Introduction
Hepatocellular carcinoma (HCC) is the fifth most
frequent cancer worldwide and is also the third leading cause of
cancer-associated mortality (1).
Currently, surgical resection or liver transplantation remains the
main and effective treatment throughout the world (2). However, the recurrence rate within 2
years in patients who have undergone tumor resection remains
>50% (3). Uncontrolled tumor
metastasis, frequent intrahepatic spread and extrahepatic
metastasis are the primary causes for the poor prognosis of HCC
(4). Therefore, investigation of
the molecular mechanism of metastasis and recurrence may improve
the prognosis of patients with HCC.
MicroRNAs (miRNAs) are 21–24 nucleotide, small,
non-coding RNAs that regulate gene expression by base pairing with
target mRNAs in the 3′-untranslated region (3′-UTR), leading to
mRNA cleavage or translational repression (5). Growing evidence suggests that miRNAs
are important in various biological processes, including cell
proliferation, development and differentiation (6–8).
Furthermore, an increasing number of miRNAs have been observed in
various types of cancer and may be involved in modulating cancer
cell behavior. For example, Li et al demonstrated that
downregulated miR-646 in clear cell renal carcinoma correlated with
tumor metastasis by targeting the nin one binding protein (9). Li et al found that miR-21
overexpression was associated with acquired resistance to the
epidermal growth factor receptor tyrosine kinase inhibitor in
non-small cell lung cancer (10).
Wang et al indicated that miRNA-497 could inhibit ovarian
cancer cell migration and invasion through targeting SMAD specific
E3 ubiquitin protein ligase (11).
These data emphasized the importance of miRNAs in cancer
development and provide new insights into understanding the
molecular mechanism of tumorigenesis. However, the expression
profile and biological role of miR-98 in HCC remains to be
elucidated.
In the present study, the potential involvement of
miR-98 in HCC was investigated. The expression level of miR-98 in
HCC tissues and cell lines was confirmed and its effects on HCC
cell proliferation, migration and invasion were assessed. In
addition, the underlying mechanism of the function of miR-98 in HCC
was examined.
Materials and methods
HCC samples and cell lines
Paired HCC and adjacent non-tumor tissues were
obtained from 30 patients were recruited into a clinical trial at
the Department of General Surgery, Huaihe Hospital of Henan
University (Kaifeng, China) between 2012 and 2013. The present
study was approved by the Ethics Committee of Henan University.
Tissues were immediately snap frozen and stored at -80°C. Three HCC
cell lines (HepG2, HuH-7 and Hep3B) and the normal human liver cell
line L02 were obtained from the American Type Culture Collection
(Manassas, VA, USA), cultured in DMEM supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and incubated in a humid atmosphere with 5%
CO2 at 37°C.
Transfection
miR-98 mimics and the miRNA mimic negative control
(miR-NC) were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). Small interfering RNA against collagen triple
helix repeat containing 1 (si-CTHRC1) and the negative control
(si-NC) were designed by Shanghai GenePharma Co., Ltd. (Shanghai,
China). The sequences of the si-CTHRC1 and si-NC were as follows:
si-CTHRC1, 5′-ACAUUCAGCUCCAUUAAAGdTdT-3′ and si-NC,
5′-ACGUGACACGUUCGGAGAAdTdT-3′. Transfection was performed using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
The final concentration was 200 nm for miR-98 mimics or miR-NC and
100 nm for si-CTHRC1 or si-NC.
Cell proliferation assay
The cell proliferation assay was performed by
3-(4,5-dimethylthiazol-2-yl)-2 5-diphenyltetrazolium bromide (MTT)
assay. Cells were plated in 96-well plates at 5×103 per
well 24 h after transfection and cultured for 24, 48 and 72 h,
after which 20 µl MTT was added to each well. Subsequently,
the cells were incubated for 4 h prior to adding 150 µl
dimethyl sulfoxide. Once the insoluble crystals were completely
dissolved, the absorbance values at 570 nm were measured using a
microplate reader (Bio-Tek Instruments, Inc., Winooski, VT,
USA).
Cell migration and invasion assay
Cell migration and invasive ability was examined
using a 24-well transwell plate with 8 mm pore polycarbonate
membrane inserts, according to the manufacturer's protocol (Corning
Inc., Corning, NY, USA). The Matrigel (Sigma-Aldrich, St. Louis,
MO, USA) employed for the invasion assays was applied to the upper
surface of the membranes. A total of 5×104 cells per
well were seeded into the top chamber in serum-free media 24 h
after transfection and this was replaced with complete growth media
for 12 h. Cells that migrated or invaded through the surface of the
membrane were fixed with methanol and stained with hematoxylin
(Sigma-Aldrich). Migrating or invasive cells from three random
microscope fields per filter were selected for cell counting under
a microscope (Olympus CKX41; Olympus Corporation, Tokyo,
Japan).
Luciferase reporter assay
The 3′-UTR of CTHRC1 containing the predicted miR-98
binding site was constructed by Guangzhou RiboBio Co., Ltd. The
mutant CTHRC1 3′-UTR was created by mutating multiple nucleotides
complementary to the miR-98 seed region. HEK 293T cells were
cultured in 96-well plates with 50–70% confluence 24 h prior to
transfection. A mixture of 100 ng pmiR-RB-Report™ h-CTHRC1 wild
type (Wt) or mutant (Mut) reporter plasmid vector together with 50
nM miR-98 mimics or miR-NC (Guangzhou RiboBio Co., Ltd.) were
co-transfected. The luciferase activity was measured 48 h
post-transfection using a Dual-Glo Luciferase Assay System (Promega
Corporation, Madison, WI, USA) with Renilla luciferase
activity as the reporter gene and firefly luciferase as the
reference gene.
RNA isolation and reverse transcription
quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturers instructions. To quantitate miR-98 expression, total
RNA was polyadenylated and reverse transcribed to cDNA using an
NCode miRNA First-Strand cDNA Synthesis kit (Invitrogen; Thermo
Fisher Scientific, Inc.). To measure the mRNA levels of CTHRC1,
total RNA was reverse transcribed using a PrimeScript RT reagent
kit with gDNA Eraser (Takara Bio Inc., Shiga, Japan). qPCR was
performed using SYBR Premix Ex Taq II (Takara Bio Inc.) in an
Agilent Mx3005P qPCR system (Agilent Technologies, Inc., Santa
Clara, CA, USA) with an initial denaturation at 95°C, followed by
40 cycles at 95°C for 15 sec and 60°C for 1 min.. The following
primers were used: miR-98, forward 5′-CGGCTGAGGTAGTAGATTGT-3′ and
reverse 5′-GTCGTATCCAGTGCAGGGTCC-3′; CTHRC1, forward
5′-TGGACACCCAACTACAAGCA-3′ and reverse 5′-GAACAAGTGCCAACCCAGAT-3′.
GAPDH was used as an internal control with the following primers:
GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′, and reverse
5′-GAAGATGGTGATGGGATTTC-3′. All samples were normalized to internal
controls and fold changes were calculated through relative
quantification (2−ΔΔCq) (12).
Western blot analysis
Cultured cells were lysed in
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) with 1% phenylmethanesulfonyl fluoride. Protein was loaded
and separated by 10% SDS-PAGE gel (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and transferred onto a polyvinylidene fluoride
membrane (EMD Millipore, Billerica, MA, USA). The blots were probed
with the following primary antibodies at 4°C overnight: Rabbit
anti-CTHRC1 (1:1,000; ab85739; Abcam, Cambridge, MA, USA), and
rabbit anti-GAPDH (1:5,000; cat. no. ab9485; Abcam). The blots were
subsequently incubated with goat anti-rabbit horseradish
peroxidase-conjugated IgG secondary antibodies (1:3000; cat. no.
sc-2004; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Signals
were visualized using ECL substrates (Pierce Biotechnology Inc.,
Rockford, IL, USA). GAPDH was used as an endogenous control.
Statistical analysis
All statistical analyses were performed using SPSS
version 18.0 software (SPSS, Inc., Chicago, IL, USA). Data are
presented as the mean ± standard deviation. Statistical differences
were determined by analysis of variance or Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-98 expression in HCC tissues and cell
lines
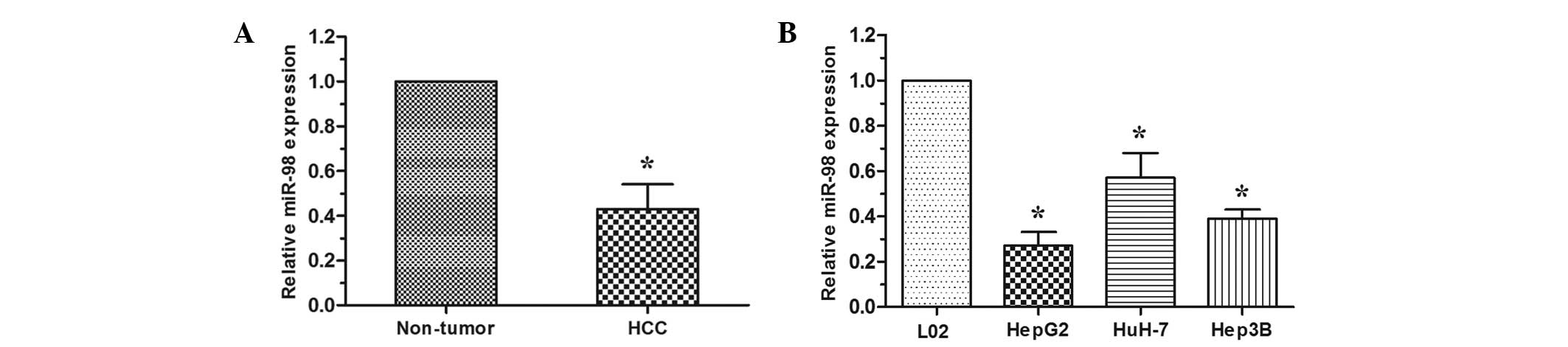
RT-qPCR analysis was performed to examine the
expression level of miR-98 in HCC tissues and cell lines. In 30
cases of HCC tissues and adjacent non-tumor tissues, the data
revealed that miR-98 was downregulated in HCC tissues when compared
with that in the paired adjacent non-tumor tissues (P<0.05;
Fig. 1A). The expression level of
miR-98 was then detected in HCC cell lines (HepG2, HuH-7 and Hep3B)
and the normal human liver cell line L02. The expression of miR-98
was decreased in HCC cell lines compared with the L02 cell line
(P<0.05; Fig. 1B). These
results indicated that reduced expression of miR-98 may be critical
in HCC progression and development.
Effect of miR-98 on HCC cell
proliferation, migration and invasion
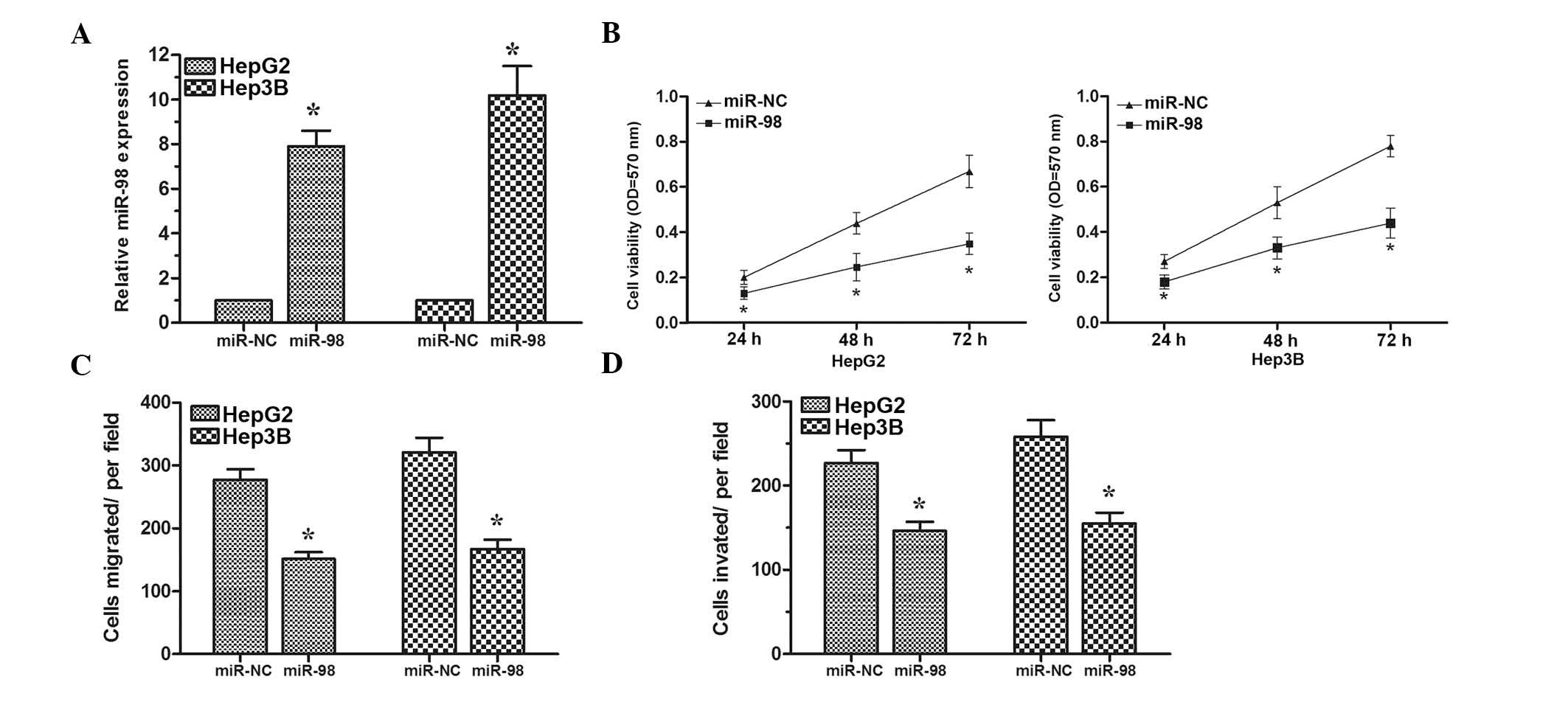
To assess the biological role of miR-98 in HCC,
HepG2 and Hep3B cells were transfected with the miR-98 mimics or
miR-NC. Transfection efficiency was confirmed by RT-qPCR (Fig. 2A). MTT assay revealed that cell
proliferation was significantly inhibited in HepG2 and Hep3B cells
transfected with miR-98 mimics compared with cells transfected with
miR-NC (Fig. 2B). Furthermore,
transwell assays demonstrated that overexpression of miR-98 could
inhibit the migratory and invasive abilities of HepG2 and Hep3B
cells (Fig. 2C and D). Taken
together, these results demonstrated that miR-98 could suppress
cell proliferation, migration and invasion in HCC cells.
CTHRC1 is a target of miR-98 in HCC
cells
To examine the molecular mechanism by which miR-98
suppresses the proliferation and invasion of HCC cells, TargetScan
(http://www.targetscan.org/) was adopted
and CTHRC1 was found to be the putative target of miR-98 (Fig. 3A). In order to confirm that CTHRC1
is a direct target of miR-98, luciferase reporter constructs
containing the wild-type (Wt) or mutant (Mut) 3′-UTR of the CTHRC1
gene were engineered. The luciferase reporter assay demonstrated
that miR-98 significantly inhibited the Wt but not Mut luciferase
activity in HEK 293T cells (Fig.
3B). In addition, western blot analyses demonstrated that
overexpression of miR-98 significantly inhibited CTHRC1 expression
in HepG2 and Hep3B cells (Fig.
3C). These results indicated that miR-98 directly modulates
CTHRC1 expression through binding to the 3′UTR of CTHRC1 mRNA.
Knockdown of CTHRC1 suppresses HCC cell
proliferation, migration and invasion
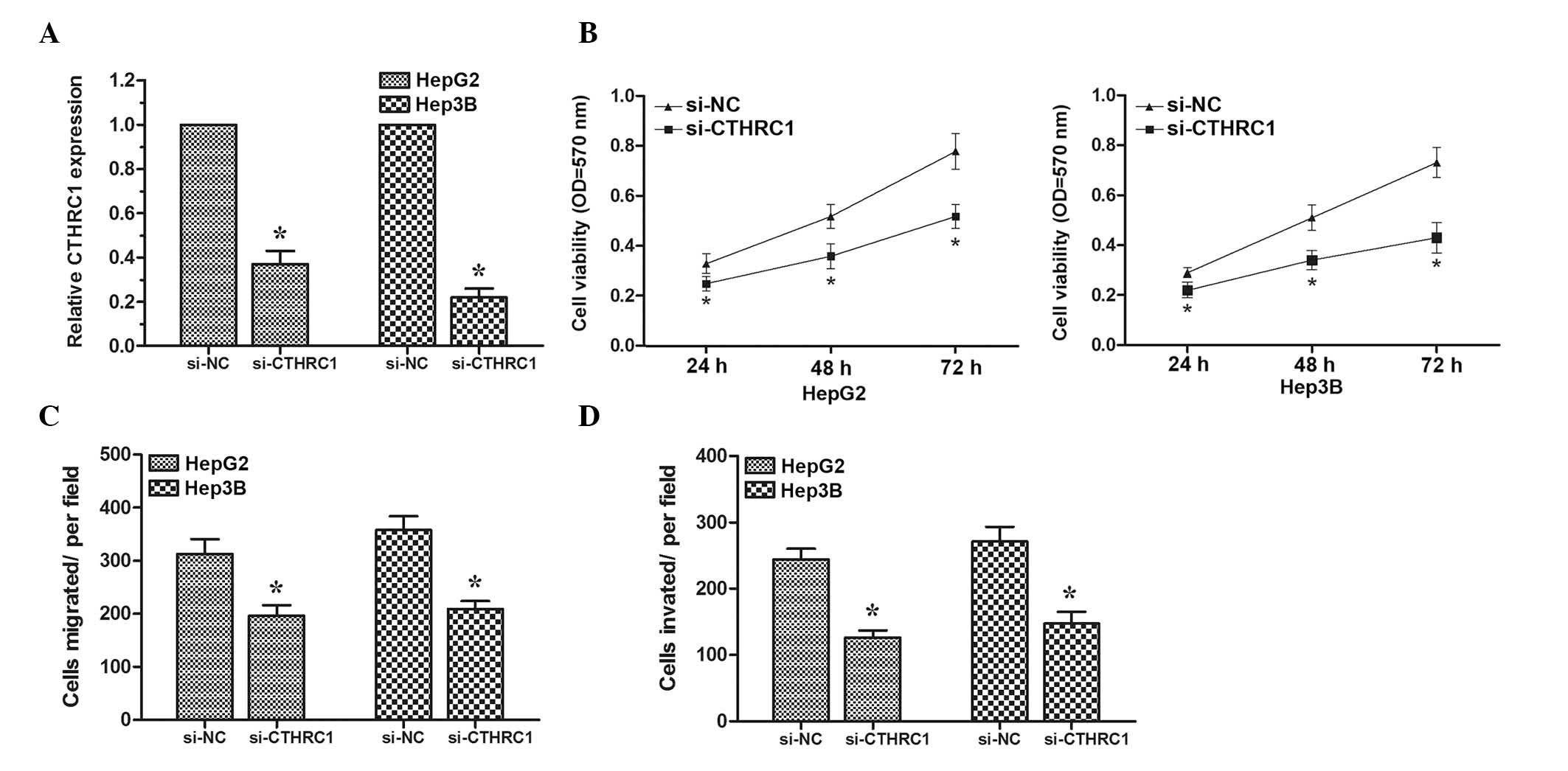
To determine whether CTHRC1 could also inhibit HCC
cell proliferation, migration and invasion, targeted knockdown of
CTHRC1 expression using RNA interference was performed in HepG2 and
Hep3B cells. The expression levels of CTHRC1 were determined by
RT-qPCR (Fig. 4A). MTT assay
revealed that HCC cells transfected with si-CTHRC1 exhibited a
significantly reduced proliferation ability compared with cells
transfected with si-NC (Fig. 4B).
Subsequently, transwell migration and invasion assays were
performed. The results demonstrated that knock down of CTHRC1 could
inhibit HCC cell migration and invasion (Fig. 4C and D). These data suggested that
decreased expression of CTHRC1 could inhibit HCC cell
proliferation, migration and invasion, demonstrating that CTHRC1
may act as a direct target gene of miR-98 in HCC.
Discussion
miRNAs serve as crucial regulators of gene
expression, regulating cellular physiology and development
(13). Increasing evidence
suggests that dysregulated expression of miRNAs is associated with
tumorigenesis and the progression of various types of human cancer,
including HCC (14). For example,
Xu et al found that miRNA-195 was significantly reduced in
HCC tissues and upregulation of miR-195 suppressed tumorigenicity
and regulated G1/S transition of HCC cells (15). Wang et al demonstrated that
miR-302b was decreased in HCC and suppressed HCC cell proliferation
and G1-S transition via targeting AKT2 expression (16). Thus, identifying and elucidating
the exact roles and underlying mechanisms of miRNAs in HCC may aid
our understanding of the pathogenesis of HCC. The present study
focused on the potential tumor suppressor miR-98.
In the current study, it was confirmed that miR-98
was downregulated in HCC tissues and cell lines. In addition,
overexpression of miR-98 suppressed HCC cell proliferation,
migration and invasion. CTHRC1 was identified as a target of miR-98
in HCC cells. Knock down of CTHRC1 expression inhibited HCC cell
proliferation, migration and invasion, phenocopying the
overexpression of miR-98 in HCC cells. These data demonstrated that
miR-98 acts as a tumor suppressor in HCC via downregulating CTHRC1
expression.
miR-98 has been demonstrated to be decreased in
various types of cancer and functions as a tumor suppressor. For
example, Chen et al found that the expression of miR-98 was
decreased in glioma tissues and overexpression of miR-98 inhibited
glioma cell invasion (17).
Siragam et al demonstrated that overexpression of miR-98
could inhibit breast cancer cell proliferation, survival, tumor
growth, invasion and angiogenesis (18). Furthermore, Huang et al
found that miR-98 was reduced in esophageal squamous cell carcinoma
(ESCC) and overexpression of miR-98 significantly inhibited the
migration and invasion of ESCC cells, which was reversed by
transfection of EZH2 (19). The
present data expanded our understanding of the tumor suppressive
role of miR-98 in HCC. Overexpression of miR-98 inhibited HCC cell
proliferation, migration and invasion.
CTHRC1 is a 30-kDa glycosylated secreted protein
containing a short collagen-like motif with 12 Gly-X-Y repeats
similar to the collagen domains present in the C1q/tumor necrosis
factor α-related protein (20).
Accumulating evidence suggested that CTHRC1 was increased in
various types of tumor. For example, Ke et al revealed that
overexpression of CTHRC1 was associated with tumor aggressiveness
and poor prognosis in human non-small cell lung cancer (21). Ma et al suggested that
CTHRC1 acted as a prognostic factor and promoted invasiveness of
gastrointestinal stromal tumors by activating Wnt/PCP-Rho signaling
(22). Kim et al found that
CTHRC1 was highly expressed in colorectal cancer and CTHRC1 acted
via extracellular-signal-regulated kinase-dependent induction of
matrix metalloproteinase-9 to promote invasion of colorectal cancer
cells (23). In the present study,
miR-98 suppressed the proliferation, migration and invasion of HCC
cells by targeting CTHRC1, elucidating the role of CTHRC1 in HCC
development.
In conclusion, the current study demonstrated that
miR-98 could act as a tumor suppressor in HCC cells, which was
mediated through inhibiting CTHRC1 expression. These findings
suggest that miR-98 could serve as a therapeutic agent in HCC
treatment.
Acknowledgments
The present study was supported by the Scientific
and Technological Research Projects of the Henan Provincial
Education Department (grant no. 12B320003).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy KR: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruix J and Sherman M; American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sangiovanni A, Del Ninno E, Fasani P, De
Fazio C, Ronchi G, Romeo R, Morabito A, De Franchis R and Colombo
M: Increased survival of cirrhotic patients with a hepatocellular
carcinoma detected during surveillance. Gastroenterology.
126:1005–1014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Plasterk RHA: Micro RNAs in animal
development. Cell. 124:877–881. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li W, Liu M, Feng Y, Xu YF, Huang YF, Che
JP, Wang GC, Yao XD and Zheng JH: Downregulated miR-646 in clear
cell renal carcinoma correlated with tumour metastasis by targeting
the nin one binding protein (NOB1). Br J Cancer. 111:1188–1200.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li B, Ren S, Li X, Wang Y, Garfield D,
Zhou S, Chen X, Su C, Chen M, Kuang P, et al: MiR-21 overexpression
is associated with acquired resistance of EGFR-TKI in non-small
cell lung cancer. Lung Cancer. 83:146–153. 2014. View Article : Google Scholar
|
|
11
|
Wang W, Ren F, Wu Q, Jiang D, Li H, Peng
Z, Wang J and Shi H: MicroRNA-497 inhibition of ovarian cancer cell
migration and invasion through targeting of SMAD specific E3
ubiquitin protein ligase 1. Biochem Biophys Res Commun.
449:432–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo H, Li W, Zheng T and Liu Z: MiR-195
targets HDGF to inhibit proliferation and invasion of NSCLC cells.
Tumour Biol. 35:8861–8866. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gurtan AM and Sharp PA: The role of miRNAs
in regulating gene expression networks. J Mol Biol. 425:3582–3600.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Yao J, Shi X, Hu L, Li Z, Song T
and Huang C: MicroRNA-302b suppresses cell proliferation by
targeting EGFR in human hepatocellular carcinoma SMMC-7721 cells.
BMC Cancer. 13:4482013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Z, Cheng Q, Ma Z, Xi H, Peng R and
Jiang B: Overexpression of RKIP inhibits cell invasion in glioma
cell lines through upregulation of miR-98. Biomed Res Int.
2013:6951792013.
|
|
18
|
Siragam V, Rutnam ZJ, Yang W, Fang L, Luo
L, Yang X, Li M, Deng Z, Qian J, Peng C and Yang BB: MicroRNA
miR-98 inhibits tumor angiogenesis and invasion by targeting
activin receptor-like kinase-4 and matrix metalloproteinase-11.
Oncotarget. 3:1370–1385. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang SD, Yuan Y, Zhuang CW, Li BL, Gong
DJ, Wang SG, Zeng ZY and Cheng HZ: MicroRNA-98 and microRNA-214
post-transcriptionally regulate enhancer of zeste homolog 2 and
inhibit migration and invasion in human esophageal squamous cell
carcinoma. Mol Cancer. 11:512012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pyagay P, Heroult M, Wang Q, Lehnert W,
Belden J, Liaw L, Friesel RE and Lindner V: Collagen triple helix
repeat containing 1, a novel secreted protein in injured and
diseased arteries, inhibits collagen expression and promotes cell
migration. Circ Res. 96:261–268. 2005. View Article : Google Scholar
|
|
21
|
Ke Z, He W, Lai Y, Guo X, Chen S, Li S,
Wang Y and Wang L: Overexpression of collagen triple helix repeat
containing 1 (CTHRC1) is associated with tumour aggressiveness and
poor prognosis in human non-small cell lung cancer. Oncotarget.
5:9410–9424. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma MZ, Zhuang C, Yang XM, Zhang ZZ, Ma H,
Zhang WM, You H, Qin W, Gu J, Yang S, et al: CTHRC1 acts as a
prognostic factor and promotes invasiveness of gastrointestinal
stromal tumors by activating Wnt/PCP-Rho signaling. Neoplasia.
16:265–278. 278.e1–e13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim HC, Kim YS, Oh HW, Kim K, Oh SS, Kim
JT, Kim BY, Lee SJ, Choe YK, Kim DH, et al: Collagen triple helix
repeat containing 1 (CTHRC1) acts via ERK-dependent induction of
MMP9 to promote invasion of colorectal cancer cells. Oncotarget.
5:519–529. 2014. View Article : Google Scholar : PubMed/NCBI
|