Annexin V staining kit (BestBio Co. Ltd.)
for the detection of apoptotic cells and the assays were performed
according to the manufacturer's protocols. Eca109 and KYSE150 cells
were directly incubated, at 37°C for 48 h, in 6-well plates and
collected 48 h following transfection. For the cell cycle analysis,
the cells were washed with PBS for 5 min and subsequently
centrifugation at 900 × g. The cells were collected and fixed in
ice-cold 70% ethanol (Saihongrui Biotechnology Co., Ltd., Nanjing,
China) for a minimum of 2 h at 4°C, followed by treatment with 0.2
mg/ml RNase A (EMD Millipore) in PBS for 30 min at 37°C. PI was
added (final concentration, 25 μg/ml) and the cells were
incubated for 30 min at 4°C in the dark. Analysis of the samples
was conducted within 24 h. For the apoptosis assay, the transfected
cells were washed twice with ice-cold PBS, and resuspended in 195
μl 1X Binding Buffer (EMD Millipore) to a concentration of
1×104 cells/ml. Annexin V (5 μl) and PI were
gently mixed with the cells and incubated for 15 min at room
temperature in the dark. The dyes were washed out by centrifugation
for 5 min at 94 × g and the cells were resuspended in 190 μl
1X Binding Buffer. PI staining solution (10 μl) was gently
mixed in and incubated on ice and in the dark. The samples were
analyzed within 1 h. All samples for the two assays consisted of
10,000 cells and were analyzed by fluorescence-activated cell
sorting with a BD FACSMicroCount™ system (BD Biosciences, Franklin
Lakes, NJ, USA).
Statistical analysis
SPSS version 17.0 for Windows (SPSS, Inc., Chicago,
IL, USA) was used for all statistical analyses. All data were
expressed as the mean ± standard deviation from three experiments.
The two-tailed Student's t-test was used to analyze the difference
between groups and Fisher's exact test was used to analyze
correlation between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Survivin expression is positively
correlated with IKKα and IKKβ expression in ESCC tissue
samples
The expression levels of survivin, IKKα, IKKβ and
NF-κB p65 were evaluated by RT-qPCR in 40 paired ESCC and healthy
tissue samples, as presented in Table
V. In the present study, the expression of survivin was
observed to be positively correlated with IKKα (r =0.370;
P<0.05) and IKKβ mRNA expression levels (r =0.341; P<0.05) in
ESCC samples.
 | Table VAssociation between survivin and
NF-κB p65, IKKα, and IKKβ mRNA expression levels in esophageal
squamous cell carcinoma samples. |
Table V
Association between survivin and
NF-κB p65, IKKα, and IKKβ mRNA expression levels in esophageal
squamous cell carcinoma samples.
| Parameter | IKKα
| IKKβ
| NF-κB p65
|
|---|
| + | − | + | − | + | − |
|---|
| Survivin |
| + | 22 | 4 | 20 | 6 | 16 | 10 |
| − | 7 | 7 | 6 | 8 | 10 | 4 |
| Statistical
value |
| r | 0.370 | 0.341 | 0.154 |
| P-value | 0.019 | 0.031 | 0.350 |
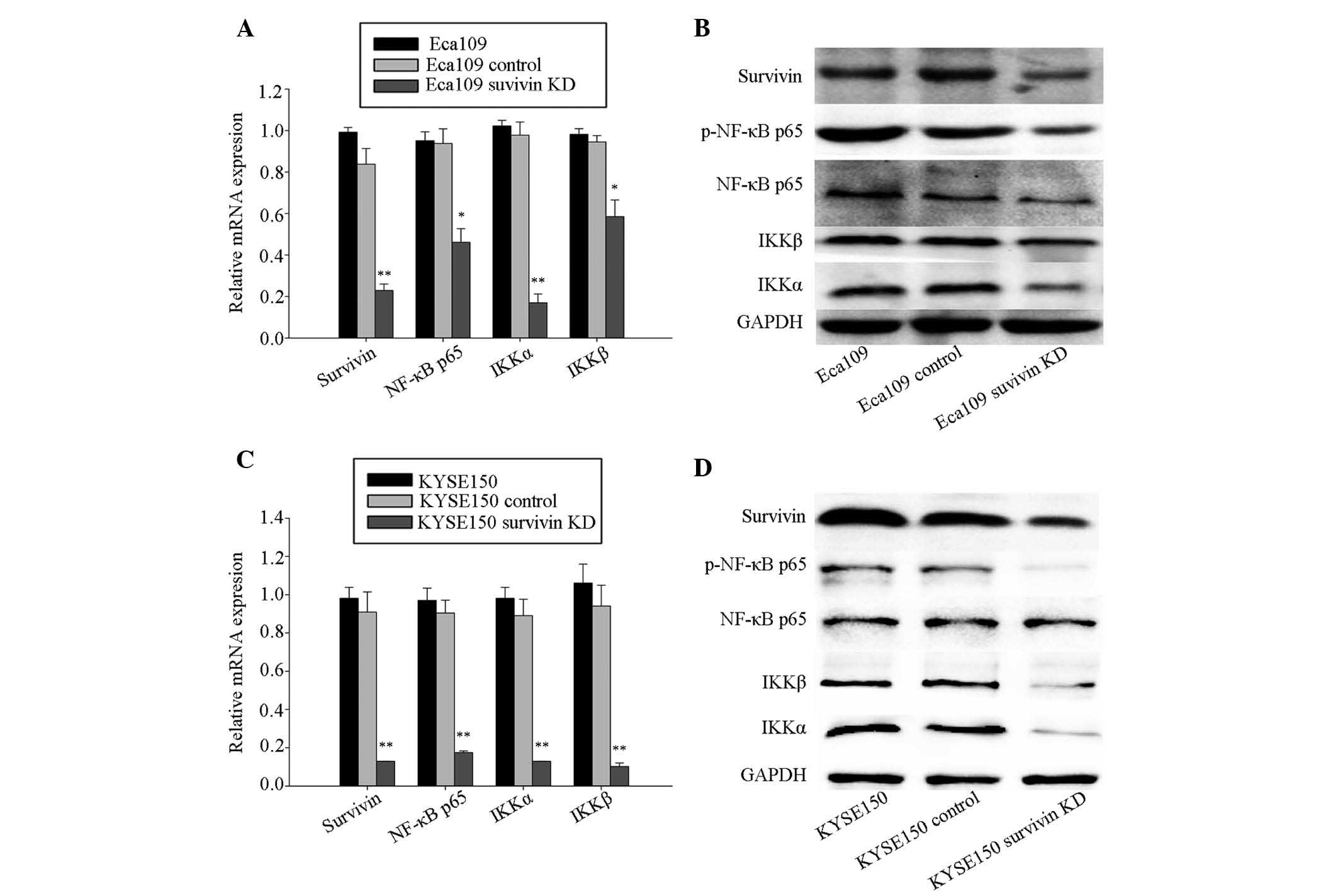
Survivin knockdown deactivates NF-κB
signaling in ESCC cells
Cells transfected with LV3-survivin shRNA and
LV3-control shRNA plasmids were designated the survivin knockdown
group and control group, respectively. The cells cultured with the
transfection reagent only were considered as a blank group. At 48 h
after transfection, cells were harvested for RT-qPCR and western
blotting assays. The RT-qPCR analysis (Fig. 1) demonstrated that expression
levels of survivin, NF-κB p65, IKKα and IKKβ were significantly
reduced in the survivin knockdown group, compared with the control
and blank group in Eca109 (Fig.
1A) and KYSE150 cells (Fig.
1C). Western blotting analysis also demonstrated that protein
expression levels of survivin, the p-NF-κB p65, IKKα and IKKβ
expression levels were similarly reduced in the survivin knockdown
group of the two cell lines (Fig. 1B
and D).
YM155 reduces cell viability and
downregulates survivin, NF-κB p65, IKKα and IKKβ
YM155, an inhibitor of survivin, was used to
investigate the effect of survivin on cell viability, as well as
the interaction between survivin and NF-κB p65, IKKα, and IKKβ
expression levels in Eca109 and KYSE150 cells. YM155 was
administered at concentrations from 0.005 to 50 μM, which
significantly reduced cell viability in a dose-dependent manner in
the Eca109 and KYSE150 cells at 48 h following administration.
According to the logarithmic curve, the IC50 of YM155
for Eca109 and KYSE150 cells was ~0.5 μM (Fig. 2A). Concentrations at, above, and
below the IC50 (0.05, 0.5 and 5 μM) were selected
for further experiments.
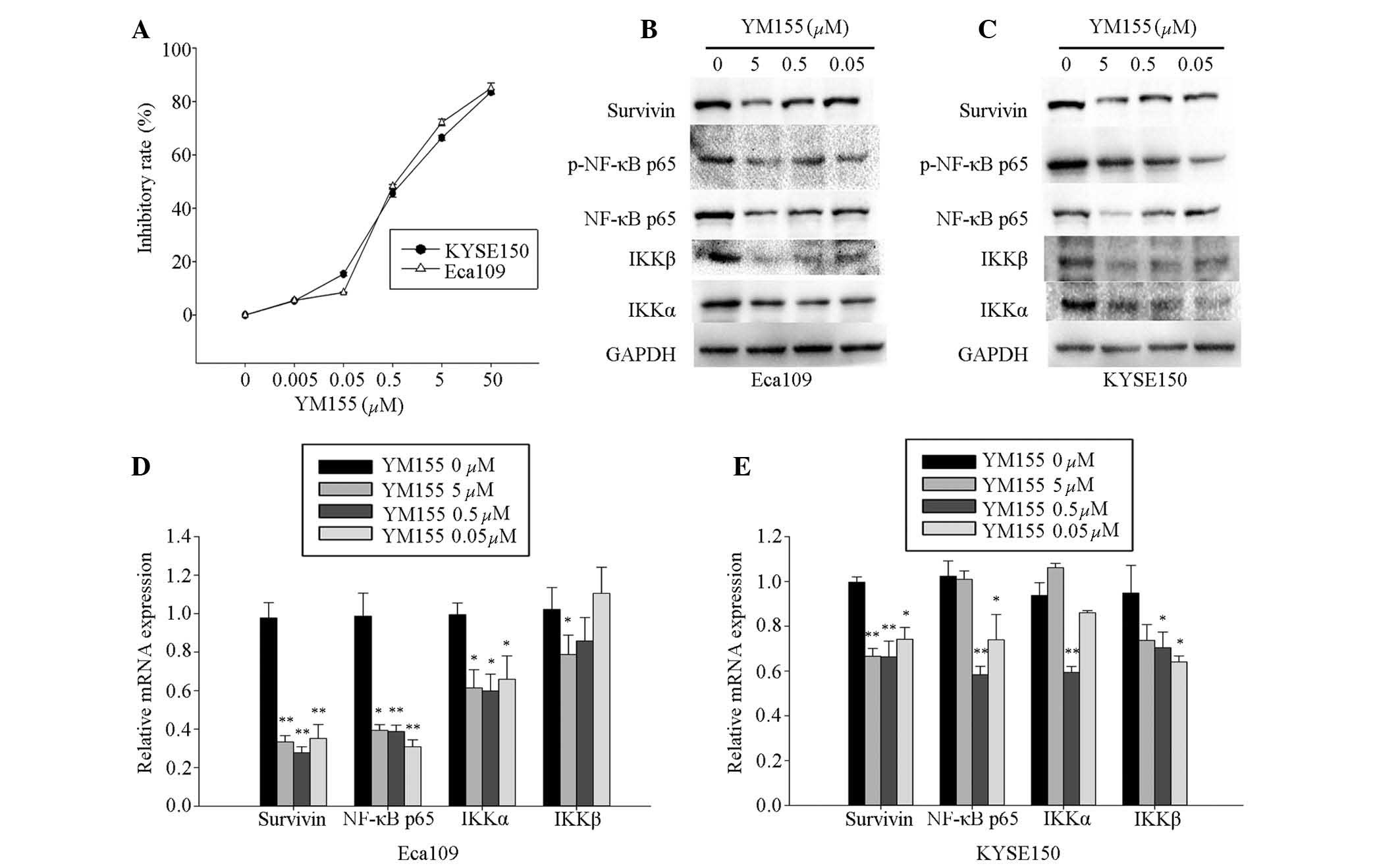 | Figure 2YM155 inhibited the expression of
survivin, NF-κB p65, IKKα and IKKβ in Eca109 and KYSE150 cells. (A)
Eca109 and KYSE150 cells were incubated with 0.005, 0.05, 0.5, 5
and 50 μM YM155 for 48 h. Cells incubated with the culture
medium served as the control group. Experiments were performed in
triplicate. Eca109 and KYSE150 cells were treated with 0 (blank),
0.05, 0.5, and 5.0 μM YM155. The expression of survivin,
NF-κB p65, IKKβ and IKKα were analyzed by (B and C) western
blotting and (D and E) RT-qPCR. Western blotting of survivin,
p-NF-κB p65, NF-κB p65, IKKβ and IKKα proteins was conducted using
total protein isolated from cells, with GADPH serving as a loading
control. Transcript levels were measured by RT-qPCR of total
isolated RNA, with GADPH serving as an internal control. YM155
inhibited expression of survivin and NF-κB p65, IKKα and IKKβ in (B
and D) Eca109 and (C and E) KYSE150 cells. Columns demonstrate the
mean values from triplicate experiments and the error bars indicate
standard deviation. *P<0.05; **P<0.01
vs. control. RT-qPCR, quantitative reverse transcription-polymerase
chain reaction; NF-κB p65, transcription factor p65; IKKα,
inhibitor of nuclear factor κB kinase subunit α; IKKβ, inhibitor of
nuclear factor κB kinase subunit β; p, phosphorylated; mRNA,
messenger RNA. |
Eca109 and KYSE150 cells were treated with YM155 at
the selected concentrations for 48 h, and the expression levels of
survivin and NF-κB p65, IKKα and IKKβ were measured by RT-qPCR and
western blotting assays. The results demonstrated that YM155
effectively inhibited mRNA expression levels of survivin and NF-κB
p65, IKKα and IKKβ, and protein expression levels of survivin and
the p-NF-κB p65, IKKα and IKKβ in Eca109 (Fig. 2B and D) and KYSE150 (Fig. 2C and E) cells, which was comparable
with the survivin shRNA knockdown.
Survivin overexpression activates NF-κB
signaling in ESCC cells
Cells transfected with GV142-survivin overexpression
plasmid and GV142-control plasmid were designated the survivin
overexpression group and the control group, respectively. The cells
with the transfection reagent only served as the blank group. At 48
h after transfection, cells were harvested for RT-qPCR and western
blotting assays. RT-qPCR analysis demonstrated increased mRNA
expression levels of survivin, NF-κB p65, IKKα and IKKβ in the
survivin overexpres-sion group, when compared with the control
group and the blank control group in Eca109 (Fig. 3A and B) and KYSE150 (Fig. 3C and D) cells. In addition, western
blotting analysis indicated that the protein levels of survivin,
and phosphorylation of NF-κB p65 and IKKβ were also increased in
the survivin overexpression group in the Eca109 (Fig. 3B) and KYSE150 (Fig. 3D) cells. Furthermore, KYSE150 cells
demonstrated increased expression levels of IKKα protein (Fig. 3D).
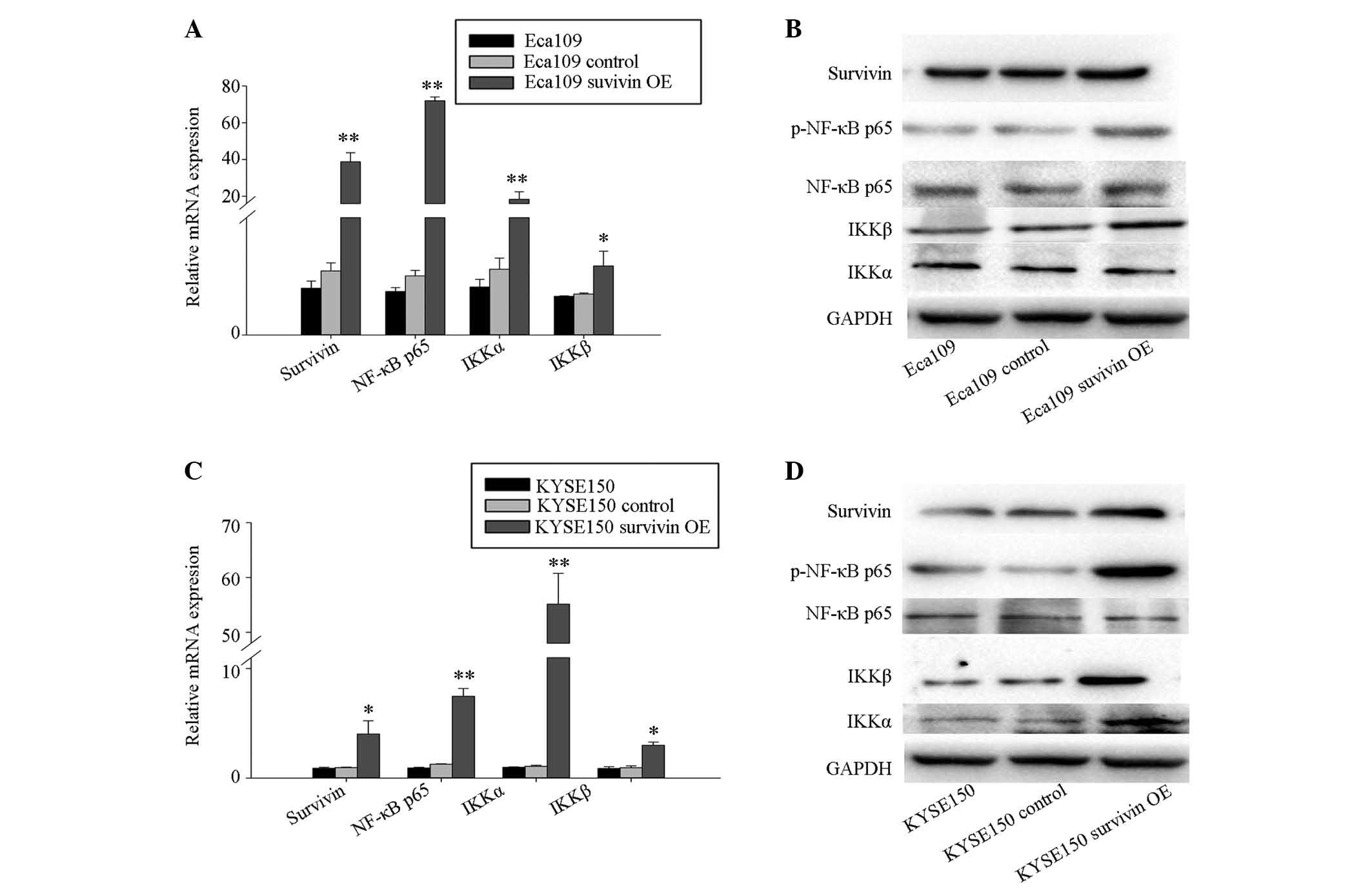 | Figure 3Survivin overexpression results in
upregulation of NF-κBp65, IKKα, IKKβ in Eca109 and KYSE150cells. (A
and B) Eca109 and (C and D) KYSE150 cells were transfected with
GV142-survivin overexpression plasmid (Eca109 survivin OE and
KYSE150 survivin OE), GV142-control plasmid (Eca109 control and
KYSE150 control), and mock transfection. Expression levels of
survivin, NF-κB p65, IKKβ, and IKKα were analyzed by (A and C)
RT-qPCR and (B and D) western blotting of total protein. Transcript
levels were measured by RT-qPCR of total isolated RNA, with GADPH
serving as an internal control. Columns indicate the mean values
from triplicate experiments and the error bars indicate standard
deviation. *P<0.05; **P<0.01 vs.
control. OE, overexpression; RT-qPCR, quantitative reverse
transcription-polymerase chain reaction; NF-κB p65, transcription
factor p65; IKKα, inhibitor of nuclear factor κB kinase subunit α;
IKKβ, inhibitor of nuclear factor κB kinase subunit β; p,
phosphorylated; mRNA, messenger RNA. |
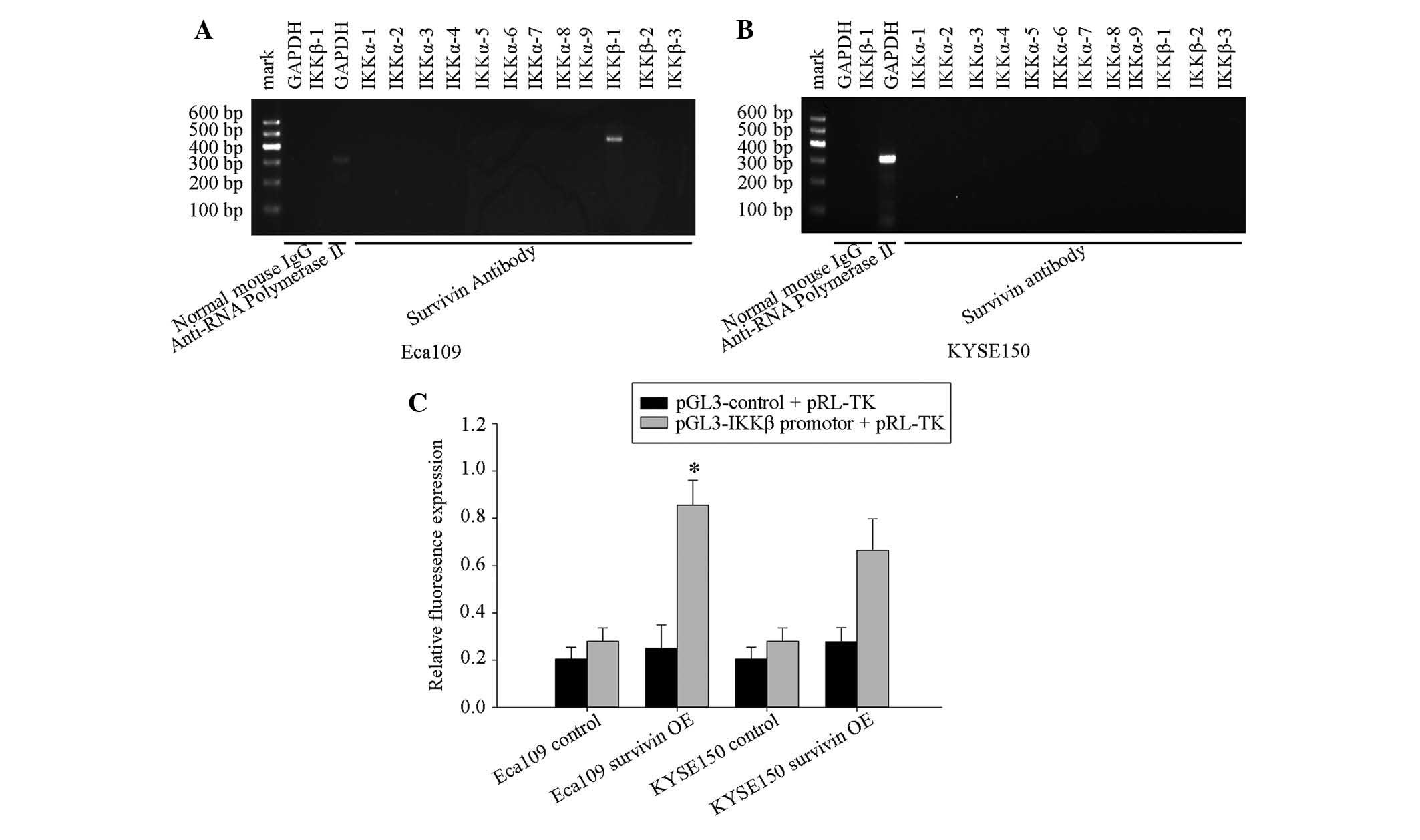
Survivin binds to IKKβ promoter and
increases the transcriptional activity of the IKKβ promoter in
Eca109 cells
To further determine whether survivin recognized and
bound to the IKKβ promoter in vivo, and increased its
transcriptional activity, the ChIP assay was performed in Eca109
cells. DNA was immunoprecipitated using anti-survivin polyclonal
antibodies and subjected to PCR with promoter-specific primers for
IKKβ. Positive amplification in Eca109 cell lines demonstrated that
survivin may bind the upstream 700 bp IKKβ promoters (Fig. 4A and B).
To investigate whether overexpression of survivin
affects the promoter transcriptional activities of IKKβ in Eca109
and KYSE150 cells, a Luciferase reporter gene assay was performed
and overexpressed survivin was observed to significantly increase
the transcriptional activity of the IKKβ promoter in Eca109 cells,
but not in the KYSE150 cells (Fig.
4C).
Survivin knockdown induces apoptosis and
G2/M phase arrest in vitro
To analyze the effect of survivin on apoptosis, flow
cytometric analysis with PI and Annexin V staining was performed
(Fig. 5). Results demonstrated
that surviving knockdown and subsequent reduction in activation of
NF-κB p65 increased apoptosis in Eca109 (Fig. 5C and D) and KYSE150 (Fig. 5G and H) cells. Conversely, survivin
overexpression and concomitant activation of NF-κB p65 decreased
apoptosis in Eca109 (Fig. 5A and
B) and KYSE150 (Fig. 5E and F)
cells.
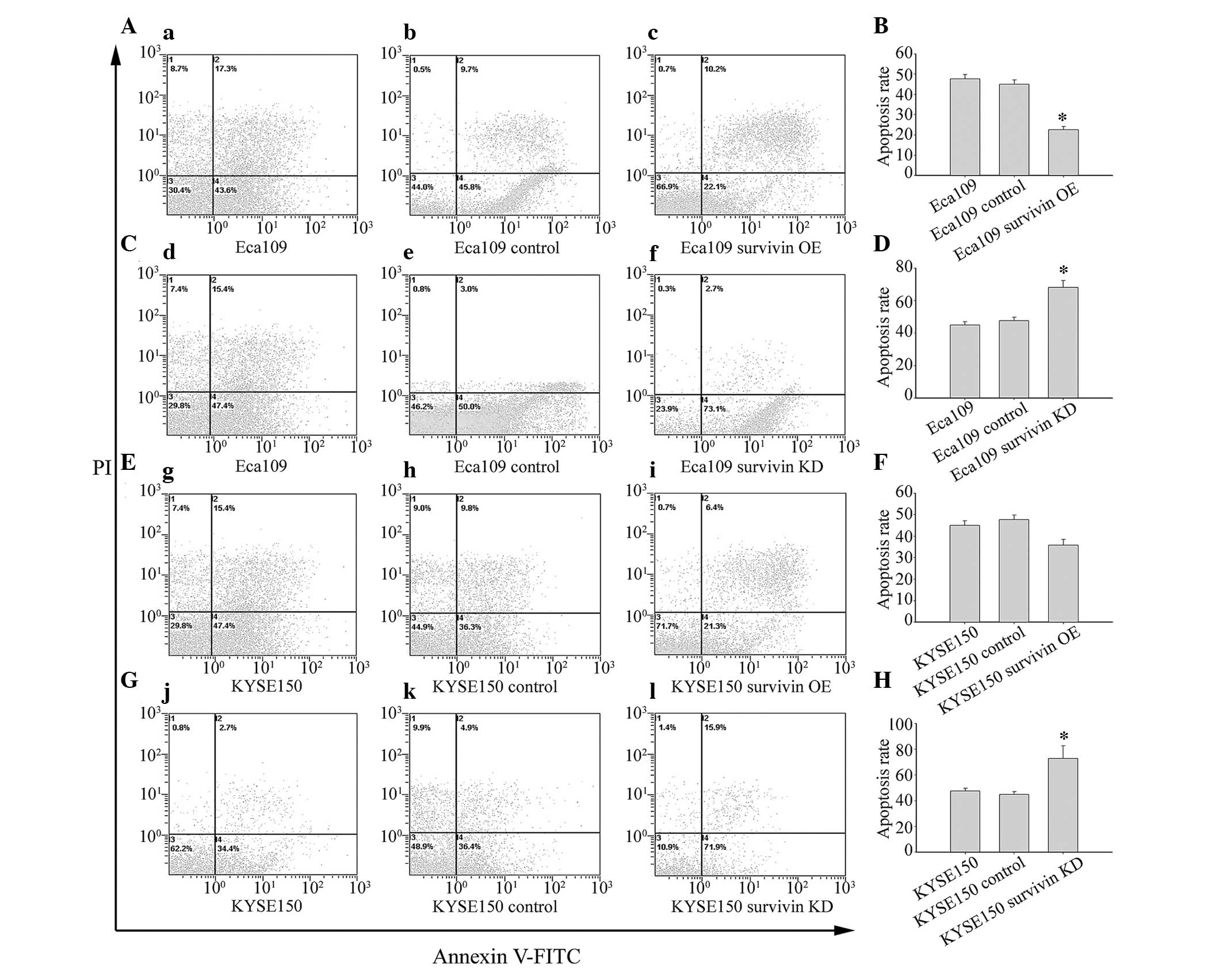 | Figure 5Survivin overexpression significantly
inhibited, whereas survivin knockdown significantly induced,
apoptosis in Eca109 cells. (A–D) Eca109 and (E–H) KYSE150 cells
were transfected with GV142-survivin overexpression plasmid (c,
Eca109 survivin OE; i, KYSE150 survivin OE), GV142-control plasmid
(b, Eca109 control; h, KYSE150 control), and transfection reagent
alone (a, Eca109; g, KYSE150); or LV3-survivin shRNA plasmid (f,
Eca109 survivin KD; l, KYSE150 survivin KD), LV3-control plasmid
(e, Eca109 control; k, KYSE150 control), and transfection reagent
alone (d, Eca109; j, KYSE150). Cells were stained with Annexin V
and analyzed by FACScan flow cytometry. Columns demonstrate the
mean values from triplicate experiments and the error bars indicate
standard deviation. *P<0.05; **P<0.01
vs. control. OE, overexpression; FITC, fluorescein isothiocyanate;
PI, propidium iodide; KD, knockdown. |
To analyze the effect of survivin on the cell cycle,
flow cytometry was performed. The results indicated that survivin
knockdown increased the fraction of cells arrested in the
G2/M phase in Eca109 and KYSE150 cells (Fig. 6).
 | Figure 6Survivin overexpression significantly
increased the cell number in the G2/M phase, whereas
survivin knockdown significantly delayed the cell cycle in the
G2/M phase in Eca109 and KYSE150 cells. (A–D) Eca109 and
(E-H) KYSE150 cells were transfected with GV142-survivin
overexpression plasmid (c, Eca109 survivin OE; i, KYSE150 survivin
OE), GV142-control plasmid (b, Eca109 control; h, KYSE150 control),
and transfection reagent alone (a, Eca109; g, KYSE150); or
LV3-survivin shRNA plasmid (f, Eca109 survivin KD; l, KYSE150
survivin KD), LV3-control plasmid (e, Eca109 control; k, KYSE150
control), and transfection reagent alone (d, Eca109; j, KYSE150).
The cell cycle distribution was assessed by FACScan flow cytometry.
Columns demonstrate mean values from triplicate experiments and the
bars indicate standard deviation. *P<0.05,
**P<0.01 vs. control group. KD, knockdown; OE,
overexpression. |
Discussion
Survivin, the smallest member of the IAP family, is
overexpressed in ESCC. Survivin detection is correlated with the
clinical stage, metastasis, relapse rate and the overall survival
of ESCC patients, and provides valuable information to predict the
prognosis of ESCC (48,49). The present study demonstrated that
survivin overexpression inhibited cell apoptosis and induced cell
proliferation. Conversely, survivin knockdown increased cell
apoptosis; thus, as survivin inhibits apoptosis it is involved in
the progression of ESCC.
Activated NF-κB has been associated with
acid-induced esophageal epithelial cell transformation (50). NF-κB nuclear expression was
significantly increased in ESCC tissue samples compared with
healthy esophageal tissues (51–53).
NF-κB activation indicates a poorly differentiated cancer and is
associated with a low survival rate in ESCC patients (54). Interference with the NF-κB
signaling pathway increases the chemotherapeutic sensitivity in
ESCC, and suppresses metastasis (55), as activated NF-κB p65 is involved
in the progression of ESCC via activating multiple
apoptosis-associated genes, including survivin (56,57).
However, the underlying mechanism of NF-κB p65 activation in ESCC
remains unclear. In the present study, it was observed that
survivin regulated the expression of IKKα, IKKβ, and NF-κB p65 in
Eca109 and KYSE150 cells. Knockdown of survivin was demonstrated to
deactivate NF-κB p65, which induced cell apoptosis and arrested
cells in the G2/M phase. Notably, survivin
overexpression activated NF-κB p65, inhibiting cell apoptosis and
indicating that activation of NF-κB p65 by survivin is potentially
important in cell apoptosis, cell proliferation and the progression
of ESCC.
NF-κB dimers are observed in the majority of resting
cells and retained in the cytosol via interaction with one of the
prototypical IκB proteins (58).
Extracellular stimulating factor induces degradation of IκB kinase
(IKK) proteins upon their phosphorylation by the IKK complex, and
inducing NF-κB dimer translocation into the nucleus and resulting
in target gene transcription (59). IKKα and IKKβ are important IKKs,
which phosphorylate nuclear factor of κ light polypeptide gene
enhancer in B-cells inhibitor, α (IκBα) proteins and determine
NF-κB cytosolic localization (60). The present study demonstrated that
overexpression of survivin increased the expression and
transcriptional activity of IKKβ in Eca109 cells by binding to the
IKKβ promoter. Overexpression of survivin may result in binding of
survivin to the IKKβ promoter and increase the transcriptional
activity of IKKβ, which phosphorylates IκBα and releases NF-κB p65
to translocate into the nucleus.
In conclusion, survivin performs its biological
functions by affecting cell apoptosis and proliferation, and
increases the activity of the inducible transcription factor, NF-κB
p65 via maintaining a high expression level of IKKβ and
upregulating the phosphorylation level of IκBα via IKKβ, and
finally releasing NF-κB p65 from the cytoplasm to the nucleus in
ESCC cells.
The present study provides valuable data toward an
increased understanding of constant high expression and activation
of NF-κB p65 in ESCC. In addition, investigation into the
underlying mechanisms of survivin/NF-κB p65 regulation in the
tumorigenesis and progression of ESCC may result in the development
of a novel biomarker for the early diagnosis and personalized
therapeutic strategies for the treatment of ESCC.
Acknowledgments
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81460359).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay vvJ,
Ward E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holmes RS and Vaughan TL: Epidemiology and
pathogenesis of esophageal cancer. Semin Radiat Oncol. 17:2–9.
2007. View Article : Google Scholar
|
|
3
|
Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y,
Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal
cancer in Japan and China. J Epidemiol. 23:233–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu KS, Huo X and Zhu GH: Relationships
between esophageal cancer and spatial environment factors by using
Geographic Information System. Sci Total Environ. 393:219–225.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey
SM, Dong ZW, Mark SD, Qiao YL and Taylor PR: Prospective study of
risk factors for esophageal and gastric cancers in the Linxian
general population trial cohort in China. Int J Cancer.
113:456–463. 2005. View Article : Google Scholar
|
|
6
|
Muñoz N, Crespi M, Grassi A, Qing WC,
Qiong S and Cai LZ: Precursor lesions of oesophageal cancer in
high-risk populations in Iran and China. Lancet. 1:876–879. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blot WJ, Li JY, Taylor PR, Guo W, Dawsey
SM and Li B: The Linxian trials: Mortality rates by vitamin-mineral
intervention group. Am J Clin Nutr. 62(Suppl): 1424S–1426S.
1995.PubMed/NCBI
|
|
8
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen CP, Braunstein S, Mourad M, Hsu IC,
Haas-Kogen D, Roach M III and Fogh SE: Quality improvement of
International Classification of Diseases, 9th revision, diagnosis
coding in radiation oncology: Single-institution prospective study
at University of California, San Francisco. Pract Radiat Oncol.
5:e45–e51. 2015. View Article : Google Scholar
|
|
10
|
Macefield RC, Avery KN and Blazeby JM:
Integration of clinical and patient-reported outcomes in surgical
oncology. Br J Surg. 100:28–37. 2013. View
Article : Google Scholar
|
|
11
|
Daly JM, Fry WA, Little AG, Winchester DP,
McKee RF, Stewart AK and Fremgen AM: Esophageal cancer: Results of
an American College of Surgeons Patient Care Evaluation Study. J Am
Coll Surg. 190:562–573. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lepage C, Rachet B, Jooste V, Faivre J and
Coleman MP: Continuing rapid increase in esophageal adenocarcinoma
in England and Wales. Am J Gastroenterol. 103:2694–2699. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pohl H and Welch HG: The role of
overdiagnosis and reclassification in the marked increase of
esophageal adenocarcinoma incidence. J Natl Cancer Inst.
97:142–146. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kranzfelder M, Büchler P and Friess H:
Surgery within multimodal therapy concepts for esophageal squamous
cell carcinoma (ESCC): the MRI approach and review of the
literature. Adv Med Sci. 54:158–169. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lightdale CJ; American College of
Gastroenterology: Esophageal cancer. Am J Gastroenterol. 94:20–29.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iizuka T, Isono K, Kakegawa T and Watanabe
H; Japanese Committee for Registration of Esophageal Carcinoma
Cases: Parameters linked to ten-year survival in Japan of resected
esophageal carcinoma. Chest. 96:1005–1011. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Toh Y, Egashira A and Yamamoto M:
Epigenetic alterations and their clinical implications in
esophageal squamous cell carcinoma. Gen Thorac Cardiovasc Surg.
61:262–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shang L and Wang M: Molecular alterations
and clinical relevance in esophageal squamous cell carcinoma. Front
Med. 7:401–410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takeno S, Yamashita S, Takahashi Y, Ono K,
Kamei M, Moroga T and Kawahara K: Survivin expression in
oesophageal squamous cell carcinoma: Its prognostic impact and
splice variant expression. Eur J Cardiothorac Surg. 37:440–445.
2010.
|
|
21
|
Akyürek N, Memiş L, Ekinci O, Köktürk N
and Oztürk C: Survivin expression in pre-invasive lesions and
non-small cell lung carcinoma. Virchows Arch. 449:164–170. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Altieri DC: Validating survivin as a
cancer therapeutic target. Nat Rev Cancer. 3:46–54. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang X, Xiong G, Chen X, Xu X, Wang K, Fu
Y, Yang K and Bai Y: Polymorphisms of survivin promoter are
associated with risk of esophageal squamous cell carcinoma. J
Cancer Res Clin Oncol. 135:1341–1349. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kato J, Kuwabara Y, Mitani M, Shinoda N,
Sato A, Toyama T, Mitsui A, Nishiwaki T, Moriyama S, Kudo J and
Fujii Y: Expression of survivin in esophageal cancer: Correlation
with the prognosis and response to chemotherapy. Int J Cancer.
95:92–95. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ikeguchi M and Kaibara N: Survivin
messenger RNA expression is a good prognostic biomarker for
oesophageal carcinoma. Br J Cancer. 87:883–887. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grabowski P, Kühnel T, Mühr-Wilkenshoff F,
Heine B, Stein H, Höpfner M, Germer CT and Scherübl H: Prognostic
value of nuclear survivin expression in oesophageal squamous cell
carcinoma. Br J Cancer. 88:115–119. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dabrowski A, Filip A, Zgodziński W,
Dabrowska M, Polańska D, Wójcik M, Zinkiewicz K and Wallner G:
Assessment of prognostic significance of cytoplasmic survivin
expression in advanced oesophageal cancer. Folia Histochem
Cytobiol. 42:169–172. 2004.PubMed/NCBI
|
|
28
|
Mita AC, Mita MM, Nawrocki ST and Giles
FJ: Survivin: Key regulator of mitosis and apoptosis and novel
target for cancer therapeutics. Clin Cancer Res. 14:5000–5005.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li C, Li Z, Zhu M, Zhao T, Chen L, Ji W,
Chen H and Su C: Clinicopathological and prognostic significance of
survivin over-expression in patients with esophageal squamous cell
carcinoma: A meta-analysis. PLoS One. 7:e447642012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Altieri DC: Survivin, cancer networks and
pathway-directed drug discovery. Nat Rev Cancer. 8:61–70. 2008.
View Article : Google Scholar
|
|
31
|
Altieri DC: Survivin and IAP proteins in
cell-death mechanisms. Biochem J. 430:199–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kanwar JR, Kamalapuram SK and Kanwar RK:
Targeting survivin in cancer: The cell-signalling perspective. Drug
Discov Today. 16:485–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ryan BM, O'Donovan N and Duffy MJ:
Survivin: A new target for anti-cancer therapy. Cancer Treat Rev.
35:553–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saito T, Hama S, Izumi H, Yamasaki F,
Kajiwara Y, Matsuura S, Morishima K, Hidaka T, Shrestha P, Sugiyama
K and Kurisu K: Centrosome amplification induced by survivin
suppression enhances both chromosome instability and
radiosensitivity in glioma cells. Br J Cancer. 98:345–355. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sharma H, Sen S, Lo Muzio L, Mariggiò A
and Singh N: Antisense-mediated downregulation of anti-apoptotic
proteins induces apoptosis and sensitizes head and neck squamous
cell carcinoma cells to chemotherapy. Cancer Biol Ther. 4:720–727.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Islam A, Kageyama H, Takada N, Kawamoto T,
Takayasu H, Isogai E, Ohira M, Hashizume K, Kobayashi H, Kaneko Y
and Nakagawara A: High expression of Survivin, mapped to 17q25, is
significantly associated with poor prognostic factors and promotes
cell survival in human neuroblastoma. Oncogene. 19:617–623. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hattori M, Sakamoto H, Satoh K and
Yamamoto T: DNA demethylase is expressed in ovarian cancers and the
expression correlates with demethylation of CpG sites in the
promoter region of c-erbB-2 and survivin genes. Cancer Lett.
169:155–164. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li F and Altieri DC: Transcriptional
analysis of human survivin gene expression. Biochem J. 344:305–311.
1999.PubMed/NCBI
|
|
39
|
Kawakami H, Tomita M, Matsuda T, Ohta T,
Tanaka Y, Fujii M, Hatano M, Tokuhisa T and Mori N: Transcriptional
activation of survivin through the NF-kappaB pathway by human
T-cell leukemia virus type I tax. Int J Cancer. 115:967–974. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schreck R, Albermann K and Baeuerle PA:
Nuclear factor kappa B: An oxidative stress-responsive
transcription factor of eukaryotic cells (a review). Free Radic Res
Commun. 17:221–237. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Takizawa BT, Uchio EM, Cohen JJ, Wheeler
MA and Weiss RM: Downregulation of survivin is associated with
reductions in TNF receptors' mRNA and protein and alterations in
nuclear factor kappa B signaling in urothelial cancer cells. Cancer
Invest. 25:678–684. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hayden MS and Ghosh S: Signaling to
NF-kappaB. Genes Dev. 18:2195–2224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sen R and Baltimore D: Inducibility of
kappa immunoglobulin enhancer-binding protein Nf-kappa B by a
posttranslational mechanism. Cell. 47:921–928. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Scheidereit C: IkappaB kinase complexes:
Gateways to NF-kappaB activation and transcription. Oncogene.
25:6685–6705. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim HJ, Hawke N and Baldwin AS: NF-kappaB
and IKK as therapeutic targets in cancer. Cell Death Differ.
13:738–747. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Karin M: NF-kappaB and cancer: Mechanisms
and targets. Mol Carcinog. 45:355–361. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin C, Song L, Gong H, Liu A, Lin X, Wu J,
Li M and Li J: Nkx2–8 downregulation promotes angiogenesis and
activates NF-κB in esophageal cancer. Cancer Res. 73:3638–3648.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu H, Wang Q, Hu C, Zhang W, Quan L, Liu
M, Xu N and Xiao Z: High expression of survivin predicts poor
prognosis in esophageal squamous cell carcinoma following
radiotherapy. Tumour Biol. 32:1147–1153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cao M, Yie SM, Wu SM, Chen S, Lou B, He X,
Ye SR, Xie K, Rao L, Gao E and Ye NY: Detection of
survivin-expressing circulating cancer cells in the peripheral
blood of patients with esophageal squamous cell carcinoma and its
clinical significance. Clin Exp Metastasis. 26:751–758. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Debruyne PR, Witek M, Gong L, Birbe R,
Chervoneva I, Jin T, Domon-Cell C, Palazzo JP, Freund JN, Li P, et
al: Bile acids induce ectopic expression of intestinal guanylyl
cyclase C through nuclear factor-kappaB and Cdx2 in human
esophageal cells. Gastroenterology. 130:1191–1206. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Konturek PC, Nikiforuk A, Kania J, Raithel
M, Hahn EG and Mühldorfer S: Activation of NFkappaB represents the
central event in the neoplastic progression associated with
Barrett's esophagus: A possible link to the inflammation and
overexpression of COX-2, PPARgamma and growth factors. Dig Dis Sci.
49:1075–1083. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kang MR, Kim MS, Kim SS, Ahn CH, Yoo NJ
and Lee SH: NF-kappaB signalling proteins p50/p105, p52/p100, RelA,
and IKKepsilon are over-expressed in oesophageal squamous cell
carcinomas. Pathology. 41:622–625. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kausar T, Sharma R, Hasan MR, Tripathi SC,
Saraya A, Chattopadhyay TK, Gupta SD and Ralhan R: Clinical
significance of GPR56, transglutaminase 2, and NF-κB in esophageal
squamous cell carcinoma. Cancer Invest. 29:42–48. 2011. View Article : Google Scholar
|
|
54
|
Izzo JG, Malhotra U, Wu TT, Ensor J,
Luthra R, Lee JH, Swisher SG, Liao Z, Chao KS, Hittelman WN, et al:
Association of activated transcription factor nuclear factor kappab
with chemo-radiation resistance and poor outcome in esophageal
carcinoma. J Clin Oncol. 24:748–754. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li B, Li YY, Tsao SW and Cheung AL:
Targeting NF-kappaB signaling pathway suppresses tumor growth,
angiogenesis, and metastasis of human esophageal cancer. Mol Cancer
Ther. 8:2635–2644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Song L, Gong H, Lin C, Wang C, Liu L, Wu
J, Li M and Li J: Flotillin-1 promotes tumor necrosis factor-α
receptor signaling and activation of NF-κB in esophageal squamous
cell carcinoma cells. Gastroenterology. 143:995–1005. 2012.
View Article : Google Scholar
|
|
57
|
Tian F, Zhang C, Tian W, Jiang Y and Zhang
X: Comparison of the effect of p65 siRNA and curcumin in promoting
apoptosis in esophageal squamous cell carcinoma cells and in nude
mice. Oncol Rep. 28:232–240. 2012.PubMed/NCBI
|
|
58
|
Oeckinghaus A and Ghosh S: The NF-kappaB
family of transcription factors and its regulation. Cold Spring
Harb Perspect Biol. 1:a0000342009. View Article : Google Scholar
|
|
59
|
Sakurai H, Chiba H, Miyoshi H, Sugita T
and Toriumi W: IkappaB kinases phosphorylate NF-kappaB p65 subunit
on serine 536 in the transactivation domain. J Biol Chem.
274:30353–30356. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hayden MS and Ghosh S: S hared principles
in NF-kappaB signaling. Cell. 132:344–362. 2008. View Article : Google Scholar : PubMed/NCBI
|