Introduction
Aspergillus fumigatus is a filamentous
saprophyte and opportunist fungi spread ubiquitously in the
environment (1). It is a leading
cause of mold fungal infections in immune-compromised patients
(2), and is a key allergen that is
frequently accompanied by severe asthma, allergies and sinusitis
(1,3–5). The
spores are usually eradicated by the innate immune system response
in lung epithelial tissue in immune-competent host, however,
immune-compromised individuals are at risk for invasive
Aspergillosis as a result of increasing spore loads in the lungs or
other tissues (6). One of the most
important reasons why A. fumigatus is able to infect avian
and mammalian hosts is its thermotolerance capability (7). A. fumigatus is able to grow
rapidly at 37°C and has been known as thermotolerant microbial
agent (8). It has likely evolved
mechanisms and intracellular pathways of thermal resistance similar
to thermophilic fungi. A family of critically important gene
expressed at high temperatures are the heat shock proteins (HSPs),
which are a vital defense in challenged cells (9). Immediately following heat stress or
in hyperthermic conditions, heat shock transcription factor (HSF)
activates the transcription of a series of genes encoding HSPs
(10–12). HSPs have been characterized as
highly conserved molecular chaperones in evolutionary studies
(13,14). In addition, it was noted that their
expression levels are increased under various stress conditions
(15). It has been shown that the
HSP70 family of proteins contains minor molecular sequence
differences which enables them to adapt to new environmentally
imposed situations (16).
Chaperones, in particular HSP70, have been frequently reported as
an immuno-dominant response to fungal infections, and described as
a highly conserved family of ~10 proteins with a molecular mass of
approximately 70 kDa (17).
Regarding HSPs as critical microbial defense mechanisms in stressed
or high-temperature situations, such as those occurring within the
host, research in this field has great potential to increase the
clinical and scientific knowledge regarding how HSP70 is expressed
at 37°C or higher as an approach to design novel specific
therapeutics.
The present study focused on the HSP70 expression
pattern over longer durations in A. fumigatus. The aim of
the current study was to simulate the thermal conditions that occur
when fungi inhabit the host body or environment. Additionally, the
study aimed to explore the potential differences and expression
patterns of HSP70 mRNA between clinical and environmental samples
of A. fumigatus at various low and high temperatures over a
longer duration. A longer experimental duration was established in
the present study to find possible differential gene expression
patterns in comparison to shorter study durations that have been
previously reported.
Materials and methods
Strains and growth conditions
A total of 13 samples were collected by
broncho-alveolar lavages or direct blood cultures from medically
diagnosed Aspergillus infections caused by A.
fumigatus in human and animal cases. In the case of human
patients, sampling was conducted once informed consent was obtained
and for animal cases, the ethical instructions of our institutions
for animal handling and specimen collection were followed. The
present study was approved by the ethics committee of Faculty of
Veterinary Medicine, University of Tehran (Tehran, Iran).
Additionally, following a series of environmental random sampling,
six further A. fumigatus-identified isolates were
incorporated in the study for use in further experiments.
Furthermore, a standard A. fumigatus strain
(ATCC® 90906; American Type Culture Collection,
Manassas, VA, USA) was obtained and incorporated into the
experiments. This gave a total of seven samples from humans and six
samples from animals, plus six additional fresh samples from the
environment (three from food and three from air) and a standard
strain, which were cultured directly on general media in optimal
conditions for fungal growth. Briefly, to purify and enrich the
fungal isolates, fungi were cultured on sabouraud dextrose agar
(Merck Millipore, Darmstadt, Germany) and incubated at 30°C for
three days. The medium contained chloramphenicol (0.05% w/v; Merck
Millipore) and cyclohexamide (0.05% w/v; Merck Millipore) to
prevent the growth of bacteria and dermatophytic fungi. Subcultures
were avoided and immediately following the growth of fungi and
morphological, physiochemical and molecular identification, the
fungal conidia were collected using Tween 80 to trap conidia.
Briefly, the surface of the fungal culture plates were washed by
sterile phosphate-buffered saline containing 0.02% Tween 80, and
the collected fluid was centrifuged for 5 min at 5,000 × g and 4°C
to concentrate the conidia. Collected conidia were counted using a
hematocytometer. To induce the exponential/developmental phase of
fungal hyphal growth and produce a biomass of A. fumigatus
mycelia, a series of inoculations of yeast extract peptone dextrose
(YPD) liquid medium (Merck Millipore) were prepared using A.
fumigatus conidia for each clinical and environmental sample
with three biological replicates at a final concentration of
1×106 conidia/ml and incubated at 20, 30, 37 and 42°C
for five days. The standard strain was also cultured by the
described method for five days at 25°C, according to the
manufacturer's instructions.
Total RNA Extraction
Floating A. fumigatus mycelia in YPD broth
medium were harvested every 24 h for each temperature by filtration
through Whatman paper. Harvested mycelia were washed with sterile
water and then freeze-dried and ground using a mortar and pestle
containing liquid nitrogen. Total RNA was obtained using
TRIzol® Reagent RNA preparation method (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. Following precipitation, the RNA was
resuspended in a 60 µl RNAase-free tube. Contaminating
genomic DNA was removed from RNA samples by treating with 1
µl of RNAse-free DNase (SinaClon, Co., Tehran, Iran) for 30
min at room temperature. DNase was removed using a clean-up kit
(SinaClon, Co.,). RNA integrity and the quantity of total RNA were
measured using a NanoDrop 2000 (Thermo Fisher Scientific,
Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The RT-qPCR reaction was carried out using EvaGreen
which is a modified version of SYBR green fluorescent DNA-binding
dye in two steps. First, 2 µg of purified RNA was used to
synthesize first strand cDNA using a cDNA synthesis kit, which
included an engineered reverse transcriptase to resist high
temperatures providing increased specificity and higher yields of
cDNA, in a final volume of 25 µl. Oligo-dT primers were used
for the reverse transcription of protein-coding mRNAs. DEPC-treated
water used to avoid RNA degradation. Prior to the addition of
reverse transcriptase, the reaction mixture was incubated at 65°C
for 5 min to denature RNA, and to avoid RNA secondary structures
reforming, immediately chilled on ice. cDNA synthesis was initiated
by an engineered Moloney Murine Leukemia Virus reverse
transcriptase (SuperScript® III First-Strand synthesis
kit; Invitrogen; Thermo Fisher Scientific, Inc.) at 50°C for 50
min. The transcription reaction was stopped by heating the tubes to
85°C for 5 min. Synthesized cDNA was immediately used to perform
the PCR amplification procedure using a routine protocol to check
the specificity of the primers and avoid homo- or hetero-dimer
problems. qPCR gene-specific primers were incorporated in the
reaction. The remaining cDNAs were stored at -20°C until further
use.
A qPCR kit (Jena Bioscience GmbH, Munich, Germany)
with EvaGreen as the fluorescent dye was used to perform the qPCR
assay. A total of 5 µl cDNA, 0.4 µM of each forward
and reverse primer, molecular-grade water and 12.5 µl ready
to use EvaGreen qPCR reagent in a final volume of 20 µl were
used to perform the qPCR reaction using a Rotor Gene Q real-time
PCR system (Qiagen GmbH, Hilden, Germany). For each sample and for
their relative reference gene, the qPCR reaction was performed in
three technical replicates. The cycling conditions were as follows:
Initial denaturation and polymerase activation, 95°C for 2 min;
denaturation, 95°C for 15 sec; annealing, 55°C for 30 sec;
extension, 72°C for 30 sec; amplification cycles, 45 cycles; final
extension, 72°C for 5 min; and melting curve analysis, a range
between 45- and 95°C with 0.2°C increments. The melting-curve
analysis cycle was set to test for the existence of possible
primer-dimers or other unspecific products. Primers for HSP70 in
A. fumigatus were designed (Table I) using Primer-BLAST online
software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) which
uses both Primer3 and BLAST algorithms. Primers were designed to
span exon-exon junctions to avoid the amplification of possible
genomic DNA contamination. Additionally, an option to allow primers
to amplify mRNA splice variants was also enabled to make the
primers capable of amplifying all transcript variants and isoforms
of the HSP70 gene. A reference mRNA sequence retrieved from the
GenBank database (XM_745397) was selected to design the A.
fumigatus HSP70 specific primers, as it is near identical to
the Afu1g07440 sequence from the Aspergillus Genome Database
(AspGD). The BLAST online tool was used to check the similarity of
the XM_745397 reference sequence to the orthologous heat inducible
HSP70 genes in Aspergillus and other fungal species. Primers
were designed for 18S ribosomal RNA (rRNA) to be used as the
reference gene, using Primer-BLAST with its default settings
(Table I). The designed primers
were also checked for possible secondary structures in order to
avoid unexpected structures such as homo- & hetero-dimers,
using an online oligo-nucleotide analysis tool (IDT OligoAnalyzer
3.1; https://www.idtdna.com/calc/analyzer) (18). In addition, to check whether
primers were able to amplify the target and reference gene cDNAs, a
series of conventional PCR assays was performed. The PCR products
were run on 1.5% agarose electrophoresis gels in TBA buffer (0.45 M
Tris-Borate and 0.001 M ethylenediaminetetraacetic acid) at 80 V
for 90 min. Gels were stained with ethidium bromide (0.5
µg/ml) and photographed using UV light in a gel
documentation system (G:Box; Syngene, Cambridge, UK).
 | Table IPrimer sequences used. |
Table I
Primer sequences used.
| Gene | Direction | Sequence | Length (bp) | Amplicon size
(bp) | Source accession
no. |
|---|
| 18S rRNA | Forward |
5′-TCAGGGAACGAAAGTTAGGG-3′ | 20 | 107 | AB008401 |
| Reverse |
5′-CGAGCGGGTCATCATAGAAA-3′ | 20 | | |
| HSP70 | Forward |
5′-GCCATTGCCTACGGTCTTG-3′ | 19 | 225 | XM_745397 |
| Reverse |
5′-GGTGAGATCCTTCTTGTGCTT-3′ | 21 | | |
Statistical analysis and comparative
Cq method
MedCalc software, version 12 (MedCalc Software,
Oostende, Belgium) (19) was used
for statistical analysis. The Kolmogorov-Smirnov and normal
probability plot indicated the data was normally distributed thus,
two-tailed Student's t-test was used to test for significant
differences between means. Tukey's test was used for multiple
comparisons among several time points. The α-level was set at 0.05,
thus P<0.05 was considered to indicate a statistically
significant difference. The mean, median and 95% confidence
interval (CI) of data were calculated using MedCalc software,
version 12 (MedCalc Software bvba, Ostend, Belgium). Three
specimens were collected from air and three from food and, to avoid
statistical bias due to small sample size, all six were considered
together as samples in a single environmental group. Expression
fold changes and relative expression differences between
experimental groups were calculated using 2–ΔΔCq method,
which is additionally known as the comparative Cq or
Livak method (20). In this
method, the efficiency of both the target gene and reference gene
are assumed to be 100%. The mathematical calculations of the method
are based on the normalization of the quantification cycle (Cq)
values of both the sample and control groups to a reference gene,
to prevent a cDNA amount input error, then the subtraction of the
Cq values of the two experimental groups from each other, prior to
the logarithmic transform of the resulting fold change values, to
avoid misinterpreting the expression differences between the
evaluated groups. The fold change values for each temperature point
and experimental day in the sample groups were presented as graphs
relative to the control group (A. fumigatus ATCC 90906
standard strain) expression level on the same day. The formula for
the comparative Cq method is as follows:
Relative expression fold change=2–[(meanCq of
target gene in sample or treated group-meanCq of reference gene in
sample or treated group)–(meanCq of target gene in control or
untreated group-meanCq of reference gene in control or untreated
group)].
Results
HSP70 primers and cDNAs were used in conventional
PCR reactions, resulting in the detection of HSP70 expression in
the different samples. The clinical samples exhibited clearly
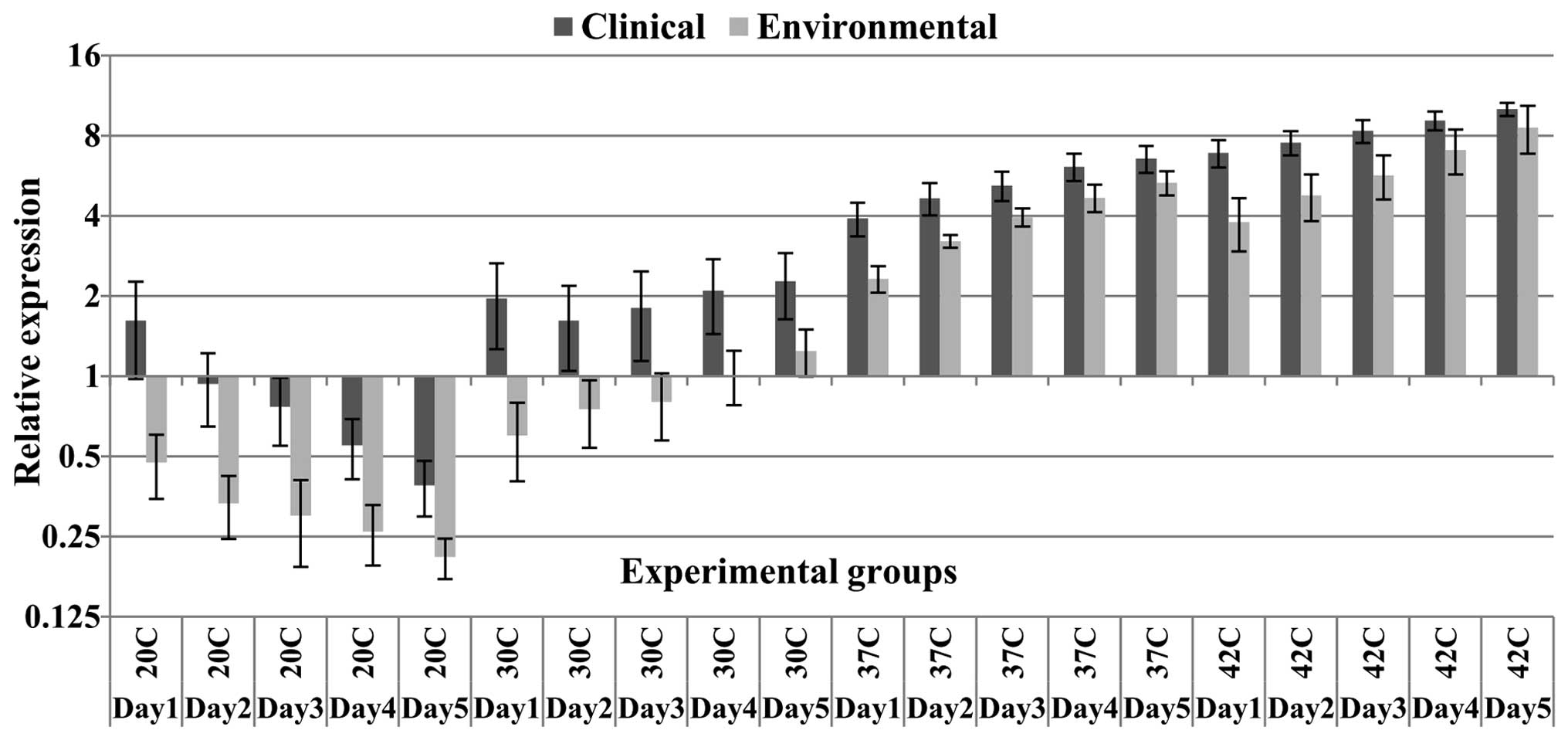
detectable bands following gel electrophoresis (Fig. 1). Additionally, at all the
temperatures except, at the fifth day of incubation at 42°C,
relative expression of HSP70 mRNA in clinical samples was
significantly higher compared with the environmental samples
(Table II). There was a ~2-fold
change in the expression levels normalized to the 18S RNA reference
gene in the comparison of clinical and environmental samples at all
temperatures during all experiment days, except at 37°C and 42°C,
where the majority of environmental samples exhibited smaller
differences in expression levels compared with expression levels in
the clinical samples (Fig. 2).
There was no significant difference in HSP70 mRNA levels between
clinical and environmental samples at the fifth day of incubation
at 42°C. Among all the experimental temperature points, the least
significant difference in HSP70 expression levels was observed
between clinical and environmental groups at 42°C (Table II). In detail, from the six
environmental samples, four samples (66.6%) at 42°C exhibited
relatively competitive levels of expression compared with clinical
samples. However, except for the fifth day, significant differences
in the mean expression levels compared with the clinical samples at
42°C were observed for all incubation days (subgroup data not
shown), however, the P-values at 42°C were higher than the P-values
at the other higher temperature points (Table II). HSP70 mRNA levels in the
environmental samples at the fourth and fifth days of incubation at
37°C and 42°C was closer to the HSP70 expression levels in the
clinical samples in comparison with the expression levels at the
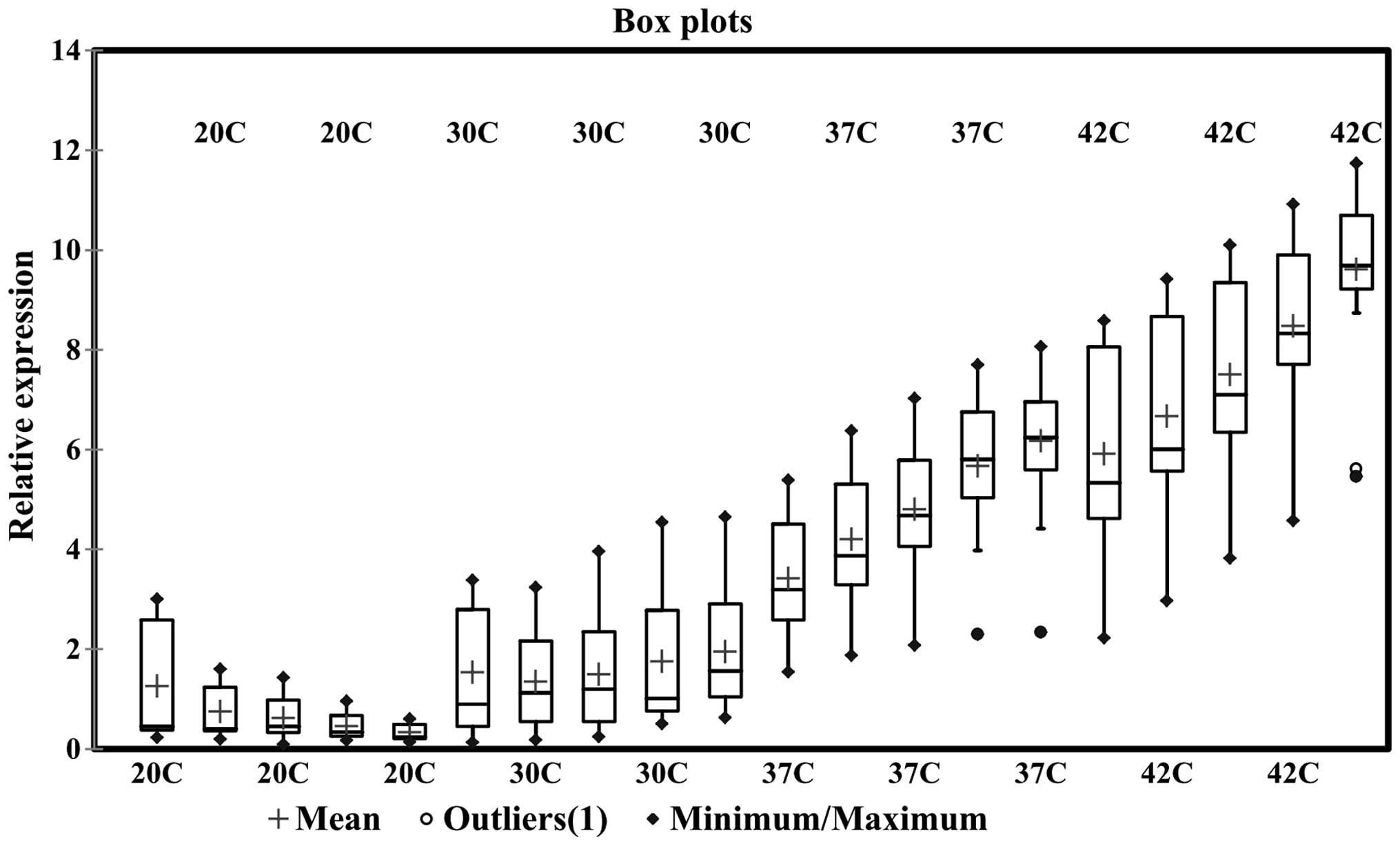
first three days of incubation. Notably, one environmental strain
demonstrated significantly lower expression levels of HSP70 mNRA at
all temperatures, even at 42°C compared with the clinical samples.
This strain was detected as an outlier in the confidence interval
calculations and is indicated in the box-whisker plot as a separate
data point (Fig. 3). Additionally,
the expression levels of the environmental group at 37 and 42°C was
significantly higher compared with the expression levels at lower
temperature points (P<0.05, Tukey's test). Furthermore, despite
the mentioned high levels of expression at 37°C in the
environmental group, differences in the expression levels between
the clinical and environmental groups at this temperature were
significant. By contrast, the clinical samples showed a marked
increase in HSP70 expression, in particular at 37 and 42°C, with an
up to 10-fold increase compared with the reference gene. Of note,
on the first day at 20°C, the clinical samples showed an
upregulation of HSP70, while over the following days, in both
clinical and environmental samples, the HSP70 gene was
downregulated (Fig. 2). However,
at 30°C, the clinical samples demonstrated an upregulation of HSP70
mRNA levels at all five days. Additionally, the clinical samples on
the first day at 20°C demonstrated expression levels greater than
on the second day. In fact, HSP70 expression at 30°C in the
clinical samples showed an initial reduction on the second day,
however, gradually increased over the subsequent days. The
environmental samples demonstrated downregulation compared with the
control group during the first three days of incubation at 30°C,
however, their expression levels were increased compared with the
mRNA levels at 20°C (Fig. 2). In
contrast to the results obtained at 20 and 30°C, a continual
ascending pattern of HSP70 expression was observed for both the
clinical and environmental samples at 37 and 42°C (Figs. 2 and 3). Furthermore, a considerable peak was
observed at 37°C in the majority of the clinical samples during the
five experimental days (Fig. 2).
The expression levels in the clinical samples at 20°C on the first
day were high, however, from day two demonstrated represented a
significant reduction in expression, following which it exhibited a
gradual reduction until the last day. By contrast, at 30°C both
groups demonstrated an increasing trend. This was additionally
observed at 37 and 42°C during the five experimental days. However,
the increase at 37°C in HSP70 expression, particularly in the
clinical samples which have the most considerable slope, indicates
an instant increase and high levels of HSP70 expression (Figs. 2 and 3). This increase had an impact on the
P-values at 37°C, resulting in the most significant P-values among
the other temperatures (Table
II). Furthermore, from the 13 clinical samples, four
demonstrated relative expression as low as the environmental
samples at 20 and 30°C. In addition, one other clinical sample
demonstrated significantly higher expression at 42°C alone, in
comparison to the environmental samples. However, seven samples
(53.8%) from the total 13 clinical samples exhibited significantly
greater expression at all temperatures when compared with the mean
expression levels of the environmental samples. Of the total 13
clinical isolates, at 42°C just one isolate expressed HSP70 with no
significant difference compared with HSP70 expression in the
environmental isolates (data not shown). These results indicate
that 37 and 42°C are major temperatures responsible for the
upregulation of HSP70 mRNA expression (Fig. 3). Furthermore, among the clinical
isolates there were no significant differences in HSP70 expression
between human- and animal- originated samples (data not shown).
Overall, a continually increasing trend in HSP70 mRNA expression
levels was observed in the increasing temperatures over the five
days (Figs. 3 and 4). Evaluating the expression trend using
a timecourse revealed that both the clinical and environmental
samples demonstrated an increasing pattern of HSP70 mRNA expression
levels at 30, 37 and 42°C, however, a reduction at 20°C (Figs. 3 and 4).
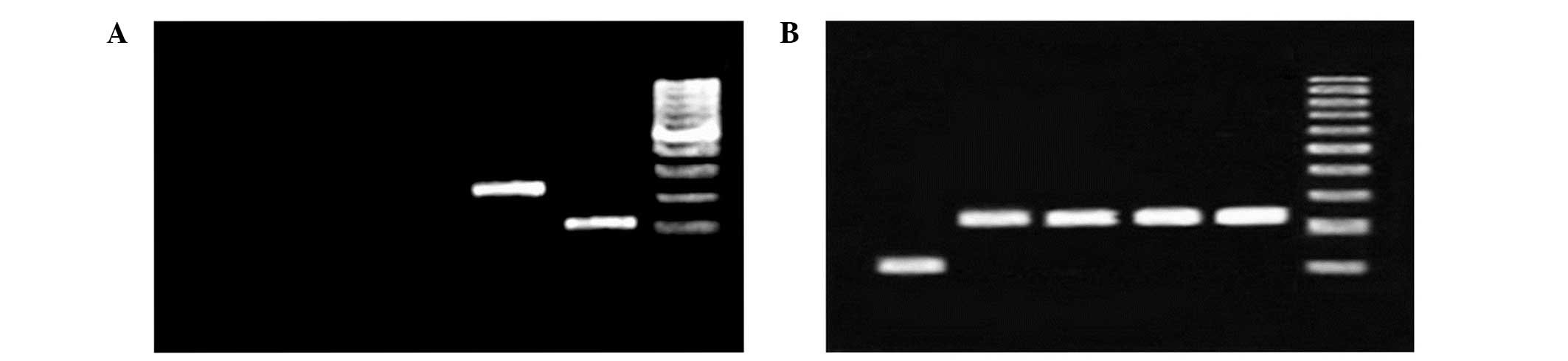 | Figure 1Gel electrophoresis image of PCR
reaction using heat shock protein 70 primers and cDNA template
demonstrate the lack of genomic DNA contamination and the correct
binding of the primers. (A) PCR reaction using cDNA from an
environmental sample at different temperatures. Only 42°C results
in a detectable band on gel electrophoresis. From left to right:
Lane 1, environmental sample at 20°C; lane 2, environmental sample
at 30°C; lane 3, environmental sample at 37°C; lane 4,
environmental sample at 42°C; lane 5, 18S rRNA; and lane 6, ladder.
(B) PCR products from a clinical sample. From left to right: Lane
1, 18S rRNA; lane 2, clincal sample at 20°C; lane 3, clincal sample
at 30°C; lane 4, clincal sample at 37°C; lane 5, clincal sample at
42°C; and lane 6, 1 kilo base ladder. PCR, polymerase chain
reaction; rRNA, ribosomal RNA. |
 | Table IIP-values from two-tailed Student's
t-test between the expression levels in the clinical and
environmental groups at each temperature point over the five
experimental days. |
Table II
P-values from two-tailed Student's
t-test between the expression levels in the clinical and
environmental groups at each temperature point over the five
experimental days.
| Culture day | Culture temperature
(°C)
|
|---|
| 20 | 30 | 37 | 42 |
|---|
| One | 0.006 | 0.0033 | 0.0002 | 0.0004 |
| Two | 0.002 | 0.0175 | 0.0011 | 0.0017 |
| Three | 0.0025 | 0.0175 | 0.0053 | 0.0046 |
| Four | 0.003 | 0.011 | 0.01 | 0.044 |
| Five | 0.0037 | 0.013 | 0.027 | 0.2 |
Discussion
Temperature fluctuations are common in the
environment, and unstable thermal conditions may result in
downstream alterations in the transcription levels of
stress-related genes such as HSPs in environmental and
opportunistic microbial agents. Microbial agents have evolved
protective mechanisms in the face of harmful heat stress induced in
diverse environmental conditions and host bodies (21). Long-term maintained high
temperatures may lead microbes or host cells towards a lethal
cascade of the denaturation of proteins which are vital for cells
and microbes. In fact, during periods of elevated temperature, HSPs
are the predominant proteins synthesized in fungal cells (22).
In yeasts and other opportunist pathogenic fungi,
thermal adaptation is not only essential to maintain thermal
homeostasis, but is also required for virulence (23,24).
High temperatures in the host body is an inducer of virulence
factors and HSPs, however, disparate observations in the virulence
ability have been reported in mutation-studies of
thermotolerance-related genes in fungal pathogens, indicating no
direct association between thermophily and virulence traits, except
in a few genes (23,25–29).
However, HSPs are essential for the survival of eukaryotic cells
even under normal conditions. In this regard, the activation of
thermal adaptation pathways in fungal cells has additionally been
observed at mild temperature variations in host bodies (30). Thus, it may be hypothesized that
the transcription of heat stress-related genes in clinically and
host body-adapted pathogens may be regulated differentially in
comparison with environmental microbial agents.
A. fumigatus is a common mold pathogen in
human, and produces large amounts of proteins and enzymes when
invading host bodies. The accumulation of damaged, unfolded or
aggregated proteins in a pathogen may threaten its survival.
Thereby, unfolded proteins act as a cellular thermometer (31,32),
and in Saccharomyces cerevisiae there are four sub-families
of genes encoding HSP70: Ssa, Ssb, Ssc and
Ssd (22). Three location-dependent protective pathways have
been described in S. cerevisiae, a yeast fungus, with all
three pathways activated in response to injured proteins. When
unfolded proteins accumulate in the endoplasmic reticulum, yeast
cells express KAR2 chaperone and HAC1. In the case of
unfolding proteins in the cytosol, SSA4, the major HSP70
chaperone, is induced. In addition, the SSA4-HSP70 pathway
is activated following heat shock (32). Another mechanism initiated by
damaged proteins is the activation of a distinct set of genes in
the Rpn4-dependent pathway (33).
It has been demonstrated that in yeast, the
Ssa1 and Ssa2 genes are constitutively expressed
while Ssa3 and Ssa4 are upregulated under stressed
situations (34). Currently, eight
HSP70 homologues have been identified, of which six are cytosolic
including Ssa1, Ssa2, Ssa3, Ssa4, Ssb1 and Ssb2 (9,35).
In comparison to the Ssa genes, Ssb1 and Ssb2 differentially
regulate Hsf1 activity in yeast (36). It has additionally been indicated
that Ssb1/2p is not induced by heat shock (37). In the current study, primers were
designed to investigate the mRNA expression levels of HSP70 in
A. fumigatus, using a reference sequence obtained from the
GenBank database. According to the information in GenBank and AspGD
(38), the sequences were similar
or orthologous to the SSA2 gene in Candida albicans, S.
cerevisiae, Schizosaccharomyces pombe and to the HSP70-1
gene in Neurospora crassa. Furthermore, BLAST searches
indicated that the sequence was also similar to its orthologous
SSA4 gene in S. cerevisiae (69% identity, 81% query
coverage) and to the HSP70 gene in C. albicans (68%
identity, 83% query coverage). Other HSP70 subfamily genes (Ssb,
Ssc, bipA, Kar2 and HscA) of A. fumigatus exhibited lower
similarities to the yeast SSA4 gene sequence (data not shown).
By contrast, investigation of the HSP70 molecular
regulation pathways rather than the genetics will indicate whether
pre-exposure of yeast cells to stress can trigger a protective
defense against subsequent exposure to the same stressor. Repeating
thermal stimulation in a sequential-continuous manner, with a short
duration of time between the two stimulations will result in a
protective response. This phenomenon is driven by the transcription
of protective stress-related genes and the accumulation of their
mRNA or protein over a specified time in the cytoplasm as a form of
molecular memory (30). In certain
yeast species, an acquired thermotolerance phenomenon following
continuous exposure to acute heat stress has been described
(39–41).
Indeed, in eukaryotes, HSP (HSP90 and HSP70)
expression is governed by HSF1 (31), which is highly conserved from yeast
to human (42). However, unlike
the well-determined Hsf1-dependent thermotolerance pathway in S.
cerevisiae, the molecular interactions in the Hsf1-HSP70 axis
in fungal pathogens such as A. fumigatus remain poorly
understood.
In the present study, HSP70 expression levels in
A. fumigatus isolates from clinical and environmental
samples at 37 and 42°C were observed to increase continuously
during the experimental days. The ascending pattern of HSP70
expression in both sample groups was relatively similar. The power
of the association between the expression data obtained from the
clinical and environmental samples was >0.9, indicating a strong
correlation (Fig. 4). In both the
sample groups, the greatest HSP70 mRNA levels were measured at
42°C, and the highest significant increases in HSP70 expression
were observed at 37°C (Table II).
In the clinical groups, the mean relative expression level at 37°C
was 2.7 times higher than the mean expression level at 30°C, while
in the environmental group this was 4.42 times. For both groups
this was greatest increase in expression levels in comparison to
the other temperature points. Furthermore, at 42°C, HSP70
expression was markedly increased in the environmental samples,
resulting in closer HSP70 levels (Table II). Overall these results indicate
that 37 and 42°C are important temperature points able to induce
HSP70 expression in A. fumigatus. These results are in
accordance with similar studies indicating induced expression of
HSP70 at high temperatures in yeast or mold fungi in particular,
such as A. fumigatus (7,11,43). It is important to note that
in contrast to the present study, these previous studies were
performed using short experimental durations, usually up to several
hours following induction of heat stress, and that almost all have
investigated heat shock-related genes within clinical or
environmental groups separately.
In a previous study, HSP expression levels were
evaluated up to several hours following a thermal shift from 30 to
37 and 48°C, with the aim to understand the coregulatory
association between metabolic and HSP genes in A. fumigatus.
At early time points following heat induction, the lowest
expression levels of HSPs were observed whereas metabolic genes
were upregulated, from which the authors concluded that there is
negative feedback between metabolic and HSP genes. However, the
coherent feedforward loop association between the two groups of
genes became weak with increasing temperature, and HSP expression
exhibited a marked increase at 48°C. From the three strains
evaluated for HSP70 expression, two strains exhibited upregulation
following 180 min at 37 and 48°C by a 2- and 4-fold increase in the
expression ratio in a Log2 scale, respectively. The one remaining
strain showed downregulation (43). In an additional extensive
investigation of A. fumigatus, HSP70 chaperone and
additional genes in the HSP70 family were evaluated for alterations
in expression levels in response to a temperature shift from 30 to
48°C, in short time durations, from 30 to 180 min. HSP70 chaperone
exhibited a 2.45-fold increase following a 180 min incubation at
48°C (11). Furthermore, a
previous study evaluated temperature-induced protein synthesis in
thermoresistant fungi including three samples of A.
fumigatus. This indicated that at 40°C, medium- and
high-molecular HSPs were synthesized while at 50°C no HSPs were
detected (44).
It has been demonstrated that clinically isolated
S. cerevisiae is able to grow at higher temperatures (41°C)
than laboratory isolates. Thermotolerance and growth ability at
physiological temperatures is a critical requirement for the
survival of S. cerevisiae strains in mice (45). This indicates that S.
cerevisiae laboratory isolates were adapted to grow at lower
temperatures rather than high temperatures (41°C), however, by
contrast clinical isolates tend to be more thermophilic than
environmental isolates. In a previous study, southern blot patterns
revealed extremely high genomic diversity and no clustering
potential between clinical and environmental strains of A.
fumigatus (46). Furthermore,
a prior study of bovine aortic endothelial cells suggested that
continuous heating at 42°C results in a continuous increasing trend
of HSP70 expression over time (25 h), although there were some
reductions certain time points on the kinetic curve of HSP70
expression (47). Despite this,
HSP expression trends remain a matter of debate, on account of
complexities which have been noted by researchers. One study
indicated that HSP expression in A. fumigatus is tightly
negatively linked to fungal cell metabolic-related gene expression
(43). When the fungus was
incubated at high temperatures for short durations (60 and 120
min), genes coding metabolic pathways were upregulated, however,
the majority of the HSPs (other than HSP70) were downregulated. At
37 and 48°C, two strains of A. fumigatus demonstrated
induced levels of HSP70, however, one did not. Despite this, at
48°C, the majority of HSPs showed a dramatic rise in expression
levels, however, results obtained at 37°C indicate that
temperatures such as those in the human body may be associated with
reduced levels of certain HSPs, such as HSP30 (43). In addition, one of the first
physiometabolic investigations on yeast HSP70 RNA levels indicated
that HSP70 expression may vary in response to different
physiological conditions of yeast cells (stationary phase at 23°C
or sporulation phase at 30°C). In the stationary phase, the
majority of yeast HSP70-related RNAs reduced, however, the SSA3 RNA
level was reversed by the addition of glucose (48).
However, the data of the current study supports the
majority of prior findings, suggesting that HSP70 expression is
increased following heat stress, in particular at 37°C or higher.
In addition, the mentioned previous studies may be linked to the
observations of the current study, which indicate low expression
levels of HSP70 at temperatures below normal mammalian and human
body temperature. Furthermore, previous observations may indicate
that A. fumigatus, a frequently isolated filamentous fungus
from extreme environments (49),
may be adapted to ambient temperatures rather than physiological
temperatures and host body conditions (43). This means that the environmental
conditions in A. fumigatus induce a normal signal while host
niches induce a stressed or abnormal signal to regulate HSPs. The
majority of HSPs are upregulated in heat shock or when in contact
with host serum components (50).
The predominant source of air-borne spores of A. fumigatus
are the decomposing remnants of plants and other organic debris,
where fungi grow and release large number of spores into the
atmosphere.
There are some obscurities in the observations of
the current study. A downward trend of HSP70 expression levels was
observed in clinical and environmental samples at 20°C, however, in
clinical samples HSP70 was upregulated on the first day of
incubation at 20°C. In addition, in the clinical group, HSP70
expression on the first day was higher than on the second day at
30°C. The higher levels of HSP70 on the first day of incubation at
20 and 30°C may result from HSP70 mRNA remaining in the fungal
cells from when they were in the host. The sudden shift from 37°C
in the human body temperature (or higher in animals) to 20 or 30°C
did not allow sufficient time for the fungus to downregulate HSP70
levels. It has been reported that fungal spores contain vacuoles
with high amounts of HSPs and stored mRNAs belonging to various
types of proteins in addition to HSPs (11,51).
Additionally, other explanation to these results may be the HSP
induction due to cold-stress (52). In this regard, there are several
reports on the induction of HSPs at low temperatures. Using S.
cerevisiae, it has been demonstrated that the expression levels
of certain HSPs, including HSP104p, HSP82p, HSP60p and HSP10p, were
reduced in the first 2 h of the temperature downshift to 10°C, in
an early cold response. However, certain HSPs. including HSP12,
HSP26, HSP42, HSP104, YRO2 and SSE2, were induced at 10°C for 12
and/or 60 h incubation, in a late cold response (52). Other similar reports have
demonstrated that following longer durations of incubation (4–24 h)
at low temperatures (10, 4 and 0°C), certain HSPs were repressed
while others were induced. It has been reported that following 4–24
h at 10°C, HSP genes including HSP12, HSP26, HSP42, HSP104, SSA4,
SSE2 and YRO2 were induced (52,53).
Additional genes in the HSP family including CIS3, HMS2, HSC82,
HSP30, HSP60, HSP78, HSP82, HSP150, SSA1 and SSA2 demonstrated a
downregulated trend when incubated at 10°C (53). However, their mRNA content
recovered at lower temperatures (54). These observations indicate
discrepancies which may be a matter of debate. For instance, the
SSA4 and SSE2 genes were observed to be induced while SSA1 and SSA2
were reduced at 10°C. It appears that each HSP70 family of proteins
may be regulated differentially in a time- and
temperature-dependent manner (55).
In the present study, elevation of HSP70 expression
in the later days at 30°C in the clinical sample was observed as
positive feedback, and HSP70 re-upregulation following the
depletion and consumption of the previously produced and
accumulated HSPs, along with their associated mRNA. Furthermore, an
explanation for the opposite HSP70 expression results in clinical
and environmental samples at 30°C may be the adaptation of the
clinical isolates to the host and therefore the loss of the host
body's stimulatory factors. Potentially, clinical isolates have
evolved to survive at high body temperatures in animals and humans,
and adapted to the contact with host serum components and adverse
conditions within the host body. It has been suggested that HSP90,
which is associated with HSP70, orchestrates C. albicans
yeast cells to transform to the filamentous form. This transition
may be induced by two signals from the host body, the exposure to
serum and the body temperature. Host body serum components
including carbohydrates, metal ions and other nutrients may
additionally be effective on HSP induction in fungal cells
(50,56–58).
These adaptive mechanisms in clinical samples, particularly in
response to temperature elevation, may explain the upregulation of
HSP70 in clinical samples but the downregulation or lower
expression levels in environmental samples, which was observed in
the current study at 30, 37 and 42°C. In addition, another
explanation for the lower HSP70 expression levels at 30°C in the
environmental samples is the fact that the control strain
(ATCC® 90906) was originally isolated from human blood.
The standard strain was incubated at 25°C for five days as the
control group. However, the environmental samples, which were
incubated at 30°C, demonstrated downregulation of HSP70. Therefore,
the standard control strain may have acquired a partially similar
gene expression profile to the clinical isolates. It should be
noted that HSP70 expression at 30°C was increased significantly in
comparison with the levels observed at 20°C.
Overall, the results of the present study on the
high expression ratios of HSP70 in A. fumigatus at 37 and
42°C are in agreement with previous reports. However, there are
some limitations in estimating HSP gene expression, such as that
HSP expression is suppressed at very high temperatures. This is
also true for housekeeping (reference) genes which generally have
been selected in gene expression quantification assays. Gene
expression of HSPs is not only dependent on heat stress or
temperature shift but is also affected by other regulatory factors
such as metabolic genes or other pathways which themselves are
affected by additional stimuli or stresses. The multifactorial
regulation of HSPs means that studies investigating stress-related
gene expression are more complicated than other simple
single-factor gene expression studies.
A. fumigatus is a known mesophilic fungus,
however, has been shown to posses thermophilic traits in a number
of studies. Regarding the reports indicating a direct association
between thermotolerance and virulence, further knowledge on A.
fumigatus pathogenesis associated with thermoresistance may
result in the development of novel therapeutic approaches against
this opportunistic pathogen. In the present study, the results
indicated the increased expression pattern of HSP70 as a vital
stress-related gene in A. fumigatus at high temperatures. In
addition, the reported higher expression levels of HSP70 in
clinical strains compared with environmental strains indicates a
potential association between HSP70 expression and virulence in
clinical strains of A. fumigatus. However, to gain further
insight into the precise underlying molecular pathways and
potential regulatory association between HSP70 and virulence,
further studies are required.
Acknowledgments
The current study was supported by the Research
Council of the University of Tehran.
References
|
1
|
Latgé JP: Aspergillus fumigatus and
aspergillosis. Clin Microbiol Rev. 12:310–350. 1999.PubMed/NCBI
|
|
2
|
Dagenais TR and Keller NP: Pathogenesis of
Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol
Rev. 22:447–465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Denning DW: Invasive aspergillosis. Clin
Infect Dis. 26:781–803; quiz 804–805. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gugnani HC: Ecology and taxonomy of
pathogenic aspergilli. Front Biosci. 8:s346–s357. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fedorova ND, Nierman WC, Turner G, Joardar
V, Maiti R, Anderson MJ, Denning DW, Wortman JR, Goldman GH and
Osmani SA: A comparative view of the genome of Aspergillus
fumigatus. The Aspergilli: Genomics, Medical Aspects, Biotechnology
and Research Methods. Goldman GH and Osmani SA: CRC Press, Taylor
& Francis Group; pp. 25–38. 2008
|
|
6
|
Lin SJ, Schranz J and Teutsch SM:
Aspergillosis case-fatality rate: Systematic review of the
literature. Clin Infect Dis. 32:358–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nierman WC, Pain A, Anderson MJ, Wortman
JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et
al: Genomic sequence of the pathogenic and allergenic filamentous
fungus Aspergillus fumigatus. Nature. 438:1151–1156. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Latgé JP: Aspergillus fumigatus and
aspergillosis. Clin Microbiol Rev. 12:310–350. 1999.PubMed/NCBI
|
|
9
|
Burnie JP, Carter TL, Hodgetts SJ and
Matthews RC: Fungal heat-shock proteins in human disease. FEMS
Microbiol Rev. 30:53–88. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Westerheide SD, Raynes R, Powell C, Xue B
and Uversky VN: HSF transcription factor family, heat shock
response, and protein intrinsic disorder. Curr Protein Pept Sci.
13:86–103. 2012. View Article : Google Scholar
|
|
11
|
Albrecht D, Guthke R, Brakhage AA and
Kniemeyer O: Integrative analysis of the heat shock response in
Aspergillus fumigatus. BMC Genomics. 11:322010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Inouye S, Katsuki K, Izu H, Fujimoto M,
Sugahara K, Yamada S, Shinkai Y, Oka Y, Katoh Y and Nakai A:
Activation of heat shock genes is not necessary for protection by
heat shock transcription factor 1 against cell death due to a
single exposure to high temperatures. Mol Cell Biol. 23:5882–5895.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feige U and Polla BS: Hsp70-a multi-gene,
multi-structure, multi-function family with potential clinical
applications. Experientia. 50:979–986. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feige U and Polla BS: Hsp70-a multi-gene,
multi-structure, multi-function family with potential clinical
applications. Experientia. 50:979–986. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jolly C and Morimoto RI: Role of the heat
shock response and molecular chaperones in oncogenesis and cell
death. J Natl Cancer Inst. 92:1564–1572. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Allendoerfer R, Maresca B and Deepe GS Jr:
Cellular immune responses to recombinant heat shock protein 70 from
Histoplasma capsulatum. Infect Immun. 64:4123–4128. 1996.PubMed/NCBI
|
|
17
|
Mayer MP and Bukau B: Hsp70 chaperones:
Cellular functions and molecular mechanism. Cell Mol Life Sci.
62:670–684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Owczarzy R, Tataurov AV, Wu Y, Manthey JA,
McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J,
McEntaggart NO, et al: IDT SciTools: A suite for analysis and
design of nucleic acid oligomers. Nucleic Acids Res. 36(Web Server
issue): W163–W169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schoonjans F, Zalata A, Depuydt CE and
Comhaire FH: MedCalc: A new computer program for medical
statistics. Comput Methods Programs Biomed. 48:257–262. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Zugel U and Kaufmann SH: Role of heat
shock proteins in protection from and pathogenesis of infectious
diseases. Clin Microbiol Rev. 12:19–39. 1999.PubMed/NCBI
|
|
22
|
Tereshina VM: Thermotolerance in Fungi:
The role of heat shock proteins and trehalose. Mikrobiologiia.
74:293–304. 2005.In Russian. PubMed/NCBI
|
|
23
|
Nicholls S, MacCallum DM, Kaffarnik FA,
Selway L, Peck SC and Brown AJ: Activation of the heat shock
transcription factor Hsf1 is essential for the full virulence of
the fungal pathogen Candida albicans. Fungal Genet Biol.
48:297–305. 2011. View Article : Google Scholar :
|
|
24
|
Brown AJ, Budge S, Kaloriti D, Tillmann A,
Jacobsen MD, Yin Z, Ene IV, Bohovych I, Sandai D, Kastora S, et al:
Stress adaptation in a pathogenic fungus. J Exp Biol. 217:144–155.
2014. View Article : Google Scholar :
|
|
25
|
Bhabhra R, Miley MD, Mylonakis E, Boettner
D, Fortwendel J, Panepinto JC, Postow M, Rhodes JC and Askew DS:
Disruption of the Aspergillus fumigatus gene encoding nucleolar
protein CgrA impairs thermotolerant growth and reduces virulence.
Infect Immun. 72:4731–4740. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang YC, Tsai HF, Karos M and Kwon-Chung
KJ: THTA, a thermotolerance gene of Aspergillus fumigatus. Fungal
Genet Biol. 41:888–896. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dirr F, Echtenacher B, Heesemann J,
Hoffmann P, Ebel F and Wagener J: AfMkk2 is required for cell wall
integrity signaling, adhesion, and full virulence of the human
pathogen Aspergillus fumigatus. Int J Med Microbiol. 300:496–502.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wagener J, Echtenacher B, Rohde M, Kotz A,
Krappmann S, Heesemann J and Ebel F: The putative
alpha-1,2-mannosyltransferase AfMnt1 of the opportunistic fungal
pathogen Aspergillus fumigatus is required for cell wall stability
and full virulence. Eukaryot Cell. 7:1661–1673. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou H, Hu H, Zhang L, Li R, Ouyang H,
Ming J and Jin C: O-Mannosyltransferase 1 in Aspergillus fumigatus
(AfPmt1p) is crucial for cell wall integrity and conidium
morphology, especially at an elevated temperature. Eukaryot Cell.
6:2260–2268. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leach MD, Tyc KM, Brown AJ and Klipp E:
Modelling the regulation of thermal adaptation in Candida albicans,
a major fungal pathogen of humans. PLoS One. 7:e324672012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leach M and Cowen L: To sense or die:
Mechanisms of temperature sensing in fungal pathogens. Curr Fungal
Infect Rep. 8:185–191. 2014. View Article : Google Scholar
|
|
32
|
Metzger MB and Michaelis S: Analysis of
quality control substrates in distinct cellular compartments
reveals a unique role for Rpn4p in tolerating misfolded membrane
proteins. Mol Biol Cell. 20:1006–1019. 2009. View Article : Google Scholar :
|
|
33
|
Geiler-Samerotte KA, Dion MF, Budnik BA,
Wang SM, Hartl DL and Drummond DA: Misfolded proteins impose a
dosage-dependent fitness cost and trigger a cytosolic unfolded
protein response in yeast. Proc Natl Acad Sci USA. 108:680–685.
2011. View Article : Google Scholar :
|
|
34
|
Satyanarayana C, Schröder-Köhne S, Craig
EA, Schu PV and Horst M: Cytosolic Hsp70s are involved in the
transport of aminopeptidase 1 from the cytoplasm into the vacuole.
FEBS Lett. 470:232–238. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Daugaard M, Rohde M and Jäättelä M: The
heat shock protein 70 family: Highly homologous proteins with
overlapping and distinct functions. FEBS Lett. 581:3702–3710. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Verghese J, Abrams J, Wang Y and Morano
KA: Biology of the heat shock response and protein chaperones:
Budding yeast (Saccharomyces cerevisiae) as a model system.
Microbiol Mol Biol Rev. 76:115–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bonner JJ, Carlson T, Fackenthal DL,
Paddock D, Storey K and Lea K: Complex regulation of the yeast heat
shock transcription factor. Mol Biol Cell. 11:1739–1751. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cerqueira GC, Arnaud MB, Inglis DO,
Skrzypek MS, Binkley G, Simison M, Miyasato SR, Binkley J, Orvis J,
Shah P, et al: The Aspergillus genome database: Multispecies
curation and incorporation of RNA-Seq data to improve structural
gene annotations. Nucleic Acids Res. 42(Database Issue): D705–D710.
2014. View Article : Google Scholar :
|
|
39
|
De Virgilio C, Simmen U, Hottiger T,
Boller T and Wiemken A: Heat shock induces enzymes of trehalose
metabolism, trehalose accumulation, and thermotolerance in
Schizosaccharomyces pombe, even in the presence of cycloheximide.
FEBS Lett. 273:107–110. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Piper PW: Molecular events associated with
acquisition of heat tolerance by the yeast Saccharomyces
cerevisiae. FEMS Microbiol Rev. 11:339–355. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Argüelles JC: Thermotolerance and
trehalose accumulation induced by heat shock in yeast cells of
Candida albicans. FEMS Microbiol Lett. 146:65–71. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu C: Heat shock transcription factors:
Structure and regulation. Annu Rev Cell Dev Biol. 11:441–469. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Do JH, Yamaguchi R and Miyano S: Exploring
temporal transcription regulation structure of Aspergillus
fumigatus in heat shock by state space model. BMC Genomics.
10:3062009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen KU and Chen ZC: Heat shock proteins
of thermophilic and thermotolerant fungi from Taiwan. Bot Bull Acad
Sin. 45:247–257. 2004.
|
|
45
|
McCusker JH, Clemons KV, Stevens DA and
Davis RW: Saccharomyces cerevisiae virulence phenotype as
determined with CD-1 mice is associated with the ability to grow at
42 degrees C and form pseudohyphae. Infect Immun. 62:5447–5455.
1994.PubMed/NCBI
|
|
46
|
Debeaupuis JP, Sarfati J, Chazalet V and
Latgé JP: Genetic diversity among clinical and environmental
isolates of Aspergillus fumigatus. Infect Immun. 65:3080–3085.
1997.PubMed/NCBI
|
|
47
|
Wang S, Diller KR and Aggarwal SJ:
Kinetics study of endogenous heat shock protein 70 expression. J
Biomech Eng. 125:794–797. 2003. View Article : Google Scholar
|
|
48
|
Werner-Washburne M, Becker J,
Kosic-Smithers J and Craig EA: Yeast Hsp70 RNA levels vary in
response to the physiological status of the cell. J Bacteriol.
171:2680–2688. 1989.PubMed/NCBI
|
|
49
|
Tepšič K, Gunde-Cimerman N and Frisvad JC:
Growth and mycotoxin production by Aspergillus fumigatus strains
isolated from a saltern. FEMS Microbiol Lett. 157:9–12. 1997.
View Article : Google Scholar
|
|
50
|
Shapiro RS, Uppuluri P, Zaas AK, Collins
C, Senn H, Perfect JR, Heitman J and Cowen LE: Hsp90 orchestrates
temperature-dependent Candida albicans morphogenesis via Ras1-PKA
signaling. Curr Biol. 19:621–629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Low SY, Dannemiller K, Yao M, Yamamoto N
and Peccia J: The allergenicity of Aspergillus fumigatus conidia is
influenced by growth temperature. Fungal Biol. 115:625–632. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schade B, Jansen G, Whiteway M, Entian KD
and Thomas DY: Cold adaptation in budding yeast. Mol Biol Cell.
15:5492–5502. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sahara T, Goda T and Ohgiya S:
Comprehensive expression analysis of time-dependent genetic
responses in yeast cells to low temperature. J Biol Chem.
277:50015–50021. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Murata Y, Homma T, Kitagawa E, Momose Y,
Sato MS, Odani M, Shimizu H, Hasegawa-Mizusawa M, Matsumoto R,
Mizukami S, et al: Genome-wide expression analysis of yeast
response during exposure to 4 degrees C. Extremophiles. 10:117–128.
2006. View Article : Google Scholar
|
|
55
|
Aguilera J, Randez-Gil F and Prieto JA:
Cold response in Saccharomyces cerevisiae: New functions for old
mechanisms. FEMS Microbiol Rev. 31:327–341. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Berman J and Sudbery PE: Candida albicans:
A molecular revolution built on lessons from budding yeast. Nat Rev
Genet. 3:918–930. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
57
|
Swoboda RK, Bertram G, Budge S, Gooday GW,
Gow NA and Brown AJ: Structure and regulation of the HSP90 gene
from the pathogenic fungus Candida albicans. Infect Immun.
63:4506–4514. 1995.PubMed/NCBI
|
|
58
|
Zeuthen ML and Howard DH: Thermotolerance
and the heat-shock response in Candida albicans. J Gen Microbiol.
135:2509–2518. 1989.PubMed/NCBI
|