Introduction
Pancreatic cancer (PC), a digestive system tumor, is
one of the most aggressive types of cancer. With a high degree of
malignancy, rapid progression and poor prognosis, the 1- and 5-year
survival rates of patients with PC are only 8 and 3%, respectively
(1). American Cancer Society
statistics reported that there were an estimated 36,800 related
fatalities and 43,140 new cases of PC in 2010. PC remains the
fourth leading cause of cancer-related mortality in the United
States, despite advances in detection, chemotherapy and surgery
(2). In developing countries, for
example, in China, the incidence of PC has also been markedly
increasing during the past several decades, and PC has been ranked
the sixth leading cause of death from malignant disease (3).
The inability to detect PC in its early treatable
stage may be the critical factor contributing to high mortality. PC
is characterized by the lack of notable clinical symptoms and
patients often present with symptoms, such as back pain, weight
loss, and digestive problems (4).
As many as 80% of newly diagnosed patients with PC are already in
the metastatic stage of the disease, which limits the potential for
therapeutic intervention (5). At
this stage, several epigenetic as well as genetic changes have
taken place and result in the silencing of tumor suppressors and
overexpression of oncogenes, ultimately leading to tumor
progression (6). In recent years,
important advances have been made to understand the molecular
biology of PC and genetic analyses have verified that the basis of
this malignant disease is heterogenous and complex (7). The occurrence and pathogenesis of PC,
however, is not yet completely understood.
Similar to the majority of tumors, the development
and growth of PC is a multistep process including initiation,
progression, invasion and ultimately metastasis. Each step in this
process is considered to be driven by the accumulation of genetic
alterations (8). Numerous studies
involving PC have been conducted in order to identify
cancer-causing genes over the past decade, and as a result several
cancer-related genes have been identified (9,10).
For instance, DPC4, which encodes SMAD family member 4
(SMAD4), is found to be inactivated in ~50% of all PCs (11). KRAS, an oncogene which is
associated with cell survival, proliferation and differentiation,
has been identified in >90% of patients with PC, with the
majority of these being point mutations at codon 12. In addition,
it has been demonstrated that the detection of the KRAS
mutation may be useful in identifying patients at high risk for
developing PC (12). The
identification and characterization of cancer-associated genes have
increased the understanding of PC development. However, the
survival rate has not improved as much in the past years due to the
lack of early diagnosis and effective chemotherapeutic treatments.
Therefore, identification of genes associated with the development
of PC is required.
To test the hypothesis that the FK506-binding
protein 51 (FKBP51) may function as a tumor suppressor, Pei
et al (13) performed
microarray analysis and submitted the expression profile, including
36 pancreatic cancer tissue samples and 16 normal samples, to the
Gene Expression Omnibus database (GEO). This previous study was
predominantly focused on the functional mechanism of the single
gene FKBP51. Based on the gene expression profile submitted
by Pei et al (13) and
bioinformatics methods, differentially expressed genes (DEGs)
between PC tissue samples and normal samples were determined in the
present study. Furthermore, functional annotation of DEGs was
conducted, followed by the construction of the protein-protein
interaction (PPI) network. This study aimed to increase the
understanding of the mechanism underlying PC development.
Materials and methods
Affymetrix microarray data
The expression profiles were accessible at the
National Center of Biotechnology Information (NCBI) Gene Expression
Omnibus database (http://www.ncbi.nlm.nih.gov/geo) using the series
accession number GSE16515, which was deposited by Pei et al
(13). This data set was based on
the GPL570 platform of [Affymetrix Human Genome U133 Plus 2.0 Array
(HG-U133_Plus_2); Affymetrix, Santa Clara, CA, USA] and updated on
Aug 22, 2014. A total of 52 chips were divided into 2 groups: PC
tissue samples (T-group, n=36) and normal samples (N-group,
n=16).
Data processing and DEG
identification
The probe-level data were firstly transformed into
gene expression data. Then background corrections and quartile data
normalization were conducted using the robust multiarray average
(RMA) in the affy package (Fred Hutchinson Cancer Research Center,
Seattle, WA, USA) with default parameters (14).
To screen DEGs between the T-group and N-group, the
Limma package (Linear Models for Microarray Data) in R language was
used (15). The raw P-value was
adjusted to the false discovery rate based on the
Benjamini-Hochberg approach (16,17)
using the Limma package (version 3.22.1; Fred Hutchinson Cancer
Research Center). DEGs were identified with the cutoff value of
FDR<0.05 and |log (fold change)|>1 (18,19).
Subnetwork analysis
Paired Fuzzy SNet (PFSNet) (20) is a powerful method to identify
smaller parts of pathways termed subnetworks. Comparison with
previously published methods shows that significant subnetworks
(and the genes therein) identified by PFSNet are up to 51% (64%)
more consistent across independent datasets of the same disease
phenotypes, even for datasets based on different platforms
(20). In order to obtain the
genes and subnetworks that may be associated with the biological
characteristics of a sample, PFSNet was used in this study to
analyze the subnetworks of the genes from the T-group as well as
the N-group based on pathways from PathwayAPI (21), which integrated Wikipathways
(22), Kyoto Encyclopedia of Genes
and Genomes (KEGG) (23) and
Ingenuity (24). Steps were
conducted as follows: i) The pathways were divided into several
subnetworks according to the genes with high expression level; ii)
the subnetwork of each group was scored as 1 or 2, based on the
equation as previously described (20); iii) the difference of scores in
each group was evaluated by t-test and the subnetworks with
significantly different scores were screened out. Parameters in the
PFSNet were set as b= 0.5, t1=0.95, and
t2= 0.8.
Gene ontology (GO) function annotation
and pathway enrichment analysis
The Database for Annotation Visualization and
Integrated Discovery (DAVID) provides a comprehensive set of
functional annotation tools to determined the biological meaning of
a large list of genes (25). DAVID
was used for GO function annotation and REACTOME pathway enrichment
analysis of the up- and downregulated genes, respectively.
P<0.05 was selected as the cut-off criterion.
PPI network construction
Search Tool for the Retrieval of Interacting Genes
(STRING, http://string-db.org/) is an online
database which includes experimental as well as predicted
interaction information and comprises >1,100 completely
sequenced organisms (26). The
protein interactions in the STRING database were shown with a
confidence score. To identify the interactive associations between
the target genes and other genes, the up- and downregulated genes
between the T-group and N-group were inputted into STRING and
protein pairs with a confidence score ≥0.7 were considered to be
significant. Cytoscape (National Institute of General Medical
Sciences of the National Institutes of Health, Bethesda, MD, USA)
was performed to visualize the PPI network.
The PPI network was complicated; thus, further
analysis was required to expose the enriched functional modules of
the PPI network using ClusterONE (Clustering with overlapping
neighborhood expansion) in Cytoscape (27). Then DAVID was used to annotate the
function of genes in each module.
Results
DEG identification
After data preprocessing, the normalized expression
profile data were analyzed using Limma package in R language. With
FDR<0.05 and |log (fold change)|>1, 1,989 DEGs including
1,461 up- and 528 downregulated genes, were screened out in the
T-group compared with the N-group.
Subnetwork analysis
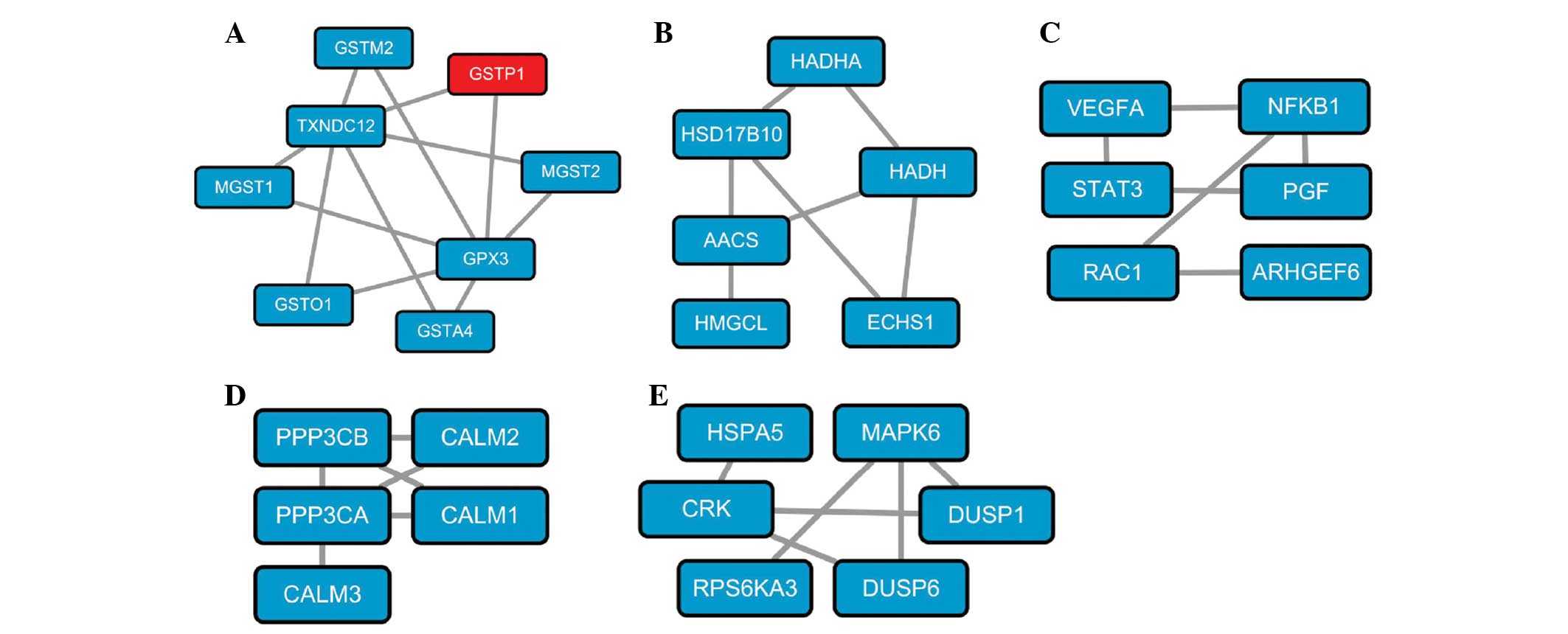
In the N-group, 5 significant subnetworks were
identified and were shown to be associated with glutathione
metabolism (Fig. 1A), leucine and
isoleucine metabolism (Fig. 1B),
pancreatic cancer (Fig. 1C),
calcium signaling pathway (Fig.
1D) and the mitogen-activated protein kinase pathway (Fig. 1E), respectively. Significant
subnetworks of genes from the T-group were in association with
galactose metabolism (Fig. 2A),
alanine, aspartic acid and glutamic acid metabolism (Fig. 2B), intercellular cell adhesion
(Fig. 2C) and contraction of
vascular smooth muscle (Fig. 2D and
E).
Function and pathway annotation
To gain further insight into the function of the
identified DEGs, the online biological classification software
DAVID was applied to annotate the DEGs. The upregulated genes were
enriched in 14 GO subcategories with the most genes enriched in the
cell adhesion pathway (Table I).
The downregulated genes were enriched in 9 subcategories with the
highest number of genes enriched in the proteolysis pathway
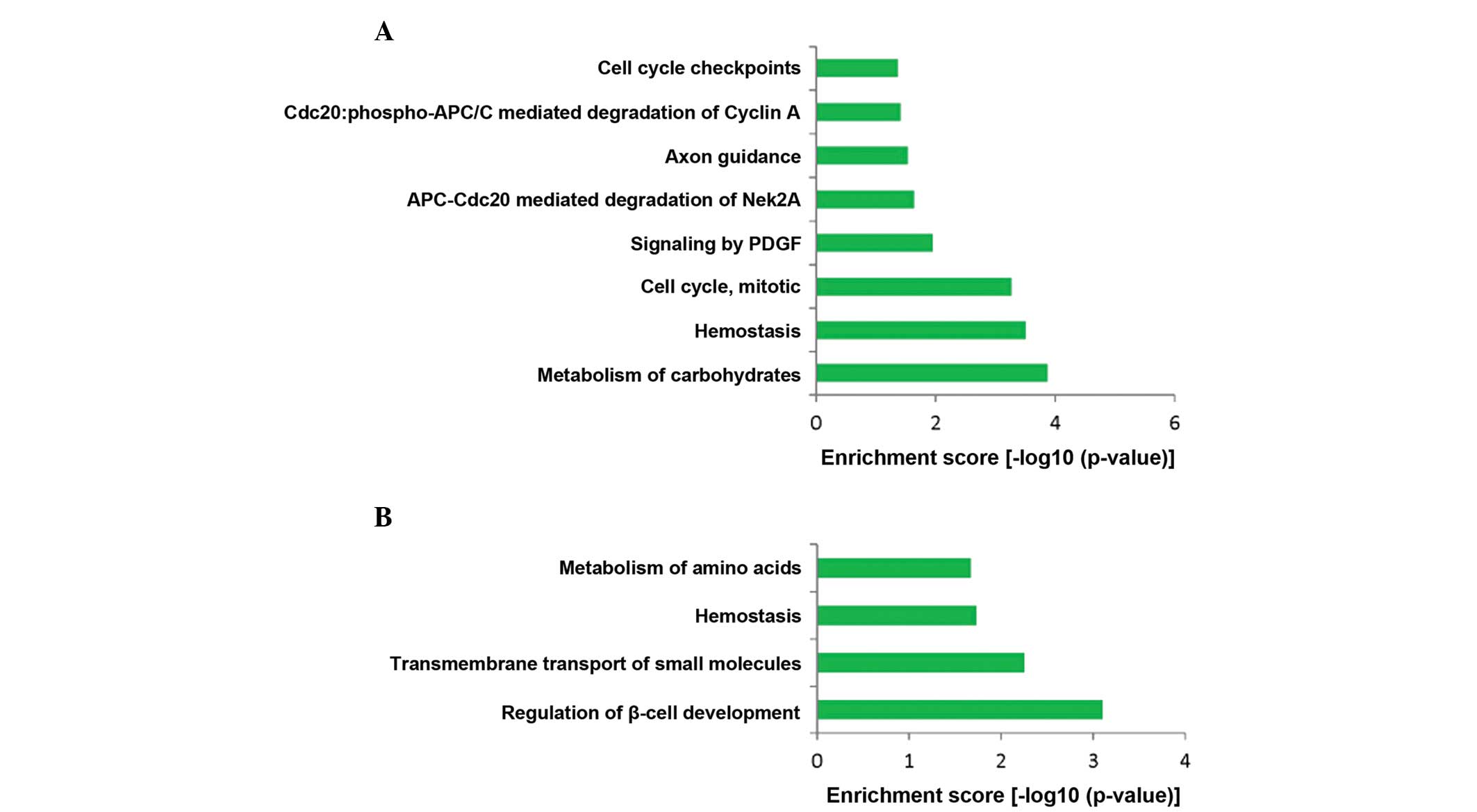
(Table I). In addition, 8
significant REACTOME pathways for upregulated genes, such as
metabolism of carbohydrates, hemostasis and cell cycle, mitotic
were identified (Fig. 3A).
Moreover, 4 significant REACTOME pathways for downregulated genes,
regulation of β-cell development, transmembrane transport of small
molecules, hemostasis and metabolism of amino acids were screened
out (Fig. 3B).
 | Table ITop gene ontology functional
enrichment of up- and downregulated genes. |
Table I
Top gene ontology functional
enrichment of up- and downregulated genes.
A, Upregulated
|
|---|
| Term | Gene count | P-value |
|---|
| GO:0007155~cell
adhesion | 73 | <0.001 |
|
GO:0022610~biological adhesion | 73 | <0.001 |
| GO:0006955~immune
response | 66 | <0.001 |
| GO:0006952~defense
response | 59 | 0.000001 |
|
GO:0042127~regulation of cell
proliferation | 58 | 0.001423 |
| GO:0007049~cell
cycle | 57 | 0.001696 |
| GO:0009611~response
to wounding | 52 | 0.000002 |
| GO:0010033~response
to organic substance | 51 | 0.006513 |
|
GO:0055114~oxidation reduction | 47 | 0.004401 |
| GO:0022402~cell
cycle process | 46 | 0.000678 |
| GO:0008219~cell
death | 46 | 0.046477 |
|
GO:0016265~death | 46 | 0.047681 |
| GO:0008283~cell
proliferation | 43 | 0.000016 |
| GO:0022403~cell
cycle phase | 41 | 0.000024 |
B, Downregulated
|
|---|
| Term | Gene count | P-value |
|---|
|
GO:0006508~proteolysis | 33 | 0.000243 |
| GO:0010033~response
to organic substance | 24 | 0.001006 |
| GO:0006811~ion
transport | 22 | 0.009722 |
|
GO:0042592~homeostatic process | 21 | 0.015010 |
| GO:0009611~response
to wounding | 20 | 0.000721 |
| GO:0048878~chemical
homeostasis | 18 | 0.003062 |
| GO:0009719~response
to endogenous stimulus | 17 | 0.000678 |
| GO:0019725~cellular
homeostasis | 17 | 0.002910 |
| GO:0009725~response
to hormone stimulus | 16 | 0.000718 |
PPI construction and module analysis
The up- and downregulated genes between the T-group
and N-group were input into the STRING database to identify the
significant interactions with a confidence score of ≥0.7. The PPI
network reveals the molecular mechanisms of pancreatic cancer, but
it contains too many nodes and interactions to select the useful
information. Therefore, the functional modules in the PPI network
were mined by ClusterONE. In the current study, the significant
module with the lowest P-value for the upregulated DEGs was
displayed in Fig. 4A, and 5 DEGs
with higher connectivity degrees including cyclin-dependent kinase
1 (CDK1), maternal embryonic leucine zipper kinase
(MELK), PDZ-binding kinase (PBK), Cyclin A2
(CCNA2) and nucleolar and spindle associated protein 1
(NUSAP1) were included in this module. GO analysis showed
that the DEGs in this module were predominantly associated with
cell-division cycle (Table II).
While, 5 hub genes with the higher degrees including albumin
(ALB), carboxypeptidase A1 (pancreatic) (CPA1),
colipase, pancreatic (CLPS), epidermal growth factor
(EGF) and complement component 5 (C5) were identified
in the significant module with the lowest P-value for the
downregulated DEGs with the lowest P-values, which is shown in
Fig. 4B. Moreover, the DEGs in
this module predominantly participated in biological processes,
such as response to wounding, endogenous stimulus and regulation of
cell proliferation (Table
II).
 | Table IITop Gene Ontology annotation of up-
and downregulated genes in the significant module with the lowest
P-value of the protein-protein interaction network. |
Table II
Top Gene Ontology annotation of up-
and downregulated genes in the significant module with the lowest
P-value of the protein-protein interaction network.
A, Upregulated
|
|---|
| Term | Gene count | P-value |
|---|
| GO:0007049~cell
cycle | 44 | <0.001 |
| GO:0022402~cell
cycle process | 38 | <0.001 |
| GO:0022403~cell
cycle phase | 37 | <0.001 |
| GO:0000279~M
phase | 36 | <0.001 |
| GO:0000278~mitotic
cell cycle | 32 | <0.001 |
| GO:0051301~cell
division | 29 | <0.001 |
B, Downregulated
|
|---|
| Term | Gene count | P-value |
|---|
|
GO:0006508~proteolysis | 15 | 0.000234 |
| GO:0009611~response
to wounding | 11 | 0.000134 |
| GO:0010033~response
to organic substance | 11 | 0.001507 |
| GO:0009725~response
to hormone stimulus | 9 | 0.000256 |
| GO:0009719~response
to endogenous stimulus | 9 | 0.000495 |
|
GO:0042127~regulation of cell
proliferation | 9 | 0.026531 |
Discussion
PC is one of the leading causes of cancer-related
mortality worldwide; however, the molecular mechanisms of PC
progression remain unclear. With the rapid expansion of knowledge
on genomics, emerging evidence suggests that the initiation,
progression, invasion and metastasis of PC are generally caused by
the differential expression of genes. In the present study, a total
of 1,989 DEGs including 1,461 up- and 528 downregulated genes were
screened out. In line with the results of the study by Pei et
al (13), FKBP5 was
identified as one of the downregulated genes in the PC samples. To
understand the interaction of these DEGs, a PPI network was
constructed and the significant module with the lowest P-value for
upregulated genes with the top 5 nodes of CDK1, MELK,
PBK, CCNA2 and NUSAP1 and the module with the
lowest P-value for downregulated genes with the top 5 nodes of
ALB, CPA1, CLPS, EGF and C5 were
identified. Among all these proteins, CDK1, ALB,
CPA1, CLPS and EGF were verified to be
associated with PC (28–31). Moreover, the association of
MELK and C5 with PC have been demonstrated in certain
studies (32–34). However, according to the present
results, CCNA2 and PBK, which have not previously
been directly associated with PC, may be pivotal for the initiation
and progression of PC. In addition, certain subnetworks may be
important in PC via the differential expression of genes involved,
such as the subnetwork directly associated with PC and the
subnetwork associated with intercellular cell adhesion.
Cyclins are a family of proteins that control the
progression of cells through the cell cycle by activating
cyclin-dependent kinases (CDK) (35). As a member of the cyclin family,
CCNA2 is produced at the onset of DNA synthesis in proliferating
somatic cells and is critical in cell cycle progression by
regulation of transition from G1 to S phase (36). Genetic variants of CCNA2,
which may affect the function of the encoded protein by changing
gene expression or by altering the protein structure, are found to
significantly increase the risk of cancer development in a
tissue-specific manner, such as colon, liver and lung cancer
(37). In addition, Gao et
al (38) reported that
CCNA2 was a biomarker for the prognosis of breast cancer and
a promising target for developing novel strategies to prevent or
even reverse tamoxifen resistance. In addition, the expression of
CCNA2 may aid in monitoring tamoxifen efficacy and directing
personalized therapies in patients with breast cancer (38). Few previous studies have focused on
the association of CCNA2 and PC, while high throughput
bioinformatics analysis in the present study indicates that
CCNA2 may be important for the initiation and development of
PC. In the present study, CCNA2 was found to be upregulated
in PC tissue samples, and functional analysis demonstrates that
CCNA2 was predominantly enriched in the cell cycle pathway
and participates in biological processes, such as regulation of
cell proliferation, regulation of cell cycle, cell cycle checkpoint
and mitosis. These findings were concordant with those of previous
studies (39,40). Therefore, it was hypothesized that
CCNA2 may be important in the pathogenesis of PC via
regulation of the cell cycle and mitosis, which may further
influence tumor occurrence.
PBK, also known as PDZ-binding kinase, is a mitotic
protein kinase and its encoding gene, PBK was found to be
upregulated in PC tissue samples. Studying upregulated kinases in
cancer may provide important clues as to the mechanism of malignant
conversion (41,42). PBK is phosphorylated in
vitro by Cdc2-cyclin B at a site in the amino terminus (Thr 9)
which is implicated in the binding of α-tubulin, and then localizes
to mitotic spindles and spindle poles during metaphase (43). Studies regarding PBK have
demonstrated that the expression of PBK is regulated by cell
cycle-specific transcription factors, such as E2F and CREB/ATF, and
knockdown expression of PBK may lead to cytokinetic
dysfunction in breast cancer (44,45).
Ayllón et al (46)
suggested that PBK is involved in DNA damage sensing and
repair via phosphorylating c-H2AX. Nandi et al (47) confirmed that with western
immunoblotting and immunoprecipation, and yeast two-hybrid
analysis, PBK can directly interact with p53, downregulate its
expression and attenuate G2/M checkpoint in fibrosarcoma cells,
which was hypothesized to be a plausible explanation for the role
of PBK in augmenting tumor cell growth. Similarly, the
GO-biological process enrichment in the present study predicted
that PBK was predominantly associated with nuclear division,
cell division, M phase of mitotic cell cycle, and PBK with
higher connectivity degree in the module with the lowest P-value of
upregulated genes was enriched in cell division cycle. Based on
these results, it was inferred that PBK may influence the
occurrence of PC by regulating the mitotic cell cycle and other
biological processes.
Intercellular cell adhesion determines the polarity
of cells and participates in the maintenance of tissues (48). Several studies have shown that
cell-cell adhesiveness is generally reduced in human cancer, which
may result in influences as follows: Reduced intercellular
adhesiveness allows loss and disruption of cell-cell adhesion,
resulting in destruction of histological structure, which is the
morphological hallmark of malignant tumors (48). Conversely, reduced intercellular
adhesiveness is also indispensable for cancer invasion and
metastasis (49). In line with the
previous studies, subnetworks associated with intercellular cell
adhesion were found to be significant in the T-group, and the
majority of the genes in this subnetwork were identified to be
upregulated in PC. Accordingly, intercellular cell adhesion may be
important in the progression of PC.
The significant subnetwork directly associated with
PC consisted of six genes, VEGFA, NFKB1, STAT3, PGF, RAC1
and ARHGEF6. Expression levels of the majority of these
genes were identified to be significantly higher in PC samples and
this subnetwork is directly involved in PC via the differential
expression of genes involved. For instance, STAT3 is
confirmed to be vital in anti-pancreatic cancer effects through its
contributions to the positive feedback loop between reactive oxygen
species and autophagy (50). The
concentration of PGF is found to be significantly increased
in pancreatic carcinoma compared with tumor-free tissue (51). Moreover, activation of
RAC1-dependent superoxide generation leads to PC cell
proliferation and inhibition of RAC1 may be a potential
therapeutic strategy (52). Hence,
as demonstrated, subnetworks directly associated with pancreatic
cancer may be crucial in the pathogenesis of PC.
In conclusion, the results of this study may
increase the understanding of the mechanism of the occurrence and
development of PC. CCNA2 and PBK of the module and
their relative pathway cell-division cycle may be pivotal for
understanding the molecular mechanism of PC. In addition, two
subnetworks (pancreatic cancer subnetwork and intercellular
adhesion subnetwork) may be highly associated with PC. However, the
whole study was conducted based on bioinformatics methods, and the
conclusions have not been verified by corresponding experiments
yet. Thus, further experiments are urgently required to confirm the
results of this study.
References
|
1
|
Moore MJ, Goldstein D, Hamm J, Figer A,
Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA, et al:
Erlotinib plus gemcitabine compared with gemcitabine alone in
patients with advanced pancreatic cancer: A phase III trial of the
national cancer institute of canada clinical trials group. J Clin
Oncol. 25:1960–1966. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Long J, Luo GP, Xiao ZW, Liu ZQ, Guo M,
Liu L, Liu C, Xu J, Gao YT, Zheng Y, et al: Cancer statistics:
Current diagnosis and treatment of pancreatic cancer in Shanghai,
China. Cancer Lett. 346:273–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan A, Diamandis EP and Blasutig IM:
Strategies for discovering novel pancreatic cancer biomarkers. J
Proteomics. 81:126–134. 2013. View Article : Google Scholar :
|
|
5
|
Pliarchopoulou K and Pectasides D:
Pancreatic cancer: Current and future treatment strategies. Cancer
Treat Rev. 35:431–436. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sato N and Goggins M: The role of
epigenetic alterations in pancreatic cancer. J Hepatobiliary
Pancreat Surg. 13:286–295. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jones S, Zhang X, Parsons DW, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et
al: Core signaling pathways in human pancreatic cancers revealed by
global genomic analyses. Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Logsdon CD, Simeone DM, Binkley C,
Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R and Hanash
S: Molecular profiling of pancreatic adenocarcinoma and chronic
pancreatitis identifies multiple genes differentially regulated in
pancreatic cancer. Cancer Res. 63:2649–2657. 2003.PubMed/NCBI
|
|
9
|
Wang B, Sun S and Liu Z: Analysis of
dysregulation of immune system in pancreatic cancer based on gene
expression profile. Mol Biol Rep. 41:4361–4367. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kern SE: Molecular genetic alterations in
ductal pancreatic adenocarcinomas. Med Clin North Am. 84:691–695.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shin SH, Kim SC, Hong SM, Kim YH, Song KB,
Park KM and Lee YJ: Genetic alterations of K-ras, p53, c-erbB-2 and
DPC4 in pancreatic ductal adenocarcinoma and their correlation with
patient survival. Pancreas. 42:216–222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fryzek JP, Garabrant DH, Schenk M, Kinnard
M, Greenson JK and Sarkar FH: The association between selected risk
factors for pancreatic cancer and the expression of p53 and K-ras
codon 12 mutations. Int J Gastrointest Cancer. 37:139–145.
2006.
|
|
13
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dorsey ER, Constantinescu R, Thompson JP,
Biglan KM, Holloway RG, Kieburtz K, Marshall FJ, Ravina BM,
Schifitto G, Siderowf A and Tanner CM: Projected number of people
with Parkinson disease in the most populous nations, 2005 through
2030. Neurology. 68:384–386. 2007. View Article : Google Scholar
|
|
15
|
Delhomme N, Padioleau I, Furlong EE and
Steinmetz LM: easyRNASeq: A bioconductor package for processing
RNA-Seq data. Bioinformatics. 28:2532–2533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Singh B, Ronghe AM, Chatterjee A, Bhat NK
and Bhat HK: MicroRNA-93 regulates NRF2 expression and is
associated with breast carcinogenesis. Carcinogenesis.
34:1165–1172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chand Y and Alam MA: Network biology
approach for identifying key regulatory genes by expression based
study of breast cancer. Bioinformation. 8:1132–1138. 2012.
View Article : Google Scholar :
|
|
18
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 289–300.
1995.
|
|
19
|
Benjamini Y: Discovering the false
discovery rate. J R Stat Soc Series B Stat Methodol. 72:405–416.
2010. View Article : Google Scholar
|
|
20
|
Lim K and Wong L: Finding consistent
disease subnetworks using PFSNet. Bioinformatics. 30:189–196. 2014.
View Article : Google Scholar
|
|
21
|
Soh D, Dong D, Guo Y and Wong L:
Consistency, comprehensiveness, and compatibility of pathway
databases. BMC Bioinformatics. 11:4492010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kelder T, van Iersel MP, Hanspers K,
Kutmon M, Conklin BR, Evelo CT and Pico AR: WikiPathways: Building
research communities on biological pathways. Nucleic Acids Res.
40(Database Issue): D1301–D1307. 2012. View Article : Google Scholar :
|
|
23
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40(Database
Issue): D109–D114. 2012. View Article : Google Scholar :
|
|
24
|
Krämer A, Green J, Pollard J Jr and
Tugendreich S: Causal analysis approaches in ingenuity pathway
analysis. Bioinformatics. 30:523–530. 2014. View Article : Google Scholar :
|
|
25
|
Hu Y, Hu Y and Liu D, Yu J and Liu D:
Screening and bioinformatics analysis of differentially expressed
genes in hyperplastic scar. Nan Fang Yi Ke Da Xue Xue Bao.
34:939–944. 2014.In Chinese. PubMed/NCBI
|
|
26
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Database Issue): D808–D815. 2013. View Article : Google Scholar :
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feldmann G, Mishra A, Bisht S, Karikari C,
Garrido-Laguna I, Rasheed Z, Ottenhof NA, Dadon T, Alvarez H,
Fendrich V, et al: Cyclin-dependent kinase inhibitor Dinaciclib
(SCH727965) inhibits pancreatic cancer growth and progression in
murine xenograft models. Cancer Biol Ther. 12:598–609. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hempen PM, Kurpad H, Calhoun ES, Abraham S
and Kern SE: A double missense variation of the BUB1 gene and a
defective mitotic spindle checkpoint in the pancreatic cancer cell
line Hs766T. Hum Mutat. 21:4452003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang P, Zou M, Wen X, Gu F, Li J, Liu G,
Dong J, Deng X, Gao J, Li X, et al: Development of serum parameters
panels for the early detection of pancreatic cancer. Int J Cancer.
134:2646–2655. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Renouf D and Moore M: Evolution of
systemic therapy for advanced pancreatic cancer. Expert Rev
Anticancer Ther. 10:529–540. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kusakai G, Suzuki A, Ogura T, Kaminishi M
and Esumi H: Strong association of ARK5 with tumor invasion and
metastasis. J Exp Clin Cancer Res. 23:263–268. 2004.PubMed/NCBI
|
|
33
|
Kokkinakis DM, Liu X and Neuner RD:
Modulation of cell cycle and gene expression in pancreatic tumor
cell lines by methionine deprivation (methionine stress):
Implications to the therapy of pancreatic adenocarcinoma. Mol
Cancer Ther. 4:1338–1348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Michl P, Buchholz M, Rolke M, Kunsch S,
Löhr M, McClane B, Tsukita S, Leder G, Adler G and Gress TM:
Claudin-4: A new target for pancreatic cancer treatment using
Clostridium perfringens enterotoxin. Gastroenterology. 121:678–684.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dun B, Sharma A, Xu H, Liu H, Bai S, Zeng
L and She JX: Transcriptomic changes induced by mycophenolic acid
in gastric cancer cells. Am J Transl Res. 6:28–42. 2013.PubMed/NCBI
|
|
36
|
Gong D, Pomerening JR, Myers JW,
Gustavsson C, Jones JT, Hahn AT, Meyer T and Ferrell JE Jr: Cyclin
A2 regulates nuclear-envelope breakdown and the nuclear
accumulation of cyclin B1. Curr Biol. 17:85–91. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim DH, Park SE, Kim M, Ji YI, Kang MY,
Jung EH, Ko E, Kim Y, Kim S, Shim YM and Park J: A functional
single nucleotide polymorphism at the promoter region of cyclin A2
is associated with increased risk of colon, liver and lung cancers.
Cancer. 117:4080–4091. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao T, Han Y, Yu L, Ao S, Li Z and Ji J:
CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen
resistance. PLoS One. 9:e917712014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kokkinakis DM, Liu XY and Neuner RD:
Modulation of cell cycle and gene expression in pancreatic tumor
cell lines by methionine deprivation (methionine stress):
Implications to the therapy of pancreatic adenocarcinoma. Mol
Cancer Ther. 4:1338–1348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Roderick HL and Cook SJ: Ca2+
signalling checkpoints in cancer: Remodelling Ca2+ for
cancer cell proliferation and survival. Nat Rev Cancer. 8:361–375.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Baselga J and Arribas J: Treating cancer's
kinase 'addiction'. Nat Med. 10:786–787. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bettencourt-Dias M, Giet R, Sinka R,
Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S,
Winter S, Carthew RW, et al: Genome-wide survey of protein kinases
required for cell cycle progression. Nature. 432:980–987. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gaudet S, Branton D and Lue RA:
Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl
Acad Sci USA. 97:5167–5172. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Park JH, Lin ML, Nishidate T, Nakamura Y
and Katagiri T: PDZ-binding kinase/T-LAK cell-originated protein
kinase, a putative cancer/testis antigen with an oncogenic activity
in breast cancer. Cancer Res. 66:9186–9195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nandi AK and Rapoport AP: Expression of
PDZ-binding kinase (PBK) is regulated by cell cycle-specific
transcription factors E2F and CREB/ATF. Leuk Res. 30:437–447. 2006.
View Article : Google Scholar
|
|
46
|
Ayllón V and O'connor R: PBK/TOPK promotes
tumour cell proliferation through p38 MAPK activity and regulation
of the DNA damage response. Oncogene. 26:3451–3461. 2007.
View Article : Google Scholar
|
|
47
|
Nandi AK, Ford T, Fleksher D, Neuman B and
Rapoport AP: Attenuation of DNA damage checkpoint by PBK, a novel
mitotic kinase, involves protein-protein interaction with tumor
suppressor p53. Biochem Biophys Res Commun. 358:181–188. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hirohashi S and Kanai Y: Cell adhesion
system and human cancer morphogenesis. Cancer Sci. 94:575–581.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Saiki I: Cell adhesion molecules and
cancer metastasis. Jpn J Pharmacol. 75:215–242. 1997. View Article : Google Scholar
|
|
50
|
Gong J, Muñoz AR, Chan D, Ghosh R and
Kumar AP: STAT3 down regulates LC3 to inhibit autophagy and
pancreatic cancer cell growth. Oncotarget. 5:2529–2541. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Heukamp I, Kilian M, Gregor JI, Kiewert C,
Schimke I, Kristiansen G, Walz MK, Jacobi CA and Wenger FA: Impact
of polyunsaturated fatty acids on hepato-pancreatic prostaglandin
and leukotriene concentration in ductal pancreatic cancer-is there
a correlation to tumour growth and liver metastasis? Prostaglandins
Leukot Essent Fatty Acids. 74:223–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Aravindan S, Delma CR, Thirugnanasambandan
SS, Herman TS and Aravindan N: Anti-pancreatic cancer deliverables
from sea: First-hand evidence on the efficacy, molecular targets
and mode of action for multifarious polyphenols from five different
brown-algae. PLoS One. 8:e619772013. View Article : Google Scholar : PubMed/NCBI
|