Introduction
Hepatocellular carcinoma (HCC) is a leading cause of
cancer-associated mortality, and is one of the most common
malignant tumors with poor prognosis worldwide (1,2). In
clinical settings only a limited number of patients with HCC are
eligible for potentially curative treatment options such as
surgical resection followed by orthotopic liver transplantation
(3,4). Therefore, the development of
effective therapeutic strategies for the treatment of greater
numbers of patients with HCC is imperative. However, the
pathogenesis of HCC remains unclear. HCC has long been considered
the result of various genetic alterations that ultimately led to
malignant transformation (5,6).
Cancer development is no longer thought to be induced by genetic
and genomic alterations alone, but is also considered to be the
result of epigenetic alterations (7,8).
MicroRNAs (miRNAs or miRs) are a type of highly
conserved non-coding small RNA that post-transcriptionally regulate
the expression of their target genes (9). miRNAs have been demonstrated to
regulate numerous aspects of cell activity, including metabolism,
differentiation and development, proliferation, apoptosis and viral
infection (10–15). Previous findings demonstrated that
miRNAs have important roles in numerous types of malignant tumors,
including HCC (16). In addition,
numerous studies have demonstrated the existence of prognostic
miRNAs in clinical tissue specimens of primary HCC, and miRNAs have
been shown to have important regulatory roles in
hepatocarcinogenesis (17–20).
The present study aimed to investigate the
expression levels of miRNA-200b as well as those of its potential
target DNA methyltransferase 3a (DNMT3a) in HCC.
Materials and methods
Patients and cell lines
Histologically normal liver (NL) tissue samples were
obtained from 10 patients with gallbladder stones during a biopsy
procedure. HCC and adjacent non-malignant para-tumorous (PT) tissue
specimens were obtained from 44 hepatitis B-positive patients by
radical hepatectomy. The patients, 33 males and 11 females, had
undergone treatment at The First People's Hospital of Yunnan
Province (mean age, 53.7±11.6 years). HCC was confirmed in the
tissue samples by pathological examination and were obtained with
written informed consent from the patients. The present study was
approved by the Institutional Review Board of the Fourth Military
Medical University (Xian, China). The HepG2 and L02 cell lines were
obtained from the Chinese Academy of Sciences (Shanghai, China) and
cultured at 37°C with 5% CO2 in RMPI-1640 medium
(Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) until cell density reached ~70–80%.
MicroRNA arrays
MicroRNA arrays were conducted as previously
described (21) on miR-200b from
ten NL tissue samples, and 44 HCC and corresponding non-malignant
PT tissue samples. Briefly, 100 ng RNA was extracted from each
tissue sample using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) and an RNeasy Mini kit (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
tissue samples were subsequently hybridized by labeling with the
miRCURY Hy3/Hy5 Power labeling kit (Exiqon A/S, Vedbæk, Denmark)
and hybridizing on the miRCURY LNA™ Array (v.11.0). They were then
scanned using an Axon GenePix 4000B microarray scanner (Thermo
Fisher Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the manually
homogenized tissue samples as well as the cell lines using
TRIzol® reagent, according to the manufacturer's
protocol. Reverse transcription of 5 ng RNA to cDNA was performed
using a QuantiMir RT kit (System Biosciences, Mountain View, CA,
USA). The following primers (Shanghai GenePharma Co., Ltd.,
Shanghai, China) were used: miR-200b forward, 5′-TCA TCA TTA CCA
GGC AGT ATTA-3′, and reverse, 5′-TCC ATC ATT ACC CGG CAG TATTA-3′;
U6 forward, 5′-CTC GCT TCG GCA GCACA-3′, and reverse, 5′-AAC GCT
TCA CGA ATT TGCGT-3′; DNMT3a forward, 5′-CAA TGA CCT CTC CAT CGT
CAAC-3′, and reverse, 5′-CAT GCA GGA GGC GGT AGAA-3′; and β-actin
forward, 5′-GAA CGG TGA AGG TGA CAG-3′, and reverse, 5′-TAG AGA GAA
GTG GGG TGG-3′. miR-200b was amplified in a MyCycler thermal cycler
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) as follows:
Denaturation at 95°C for 10 min, and then 40 cycles of 95°C for 10
sec, 60°C for 20 sec, and 72°C for 10 sec. DNMT3a was amplified as
follows: Denaturation at 95°C for 10 min, and then 40 cycles of
95°C for 15 sec and 60°C for 1 min. U6 RNA was used as an miRNA
internal control, and β-actin was used to normalize the expression
levels of total:mRNA in each sample. Values were calculated as
ratios normalized to U6 or β-actin. The expression level of miRNA
was defined based on the quantification cycle (Cq), and relative
expression levels were calculated using the 2−ΔΔCq
method (22).
Transfection of miR-200b mimics
Synthesized Dharmacon miR-200b mimics were purchased
from GE Healthcare Life Sciences (Pittsburgh, PA, USA). HepG2 cells
(2×106) were cultured in RPMI-1640 medium supplemented
with 10% FBS at 37°C in 5% CO2. When the HepG2 cells
reached 30–50% confluence, they were transfected with miR-200b
mimics (60 nM) or miRNA mimic control using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
cells were then harvested and protein expression levels were
measured by western blotting 72 h post-transfection.
Western blot analysis
Western blotting was performed as previously
described (23). Briefly, total
protein was extracted using radioimmunoprecipitation buffer
(Sigma-Aldrich) and 10 µg of each sample was separated by
SDS-PAGE (Bio-Rad Laboratories, Inc.) and transferred to
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% skimmed milk in
Tris-buffered saline with 0.1% (v/v)Tween 20 (TBST; EMD Millipore)
and then incubated with primary antibodies with gentle agitation
for 12 h at 4°C. The membranes were then washed three times with
TBST and incubated with goat anti-rabbit and goat anti-mouse
peroxidase-conjugated secondary antibody (dilution, 1:3,000;
Beyotime Institute of Biotechnology, Haimen, China; cat nos. A0208
and A0216). Rabbit anti-human monoclonal anti-DNMT3a (dilution,
1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA; cat.
no. 3598) and mouse anti-human anti-GAPDH antibodies (dilution,
1:1,000; ProMab Biotechnologies, Inc., Richmond, CA, USA; cat. no.
20035) were used. Protein bands were visualized by
chemiluminescence detection (EMD Millipore) and the quantification
of band density was conducted using Image J (version 1.5; National
Institutes of Health, Bethesda, MD, USA). GAPDH was used as an
internal control, and all values were calculated as ratios
normalized to GAPDH.
Identification of targets of
miR-200b
TargetScan (www.targetscan.org) was used to identify potential
targets of miR-200b. An important enzyme in DNA methylation,
DNMT3a, was identified as one of the potential targets.
Luciferase activity assay
The 3′-untranslated region (UTR) of DNMT3a was
amplified by PCR and inserted into a pGL3 vector (Promega
Corporation, Madison, WI, USA). The PCR was conducted on a MyCycler
thermal cycler under the following conditions: 50°C for 30 min;
95°c for 95 min; and 40 cycles of 95°C for 30 sec and 55°C for 30
sec. A pGL3 construct containing DNMT3a 3′-UTR with point mutations
in the seed sequence was synthesized using 30 reactions of the
Quik-Change Lightning Site-Directed Mutagenesis kit (Agilent
Technologies, Inc., Santa Clara, CA, USA) according to the
manufacturer's protocol. The primers used for DNMT3a were as
follows: Forward, 5′-GCT CTA GAC GAA AAG GGT TGG ACA TCAT-3′, and
reverse, 5′-GCT CTA GAG CCG AGG GAG TCT CCT TTTA-3′.
HepG2 cells (2×105) were transfected
using Lipofectamine® 2000 with the appropriate plasmids
in 24-well plates. The cells were then harvested and lysed with
reporter gene cell lysis buffer (Beyotime Institute of
Biotechnology) in order to conduct a luciferase activity assay 48 h
post-transfection using a dual luciferase reporter assay system
(Promega Corporation). Relative luciferase activity levels were
normalized to Renilla luciferase activity, which served as an
internal control.
MTT assay
An MTT assay was used to determine the effects of
ectopic miR-200b mimics on HepG2 cell proliferation. Briefly, HepG2
cells were transfected with miR-200b mimics using
Lipofectamine® 2000 and then seeded into 96-well plates
at 5×103 cells/well in 200 µl RPMI-1640 medium
for 72 h. MTT solution [0.5 mg/ml in 20 µl
phosphate-buffered saline (PBS); Sigma-Aldrich] was added to each
well and incubated for 4 h at 37°C. An enzyme-labeled instrument
(Thermo Fisher Scientific, Inc.) was used to measure the absorbance
of each well at 570 nm. Data were obtained from three independent
experiments.
TUNEL staining
DNA fragmentation of apoptotic cells was detected
using a TUNEL kit (Sigma-Aldrich) according to the manufacturer's
protocol. Briefly, 2×105 cells were cultured on cover
slips for 48 h at 37°C with 5% CO2. Following miR-200b
mimic transfection for 72 h, the cells were fixed in 4%
paraformaldehyde solution (EMD Millipore) in PBS for 30 min at room
temperature. The cells were incubated with methanol solution
containing 0.3% H2O2 for 30 min at room
temperature to block endogenous peroxidase activity, and then
incubated in the TUNEL reaction mixture for 60 min at 37°C. Nuclei
were visualized with DAPI staining (0.5–10 µg/ml; Beyotime
Institute of Biotechnology) for 10 mins. The cells were then
visualized by fluorescence microscopy (DM4000B; Leica Microsystems
GmbH, Wetzlar, Germany). Apoptotic cells were counted from four
randomly selected fields in each sample.
Flow cytometry
Apoptotic cells were detected using double-staining
with Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide
(PI; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). Briefly,
the cells were harvested 48 h post-transfection and stained with
anti-Annexin V conjugated to FITC and PI for 15 min at room
temperature. The cells were then detected using
fluorescence-activated cell sorting FACS using a FACS Calibur
obtained from BD Biosciences (Franklin Lakes, NJ, USA). The data
were analyzed using CellQuest software (version 5.1; BD
Biosciences).
Statistical analysis
All the experiments were repeated in triplicate, and
the data were expressed as the mean ± standard error of the mean.
The results were analyzed using the Student's t-test and one-way
analysis of variance. The statistical analyses were conducted using
SPSS 19.0 software (IBM SPSS, Armonk, NY, USA) and P<0.05 was
considered to indicate a statistically significant result.
Results
miR-200b expression levels are
downregulated in HCC tissue samples and HepG2 cell lines
To investigate miR-200b expression in HCC tissue
samples, RT-qPCR was used to quantify miR-200b expression levels.
miR-200b expression levels were significantly decreased in the HCC
tissue samples, as compared with those in the NL and non-malignant
PT tissue samples (P<0.05; Fig.
1A). The expression levels of miR-200b were then evaluated in
the HepG2 and L02 cells by RT-qPCR. The expression levels of
miR-200b were significantly lower in the HepG2 cells, as compared
to those in the L02 cells (P<0.05; Fig. 1B). These results indicated that
miR-200b expression levels are significantly decreased in HCC
tissue samples and cell lines, suggesting that miR-200b is
associated with hepatocellular carcinogenesis.
 | Figure 1miR-200b expression levels are
downregulated in HCC tissue samples and HepG2 hepatoma cell lines.
(A) The expression levels of miR-200b in the NL, PT, and HCC tissue
samples from 44 patients with hepatitis B-positive HCC, as
determined by RT-qPCR. *P<0.05, vs. the NL/PT groups.
(B) The expression levels of miR-200b in the HepG2 and L02 cell
lines, as determined by RT-qPCR. *P<0.05, vs. the L02
cells. The values are presented as means ± standard error of the
mean. All experiments were performed in triplicate, and similar
results were obtained from each experiment. miR, microRNA; NL,
normal liver; PT, para-tumorous; HCC, hepatocellular carcinoma;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction. |
In silico prediction of DNMT3a as an
miR-200b target
TargetScan was used to identify the potential
targets of miR-200b. An important enzyme in DNA methylation,
DNMT3a, was identified as one of the potential targets of miR-200b.
The predicted binding site of miR-200b with the DNMT3a 3′-UTR is
shown in Fig. 2A.
 | Figure 2Expression levels of the miR-200b
target gene DNMT3a are increased in HCC tissues samples compared
with NL tissue samples. (A) Bioinformatics analysis suggested that
DNMT3a was an important enzyme in DNA methylation, and a potential
target of miR-200b. (B) The mRNA expression levels of DNMT3a in NL,
PT, and HCC tissue samples were determined using reverse
transcription-quantitative polymerase chain reaction. (C and D)
Protein expression levels of DNMT3a in NL, PT and HCC tissue
samples. The data are presented as means ± standard error of the
mean. All experiments were performed in triplicate and yielded
similar results. *P<0.05, vs. the HCC group. NL,
normal liver; PT, para-tumorous; HCC, hepatocellular carcinoma;
miR, microRNA; 3′-UTR; 3′-untranslated region; DNMT3a, DNA
methyltransferase 3a. |
The expression levels of the potential
miR-200b target gene DNMT3a are significantly increased in HCC
tissue samples compared with NL tissue samples
The mRNA and protein expression levels of the
miR-200b potential target gene DNMT3a were evaluated by RT-qPCR and
western blot analysis. Compared with mRNA expression levels of
DNMT3a in the NL (0.318±0.047) or PT (0.326±0.082) tissue samples,
the mRNA expression levels of DNMT3a in the HCC (1.295±0.093)
tissue samples were significantly higher (P<0.05; Fig. 2B). In addition, compared with the
protein expression levels of DNMT3a in the NL (0.214±0.037) and PT
(0.280±0.068) tissue samples, the protein expression levels of
DNMT3a in the HCC (0.722±0.014) tissue samples were also
significantly higher (P<0.05; Fig.
2C and D).
DNMT3a is the direct target of
miR-200b
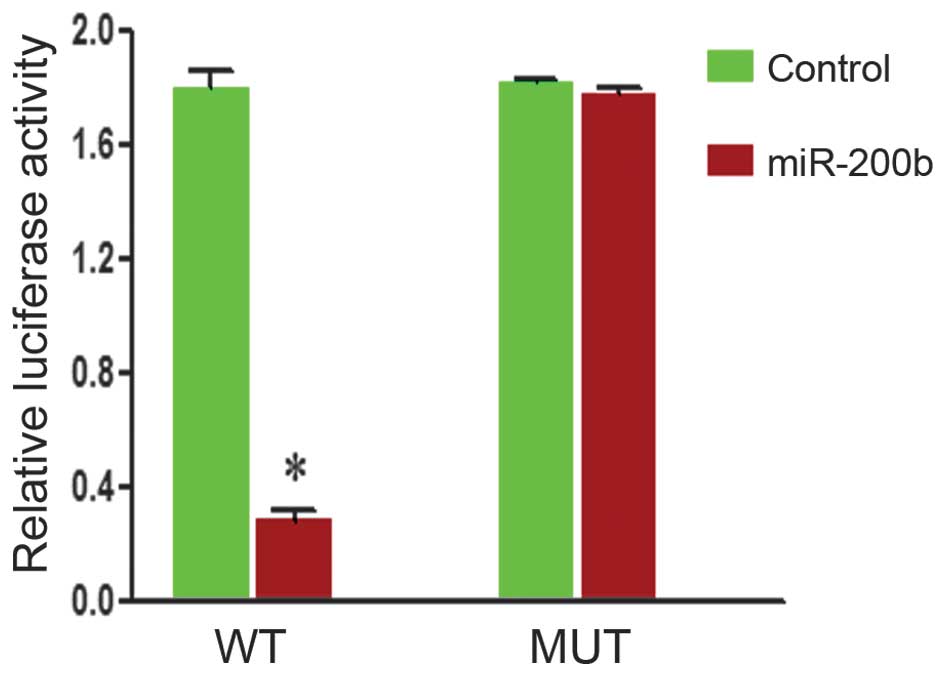
To examine miR200b-DNMT3a interactions, DNMT3a
complementary sites, with or without mutations, were cloned into
the 3′-UTR of the firefly luciferase gene and co-transfected with
miR-200b mimics or a negative control in HepG2 cells. The presence
of miR-200b led to a significant reduction in the relative
luciferase activity levels in the wild-type construct of the DNMT3a
3′-UTR in HepG2 cells (P<0.05). However, the mutant construct of
the DNMT3a 3′-UTR reversed the suppressive effect of miR-200b in
HepG2 cells (Fig. 3). These
results suggest that DNMT3a is a direct target of miR-200b.
Ectopic miR-200b decreases the expression
levels of DNMT3a and suppresses HepG2 cell proliferation
Western blotting was conducted to detect the protein
expression levels of DNMT3a following miRNA-200b mimic transfection
in HepG2 cells. DNMT3a protein expression levels were significantly
decreased in the mimic group (0.214±0.021), as compared with those
in the blank control (0.527±0.035) or mock transfection control
group (0.513±0.013; P<0.05; Fig. 4A
and B).
 | Figure 4Ectopic miR-200b decreases DNMT3a
expression levels and HepG2 cell proliferation. (A) miR-200b mimics
were transfected into HepG2 cells, and the protein expression
levels of DNMT3a were detected by western blotting. (B) At 0, 12,
24, 36, and 48 h post-miR-200b transfection, cell proliferation was
measured by MTT assay. The data are presented as means ± standard
error of the mean. All experiments were performed in triplicate,
and similar results were obtained for each experiment.
*P<0.05, vs. the blank control and mock transfection
groups. miR, microRNA; DNMT3a, DNA methyltransferase 3a. |
To determine the role of miR-200b deregulation in
hepatocarcinogenesis, an MTT assay, a TUNEL assay, and flow
cytometry were used to determine the proliferation and apoptosis
rates of HepG2 cells following miR-200b mimic exposure. The MTT
assay demonstrated that at 24, 36 and 48 h following miR-200b mimic
exposure, the proliferation rate of HepG2 cells was reduced to 68,
42 and 36.3% of the control, respectively (Fig. 4C). The differences between the
miR-200b mimic-transfected and control groups at the
above-mentioned time points were statistically significant
(P<0.05), whereas the difference between the blank and negative
control groups were not (P>0.05; Fig. 4C). The apoptosis of HepG2 cells
post-transfection with miR-200b mimics was evaluated by TUNEL assay
and flow cytometry. Apoptosis levels were markedly increased in the
miR-200b mimic group, as compared with the blank and negative
control groups (Fig. 5).
 | Figure 5Ectopic miR-200b markedly increased
apoptosis of HepG2 cells compared with the blank and negative
controls. (A) miR-200b mimics were transfected into HepG2 cells,
and apoptosis levels were determind 48 h post-transfection by TUNEL
staining. Red, apoptotic cells; blue; DAPI. Magnification, ×800. (B
and C) The apoptosis levels were also determined by flow cytometry.
The lower left quadrants represent live cells, the upper left
represent early apoptotic cells, the lower right represent late
apoptotic cells and the upper right quadrant represents dead cells.
The data are presented as means ± standard error of the mean. All
experiments were performed in triplicate, and similar results were
obtained for each experiment. *P<0.05, vs. the blank
control/mock transfection groups. miR, microRNA; DAPI,
4′,6-diamidino-2-phenylindole. |
Discussion
The present study aimed to investigate the possible
role of miR-200b in hepatocarcinogenesis and to identify its target
gene. The results demonstrated that, compared with NL and PT tissue
samples, miR-200b expression levels were significantly reduced in
HCC tissue samples. In addition, miR-200b expression levels in
HepG2 cells were significantly decreased, as compared with those in
L02 cells. Western blotting and RT-qPCR demonstrated that the
expression levels of DNMT3a, a possible target gene for miR-200b,
were significantly higher in HCC tissue samples, as compared with
NL and PT tissue samples. Furthermore, the results of the present
study demonstrated that DNMT3a was the direct target gene of
miR-200b. Upregulated miR-200b expression in HepG2 cells led to a
decrease in DNMT3a expression levels, and had an inhibitory effect
on cell proliferation. These data suggested that miR-200b is an
important factor in hepatocarcinogenesis, and acts by
downregulating DNMT3a expression. miR-200b may therefore be a
promising target for HCC treatment.
miRNAs are non-coding RNAs 19–25 nucleotides in
length that have been demonstrated to regulate gene expression by
inducing translational inhibition or cleavage of their target mRNAs
through base pairing at partially or fully complementary sites
(24–26). Numerous miRNAs have been
demonstrated to function as tumor suppressor or oncogenes by
regulating their target genes (27–29).
Previous studies have demonstrated that various miRNAs are
abnormally expressed in malignant HCC cells or tissue samples, as
compared with normal hepatocytes or tissue samples (18,30–32).
In the present study, the results revealed that miR-200b expression
levels were significantly decreased in HCC tissue samples and in
HepG2 cells, as compared with those in non-malignant liver tissue
samples and L02 cells.
Genomic stability is regulated by both genetic and
epigenetic mechanisms (33).
Promoter hypermethylation mediated by DNMTs is widely accepted as
the predominant mechanism underlying the epigenetic inactivation of
tumor suppressor genes (TSGs) (34,35).
Previous studies have demonstrated that viral genes have important
roles in regulating DNA methylation (36–38).
However, the epigenetic mechanisms underlying virus-associated
cancers are poorly understood. Numerous studies have suggested that
hypermethylation is responsible for the silencing of TSGs in
hepatocarcinogenesis (39–42). In addition, data from previous
studies support a role for miRNAs as targets and effectors in
aberrant mechanisms underlying DNA hypermethylation (43–45).
The present study demonstrated that there exists an interaction
between miR-200b and DNMT3a. Firstly, in silico analysis
suggests that DNMT3a, an important enzyme in DNA methylation, may
be one of the possible targets of miR-200b. Secondly, the results
demonstrate that the mRNA and protein expression levels of DNMT3a
are inversely correlated with miR-200b expression in HCC. Ectopic
miR-200b expression led to a reduction in the expression levels of
DNMT3a. Furthermore, DNMT3a was shown to be a direct target of
miR-200b, as demonstrated by a luciferase activity assay. The
results of the present study also demonstrate that ectopic miR-200b
expression significantly suppressed the proliferation of HepG2
cells and induced apoptosis. These data suggest that miR-200b
regulates DNMT3a expression and has a tumor suppressive role in HCC
development.
Since the number of samples used in the present
study was relatively small, further investigation with a larger
number of samples is required. Furthermore, the regulatory
mechanism underlying miR-200b expression downregulation in HCC
requires investigation. In conclusion, the results of the present
study suggest that miR-200b is an important factor in
hepatocarcinogenesis, and acts by downregulating DNMT3a expression.
Thus, miR-200b is a promising target for HCC treatment.
Acknowledgments
The authors of the present study are grateful to Dr
Wang from the Department of Pharmacology and Neurology of Harvard
Medical School (Boston, MA, USA) for the critical reading and
modification of the manuscript.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thomas MB and Zhu AX: Hepatocellular
carcinoma: The need for progress. J Clin Oncol. 23:2892–2899. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM: Updated treatment approach to
hepatocellular carcinoma. J Gastroenterol. 40:225–235. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Llovet JM and Bruix J: Novel advancements
in the management of hepatocellular carcinoma in 2008. J Hepatol.
48(Suppl 1): S20–S37. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanyal AJ, Yoon SK and Lencioni R: The
etiology of hepatocellular carcinoma and consequences for
treatment. Oncologist. 15(Suppl 4): 14–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Franco R, Schoneveld O, Georgakilas AG and
Panayiotidis MI: Oxidative stress, DNA methylation and
carcinogenesis. Cancer Lett. 266:6–11. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ziech D, Franco R, Pappa A and
Panayiotidis MI: Reactive Oxygen Species (ROS)–Induced genetic and
epigenetic alterations in human carcinogenesis. Mutat Res.
711:167–173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yao Y, Suo A-L, Li ZF, Liu LY, Tian T, Ni
L, Zhang WG, Nan KJ, Song TS and Huang C: MicroRNA profiling of
human gastric cancer. Mol Med Rep. 2:963–970. 2009.PubMed/NCBI
|
|
12
|
Chan SY, Zhang YY, Hemann C, Mahoney CE,
Zweier JL and Loscalzo J: MicroRNA-210 controls mitochondrial
metabolism during hypoxia by repressing the iron-sulfur cluster
assembly proteins ISCU1/2. Cell metabolism. 10:273–284. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen JF, Mandel EM, Thomson JM, Wu Q,
Callis TE, Hammond SM, Conlon FL and Wang DZ: The role of
microRNA-1 and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006. View Article : Google Scholar
|
|
14
|
Wang B, Cai Z, Lu F, Li C, Zhu X, Su L,
Gao G and Yang Q: Destabilization of survival factor MEF2D mRNA by
neurotoxin in models of Parkinson's disease. J Neurochem.
130:720–728. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sullivan CS and Ganem D: MicroRNAs and
viral infection. Mol Cell. 20:3–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar
|
|
19
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen RX, Xia YH, Xue TC and Ye SL:
Suppression of microRNA-96 expression inhibits the invasion of
hepatocellular carcinoma cells. Mol Med Rep. 5:800–804. 2012.
|
|
21
|
Jiang J, Zhang Y, Yu C, Li Z, Pan Y and
Sun C: MicroRNA-492 expression promotes the progression of hepatic
cancer by targeting PTEN. Cancer Cell Int. 14:1–8. 2014. View Article : Google Scholar
|
|
22
|
Tang H, Deng M, Tang Y and Xie X, Guo J,
Kong Y, Ye F, Su Q and Xie X: miR-200b and miR-200c as prognostic
factors and mediators of gastric cancer cell progression. Clin
Cancer Res. 19:5602–5612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng D, Wang B, Ma Y, Shi W, Tao K, Zeng
W, Cai Q, Zhang Z and Qin H: The Ras/Raf/Erk pathway mediates the
subarachnoid hemorrhage-induced apoptosis of hippocampal neurons
through phosphorylation of p53. Mol Neurobiol. 26–Oct;2015.Epub
ahead of print. View Article : Google Scholar
|
|
24
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
et al: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Adv Exp Med Biol.
604:17–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hammond SM: MicroRNAs as tumor
suppressors. Nat Genet. 39:582–583. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanke M, Hoefig K, Merz H, Feller AC,
Kausch I, Jocham D, Warnecke JM and Sczakiel G: A robust
methodology to study urine microRNA as tumor marker: MicroRNA-126
and microRNA-182 are related to urinary bladder cancer. Urol Oncol.
28:655–661. 2010. View Article : Google Scholar
|
|
30
|
Xu J, Wu C, Che X, Wang L, Yu D, Zhang T,
Huang L, Li H, Tan W, Wang C and Lin D: Circulating microRNAs,
miR-21, miR-122, and miR-223, in patients with hepatocellular
carcinoma or chronic hepatitis. Mol Carcinog. 50:136–142. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Karakatsanis A, Papaconstantinou I,
Gazouli M, Lyberopoulou A, Polymeneas G and Voros D: Expression of
microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c,
miR-221, miR-222, and miR-223 in patients with hepatocellular
carcinoma or intrahepatic cholangiocarcinoma and its prognostic
significance. Mol Carcinog. 52:297–303. 2013. View Article : Google Scholar
|
|
33
|
Li E: Chromatin modification and
epigenetic reprogramming in mammalian development. Nat Rev Genet.
3:662–673. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luczak MW and Jagodziński PP: The role of
DNA methylation in cancer development. Folia Histochem Cytobiol.
44:143–154. 2006.PubMed/NCBI
|
|
35
|
Rajendran G, Shanmuganandam K, Bendre A,
Mujumdar D, Goel A and Shiras A: Epigenetic regulation of DNA
methyltransferases: DNMT1 and DNMT3B in gliomas. J Neurooncol.
104:483–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Feng J, Zhou Y, Campbell SL, Le T, Li E,
Sweatt JD, Silva AJ and Fan G: Dnmt1 and Dnmt3a maintain DNA
methylation and regulate synaptic function in adult forebrain
neurons. Nat Neurosci. 13:423–430. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Viré E, Brenner C, Deplus R, Blanchon L,
Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden
JM, et al: The Polycomb group protein EZH2 directly controls DNA
methylation. Nature. 439:871–874. 2006. View Article : Google Scholar
|
|
38
|
Kuramochi-Miyagawa S, Watanabe T, Gotoh K,
Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri
TW, et al: DNA methylation of retrotransposon genes is regulated by
Piwi family members MILI and MIWI2 in murine fetal testes. Gene
Dev. 22:908–917. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xie Y, Liu J, Benbrahim-Tallaa L, Ward JM,
Logsdon D, Diwan BA and Waalkes MP: Aberrant DNA methylation and
gene expression in livers of newborn mice transplacentally exposed
to a hepatocarcinogenic dose of inorganic arsenic. Toxicology.
236:7–15. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pogribny IP, Ross SA, Wise C, Pogribna M,
Jones EA, Tryndyak VP, James SJ, Dragan YP and Poirier LA:
Irreversible global DNA hypomethylation as a key step in
hepatocarcinogenesis induced by dietary methyl deficiency. Mutat
Res. 593:80–87. 2006. View Article : Google Scholar
|
|
41
|
Zhu R, Li BZ, Li H, Ling YQ, Hu XQ, Zhai
WR and Zhu HG: Association of p16INK4A hypermethylation with
hepatitis B virus X protein expression in the early stage of
HBV-associated hepatocarcinogenesis. Pathol Int. 57:328–336. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park IY, Sohn BH, Yu E, Suh DJ, Chung YH,
Lee JH, Surzycki SJ and Lee YI: Aberrant epigenetic modifications
in hepatocarcinogenesis induced by hepatitis B virus X protein.
Gastroenterology. 132:1476–1494. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Garzon R, Liu S, Fabbri M, Liu Z, Heaphy
CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al:
MicroRNA-29b induces global DNA hypomethylation and tumor
suppressor gene reexpression in acute myeloid leukemia by targeting
directly DNMT3A and 3B and indirectly DNMT1. Blood. 113:6411–6418.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo
X, Li J, Zhou H, Tang Y and Shen N: MicroRNA-21 and microRNA-148a
contribute to DNA hypomethylation in lupus CD4+ T cells by directly
and indirectly targeting DNA methyltransferase 1. J Immunol.
184:6773–6781. 2010. View Article : Google Scholar : PubMed/NCBI
|