Introduction
Gliomas are the most common type of malignancy in
the brain, accounting for approximately 45–60% of brain tumors, and
are characterized by high incidence, recurrence and mortality rates
and a low curative rate (1). This
neoplasm is a life-threatening disease of the nervous system.
Therefore, it is of clinical importance to explore the underlying
mechanisms, which may lead to the discovery of novel treatment
modalities.
Cripto-1, otherwise known as teratocarcinoma-derived
growth factor-1, is a member of the epidermal growth
factor-cripto-1-FRL-1-cryptic protein family (2). Previous studies have demonstrated
that cripto-1 is overexpressed in numerous types of cancer, however
is under-expressed or absent in normal tissues (3–5).
This fact suggests that an increase in the expression of cripto-1
may be an early event in the development of the associated types of
cancer. An in vitro study demonstrated that overexpression
of cripto-1 accelerated the growth of human breast cancer cells and
promoted anti-apoptotic, -migratory and -invasive capabilities
(6). An in-vivo study
demonstrated that the growth of human nasopharyngeal carcinoma
CNE-2 cells was significantly inhibited in cripto-1 knockout mice
compared with the control group (7). Previous studies have demonstrated
elevated cripto-1 expression in gliomas (8,9).
Pilgaard et al (8)
suggested that cripto-1 may be a prognostic biomarker for
glioblastoma multiforme (GBM) with the potential of being a
therapeutic target. Tysnes et al (9) observed that the majority of samples
of patients with glioblastoma demonstrated significant levels of
cripto-1, as it was detected by immunohistochemistry. Thus,
numerous studies have demonstrated that cripto-1 may be a novel
tumor-specific marker, and its value in early diagnosis of tumors,
targeted treatment, drug resistance mechanisms and prognosis is of
interest (3–9). However, further research is required
to unravel the specific mechanisms of cripto-1 in gliomas.
MicroRNAs (miRNAs) are a class of short, endogenous
non-coding RNAs that function as post-transcriptional regulators
(10). The miR-15 family is
involved in the regulation of cellular functions, including
apoptosis, cell cycle, differentiation and stress, and it is
associated with a variety of human diseases, including cancer,
cardiovascular and neurodegenerative diseases (11–13).
The miR-15 family offers potential therapeutic targets. By miRNA
expression profiling analysis, Xia et al (14) demonstrated that the expression of
miR-15b/16 was downregulated in a multi-drug-resistant gastric
cancer cell line, SGC7901/VCR, compared with its non-resistant
parent cell line, SGC7901, and that overexpression of miR-15b and
miR-16 in the SGC7901/VCR cells enhanced the sensitivity of the
cells to anticancer drugs, making them more susceptible to
apoptosis. A previous study demonstrated that a lower expression
level of miR-15b was associated with a shorter overall survival
time, suggesting that miR-15b may be an intrinsic factor that
serves an important role in the malignant progression of gliomas
(15).
Prediction analysis indicated that miR-15b has
binding sites on cripto-1. It is hence possible that miR-15b may
suppress cancer by regulating cripto-1, thus cellular experiments
were performed in the current study to test the above
prediction.
Materials and methods
Clinical data and cell culture
A total of 30 glioma tissues, and matched adjacent
noncancerous tissues, were harvested from patients at the Fourth
Affiliated Hospital of Nantong University (Yancheng, China). All
included cases were newly diagnosed, which were screened and
verified by an experienced pathologist following surgical
resection. Samples were harvested and analyzed with prior written
informed consent from the patients between 2011 and 2014. The
present study was approved by the Ethics Committee of the Fourth
Affiliated Hospital of Nantong University, First Hospital of
Yancheng. Human GBM8401 and GBM glioma cells (China Center for Type
Culture Collection, Wuhan, China) and HEK293T cells (ATCC,
Manassas, VA, USA) were cultured in Gibco Dulbecco's modified
Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% heat-inactivated fetal calf serum
(Thermo Fisher Scientific, Inc.). The cells were maintained at 37°C
in an atmosphere of humidified air with 5% CO2 in a cell
culture incubator.
Over-expression vector construction and
transfection
Total RNA of GBM8401 cells was extracted using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
used to amplify the cripto-1 coding region. It was then cloned into
pcDNA3.1, using the Kpn I and EcoR I (Takara
Biotechnology Co., Ltd., Dalian, China) restriction enzymes,
sequenced and verified. The gene amplification primers are
presented in Table I. The human
miRNA (Hsa)-miR-15b and negative controls mimics were synthesized
by Shanghai GenePharma Co., Ltd (Shanghai, China). GBM8401 cells
were seeded into a 6-well plate (1×105 cells/ml) and
incubated for 24 h, and plasmid and miRNA transfection was
conducted using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), upon 70% confluency according to the
manufacturer's instructions. The concentrations of the transfection
plasmid and miRNA were 2 μg/ml and 50 nM/well, and the DMEM
medium was changed within 4–6 h subsequent to transfection. After
48 h, the transfected cells were analyzed by RT-qPCR and western
blotting.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Technique/gene | Primer sequence
(5′->3′) | Size (bp) |
|---|
| Cripto-1 CDS
amplification | S: GGGGTACCGCCACCATGGACTGCAGGAAGATGGC | 583 |
| A: CGGAATTCTTAATAGTAGCTTTGTATAGAAAGGCA | |
| Cripto-1 3′UTR
amplification | S: CCGCTCGAGTCGACATTGACCTATTTCCAG | 1,235 |
| A:
ATAAGAATGCGGCCGCGGTCAATATAGTTTCCATTTTTACTG | |
| Quantitative
PCR | | |
| GAPDH | S:
GGTATCGTGGAAGGACTC | 128 |
| A:
GTAGAGGCAGGGATGATG | 128 |
| Cripto-1 | S:
AATTTGCTCGTCCATCTC | 128 |
| A:
CTCCTTACTGTGCTGTATC | 128 |
Quantitative fluorescence PCR
Total RNA was extracted from the GBM8401 cells using
TRIzol. For the detection of mature miRNA, a stem-loop RT-qPCR
assay was conducted. Briefly, 1 μg RNA and 1 μl
specific stem-loop RT primers (10 μM) were incubated at 85°C
for 5 min, then held at 4°C. A mixture of 4 μl 5X buffer, 2
μl dNTPs (10 mM), 0.5 μl RNase inhibitor and 0.5
μl Moloney murine leukemia virus (Promega Corporation,
Madison, WI, USA) was added and incubated for 30 min at 16°C, 30
min at 42°C, 5 min at 85°C and then maintained at 4°C. qPCR was
performed using 10 μl 2X SYBR Green qPCR SuperMix
(Invitrogen; Thermo Fisher Scientific, Inc.), with 5 μl
cDNA, 0.5 μl forward primer, 0.5 μl reverse primer
and 4 μl RNase-free ddH2O contained in 20
μl reaction mixture. The reaction was performed with one
cycle of 95°C for 5 min and 40 cycles of 95°C for 15 sec, 60°C for
15 sec and 72°C for 35 sec using the Applied Biosystems 7500
Real-Time PCR system (Thermo Fisher Scientific, Inc.). The U6 small
nuclear RNA (U6 snRNA) was utilized as reference of the miRNA
examination and the primers of mature miRNA were obtained from
Thermo Fisher Scientific, Inc. Furthermore, oligo dT primers were
used in reverse transcription. The primers used for the cripto-1
examination are demonstrated in Table
I. The reaction was performed at one cycle of 95°C for 5 min
and 40 cycles of 95°C for 30 sec, 58°C for 30 sec and 72°C for 30
sec. Three independent experiments were conducted for each sample.
Data were normalized and fold changes were calculated using the
2−ΔΔCq normalization method (16).
Western blot analysis
The total cellular proteins were extracted from the
GBM8401 cells by lysis buffer (Pierce Biotechnology, Inc.,
Rockford, IL, USA). The supernatant containing proteins was
obtained following shaking at 4°C for 20 min and centrifugation at
10,000 × g at 4°C for 10 min. The Bradford protein assay (Pierce
Biotechnology, Inc.) was used to determine the protein
concentrations. An equal quantity of proteins (20 μg) was
loaded onto 8% Tris-glycine sodium dodecyl sulfate-polyacrylimide
gel electrophoresis gels (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) at 40 V for 5 h and the separated proteins were transferred
onto nitrocellulose membranes (Pierce Biotechnology, Inc.).
Membranes were then blocked in 5% non-fat milk in Tris-buffered
saline with Tween-20 (TBST; Pierce Biotechnology, Inc.) for 1 h at
4°C. Subsequent to blocking, membranes were incubated with cripto-1
(1:400; NB100-1598; Novus Biologicals, LLC, Littleton, CO, USA),
matrix metalloproteinase (MMP)-2 (1:800; 4022), MMP-9 (1:800; 2270)
polyclonal rabbit antibodies and glyceraldehyde 3-phosphate
dehydrogenase (1:1,000; 3683; all Cell Signaling Technology Inc.,
Danvers, MA, USA) monoclonal rabbit antibody overnight at 4°C.
Monoclonal anti-rabbit immunoglobulin G antibody conjugated with
horseradish peroxidase (7074; Cell Signaling Technology Inc.) at
1:7,000 dilution was added to the membranes and incubated for 1 h
at 37°C. TBST was used for washing the membranes every 10 min, for
a total of 30 min. Protein bands were detected using the West Femto
system (Pierce Biotechnology, Inc.). The light-emitting films were
scanned by a GelBlot-Pro 1.01 gel imaging system (UVP, LLC, Upland,
CA, USA) and the gray values of band were measured with the Gel-Pro
Analyzer 6.3 software (Media Cybernetics, Inc., Rockville, MD,
USA).
Dual luciferase assay
The cripto-1 3′ untranslated region (UTR) was
amplified from cDNA (Table I) and
cloned into the psiCHECK-2 vector (Promega Corporation). The
QuikChange II Site-Directed Mutagenesis kit (Stratagene; Agilent
Technologies, Inc., Santa Clara, CA, USA) was used to generate the
mutant-type cripto-1 3′UTR in which the seven mutated nucleotides
were underlined within the seed region of the miR-15b binding site.
Reporter vector psiCHECK-2 carrying the 3′UTR sequences of cripto-1
was assayed for luciferase expression using the Dual Glo Luciferase
Assay System (Promega Corporation) according to the manufacturer's
instructions. The experiment was performed in duplicate in three
independent experiments.
Cell proliferation assay
Cell proliferation was detected in 96-well plates
using a colorimetric immunoassay, based on the measurement of
5-bromo-2′-deoxyuridine (BrdU) incorporation during the DNA
synthesis (BrdU ELISA kit, Roche Diagnostics GmbH, Mannheim,
Germany). Subsequent to 48-h transfection, the medium was removed
and GBM8401 and GBM cells were incubated with BrdU (10 mM; Roche
Diagnostics GmbH) for 3 h at 37°C. Cells were fixed with 4%
paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA), and incubated
with the peroxidase-conjugated sheep anti-BrdU polyclonal antibody
(1:1,000; 11647229001; Roche Diagnostics GmbH) for 90 min at room
temperature. The peroxidase substrate
3,3′,5,5′-tetramethylbenzidine (Roche Diagnostics GmbH) was then
added to the cells and BrdU incorporation was quantitated by
differences in absorbance at wavelength 370–492 nm. Cell
proliferation was expressed as the mean percentage of the control
values (set at 100%).
Flow cytometric analysis
Subsequent to transfection with plasmid and miRNA,
the GBM8401 cells were washed with phosphate-buffered saline (PBS;
Pierce Biotechnology, Inc.) and fixed in 75% ethanol overnight at
−20°C. The fixed cells were stained with propidium iodide (PI)
solution (1.21 mg/ml Tris, 700 U/ml RNase, 50.1 μg/ml PI, pH
8.0; Sigma-Aldrich) for 4 h in the dark. Red PI fluorescence was
measured with the Epics XL flow cytometer using CXP 2.2 software
(Beckman Coulter, Inc., Brea, CA, USA). In each sample, 10,000
events were measured for apoptosis detection and cell cycle
analysis. Data were analyzed with the Expo32 analysis tool (Beckman
Coulter, Inc.). The DNA histogram was represented by the proportion
of cells in the G0/G1, S and G2/M
phases. Apoptotic cells with hypodiploid DNA content were measured
by quantifying the sub-G1 peak in the cell cycle
pattern.
Transwell matrigel invasion assay
Invasion of cells was assessed with the Transwell
matrigel invasion assay (BD Biosciences, San Jose, CA, USA).
Briefly, 200 μl of transfected GBM8401 cells
(1×106 cells/ml) and 600 μl of the complete
medium were added to the upper and lower compartments of the
chamber, respectively. Subsequent to 48-h incubation, cells
migrating to the lower side of the filter were fixed with 4%
paraformaldehyde for 15 min at room temperature, washed with PBS,
stained with crystal violet (Sigma-Aldrich) and then observed under
the CKX41 inverted microscope (Olympus Corporation, Tokyo,
Japan).
Statistical analysis
The experiments were conducted a minimum of 3 times
and results are expressed as the mean ± standard deviation. The
SPSS statistical package (SPSS software, version 17.0 for Windows;
SPSS, Inc. Chicago, IL, USA) was used for statistical analysis.
Differences between the control and treated groups were analyzed
using non-parametric statistical analysis (Mann-Whitney U test).
P<0.01 was considered to indicate a statistically significant
difference.
Results
miR-15b downregulates cripto-1 expression
in glioma cells
The expression levels of miR-15b and cripto-1 from
the collected clinical samples were examined to investigate the
association of miR-15b and cripto-1 in gliomas. The results
demonstrated that the mRNA expression levels of miR-15b were
downregulated in glioma tissues whereas cripto-1 expression was
significantly increased, suggesting a negative correlation between
the two molecules (Fig. 1A and B).
The addition of miR-15b mimics led to significantly reduced
cripto-1 expression in glioma cells (Fig. 1C and D). Although cripto-1
expression was still upregulated compared with the control
subsequent to co-transfection with miR-15b mimics and cripto-1
overexpression vector, its level was significantly lower than in
cells transfected with the cripto-1 overexpression vector alone
(Fig. 1C and D), indicating that
miR-15b can inhibit cripto-1 expression in glioma cells.
miR-15b regulates cripto-1 in a targeted
manner
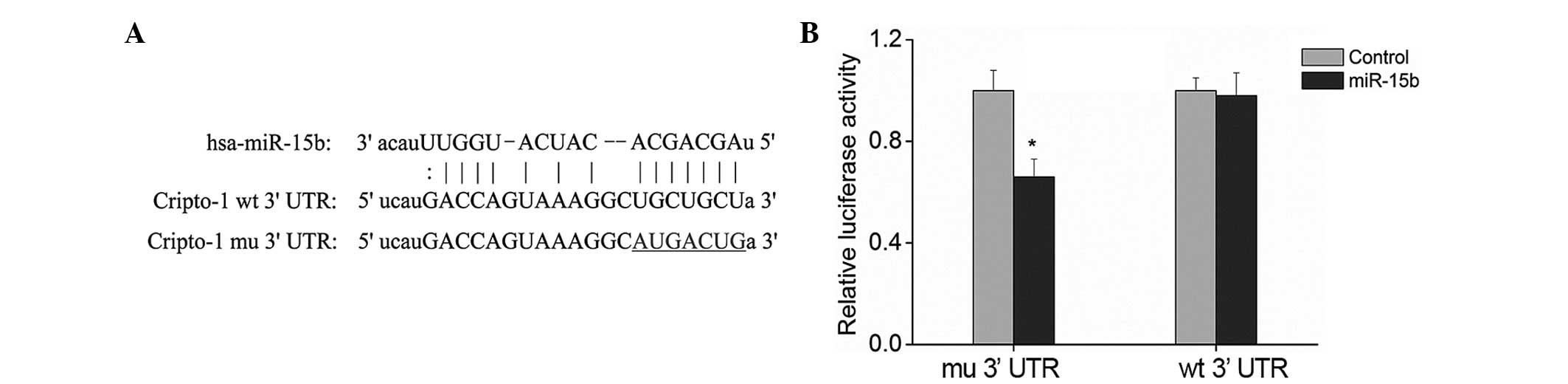
Prediction by biological software demonstrated that
miR-15b has binding sites on cripto-1 (Fig. 2A). The dual luciferase reporter
gene assay was performed to test this prediction. The 3′ UTR of
cripto-1 gene was cloned into psiCHECK-2, a dual luciferase
reporter vector, and the target sites of miR-15b were mutated
(Fig. 2A). The two plasmids were
then transfected together with the miR-15b mimics into human
embryonic kidney-293T cells and the alterations in the luciferase
expression were analyzed. Luciferase expression in the cells
transfected with plasmids containing 3′ UTR of cripto-1 was
significantly reduced by approximately 35% compared with the cells
transfected with the empty reporter vector (Fig. 2B; P<0.01). Furthermore, no
significant alterations were observed in the luciferase expression
in the cells transfected with plasmids containing the mutant 3′ UTR
of cripto-1 (Fig. 2B; P>0.05).
This demonstrated that miR-15b specifically binds to the 3′ UTR of
cripto-1 gene, suggesting that cripto-1 is a target gene of
miR-15b. Therefore, miR-15b may have an anti-tumor role by
regulating cripto-1.
miR-15b inhibits glioma cell
proliferation through targeted regulation of cripto-1
As demonstrated in Fig.
3, the cell proliferation assays with BrdU detection following
transfection with the miR-15b mimics indicated that miR-15b
significantly inhibited the proliferation of the GBM8401 and GBM
cells compared with the control group (Fig. 3; P<0.01). Transfection with the
cripto-1 overexpression vector resulted in significantly increased
cripto-1 expression and enhanced proliferation of the GBM8401 and
GBM cells compared with the control group (Fig. 3; P<0.01). In addition, cell
proliferation was significantly increased in the cells
co-transfected with the miR-15b mimics and cripto-1 overexpression
vector, compared with the cells transfected with the cripto-1
overexpression vector alone (Fig.
3). Furthermore, cells transfected with the miR-15b mimics
alone had significantly lower proliferation compared with the other
three groups (Fig. 3). The results
suggested that miR-15b and cripto-1 have opposite roles in the
glioma cell proliferation, and that miR-15b may suppress glioma
cell proliferation by regulating the expression of cripto-1.
miR-15b promotes apoptosis of glioma
cells through targeted regulation of cripto-1
Further tests on the cell cycle demonstrated that
the proportion of sub-G1 peak markedly increased in the
cells transfected with the miR-15b mimics and the cell cycle was
arrested in the G0/G1 phase (Fig. 4), which further demonstrates that
miR-15b promotes apoptosis of glioma cells. Western blot results
(Fig. 1C and D) demonstrated that
the degree of apoptosis was negatively correlated with the
expression levels of cripto-1. The pro-apoptotic effect of miR-15b
was inhibited by sustained cripto-1 expression in the cells
co-transfected with miR-15b and cripto-1, compared with the control
group. Cripto-1 expression was significantly reduced in the cells
transfected with miR-15b mimics alone, which resulted in
significantly increased apoptosis. These results demonstrated that
miR-15b promoted apoptosis by inhibiting the cripto-1
expression.
miR-15b inhibits migration of glioma
cells through targeted regulation of cripto-1
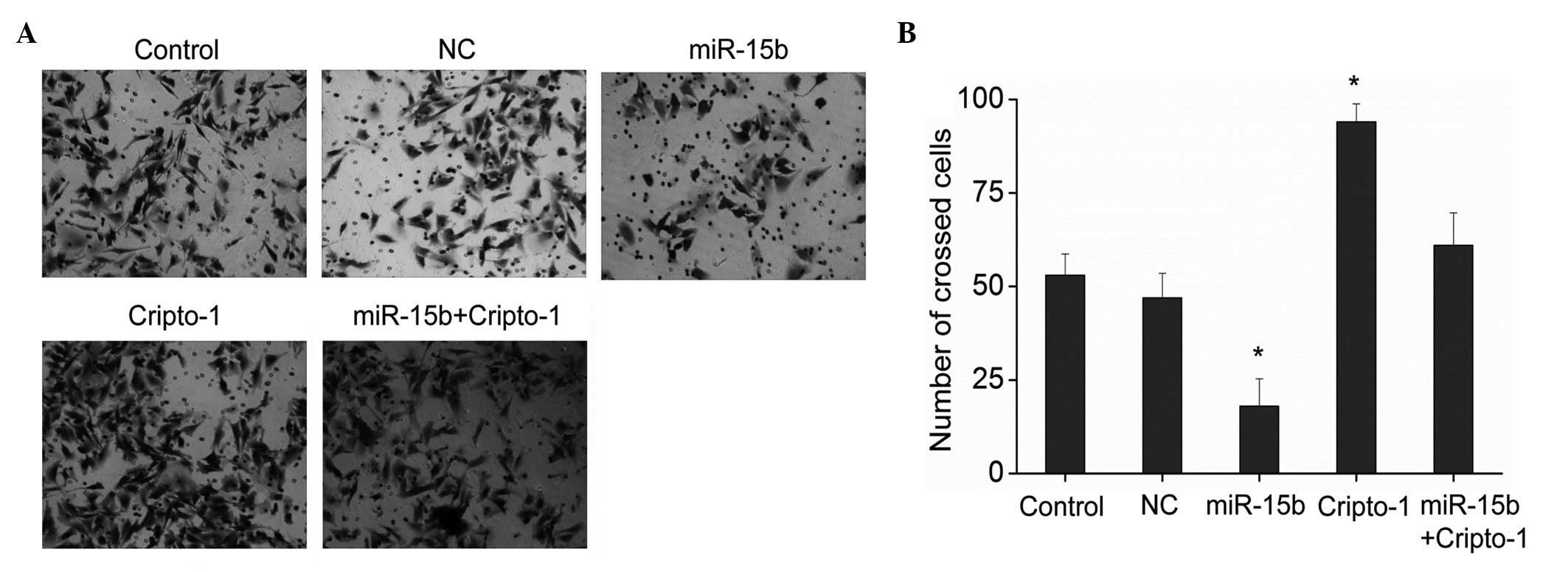
Glioma cells have invasive and metastatic abilities
(17), and cripto-1 was
demonstrated to promote cell invasion and metastasis. Therefore,
the effects of the miR-15b on the invasion and metastasis of glioma
cells were further investigated. Results from the Transwell assay
demonstrated that miR-15b significantly inhibited the invasion of
glioma cells and cripto-1 increased the invasiveness of glioma
cells compared with the control group (Fig. 5; P<0.01). Furthermore, no
significant difference was demonstrated in the ability of cell
invasion among cells co-transfected with the miR-15b and cripto-1,
and the control cells. The invasion and metastatic abilities are
associated with the expression of MMPs, of which MMP-2 and -9 are
two major members. Western blot analysis demonstrated that miR-15b
significantly reduced the expression of MMP-2 and -9, and cripto-1
promoted the expression of MMP-2 and -9 (Fig. 1C and D). In cells co-transfected
with miR-15b and cripto-1, the expression of MMP-2 and -9 remained
upregulated, but significantly reduced relative to cells
transfected with the cripto-1 overexpression vector alone. These
results demonstrated that miR-15b modulates the invasive ability of
glioma cells by regulating the expression of cripto-1, subsequently
reducing the expression of MMP-2 and -9.
Discussion
The etiology, mechanisms, diagnosis and treatment of
gliomas have been previously investigated, however their origin
remains undefined and the clinical progress is slow (18,19).
Therefore, the search for the molecular mechanisms underlying the
pathological process of gliomas has emerged as a focus for
research, to aid in the diagnosis and treatment of gliomas.
Previous in-vivo and in-vitro studies
have demonstrated the oncogenic role of human cripto-1 (20,21),
however, the role of cripto-1 in gliomas is unclear. The present
study demonstrated that cripto-1 expression was significantly
increased in glioma tissue, which is consistent with the results of
Pilgaard et al (8) and
Tysnes et al (9). In
addition, the present study demonstrated that cripto-1 promoted the
proliferation and inhibited the apoptosis of glioma cells. Strizzi
et al (3) demonstrated that
inhibition of the cripto-1 expression in colon cancer cells
suppressed their growth and tumorigenesis potential in soft agar.
In another study, the use of cripto-1 blocking antibody suppressed
the cripto-nodal signaling system and inhibited tumor growth by 70%
in in-vivo models of testicular and colon cancer (22). Therefore, cripto-1 has a similar
growth-promoting and neoplastic role in gliomas as in other types
of cancer.
In the present study, the results of the cell
invasion assays demonstrated that cripto-1 promoted the invasion of
glioma cells and enhance the expression of MMP-2 and -9. Wu et
al (7) demonstrated that the
use of lentivirus-mediated small interfering RNA inhibited cripto-1
expression in nasopharyngeal carcinoma cell lines, suppressed cell
growth, and significantly reduced the invasion potential of
nasopharyngeal carcinoma cell lines. Strizzi et al (23) demonstrated that cripto-1 promoted
invasion and metastasis by regulating epithelial-mesenchymal
transition in mouse mammary epithelial cells cultured
in-vitro and mouse mammary tumor cells grown in-vivo.
Normanno et al (6)
indicated that MCF-7 breast cancer cells with increased expression
of teratocarcinoma-derived growth factor 1 exhibited significantly
improved apoptosis resistance, growth and proliferation
capabilities, and invasion and migration potential in-vitro.
These results are consistent with the observations of the current
study, supporting the oncogenic role of cripto-1.
Numerous studies have demonstrated that miRNAs serve
an important role in regulating numerous physiological and
pathological processes. Abnormally increased onco-miRNA expression
and abnormally reduced tumor suppressor miRNA expression are
associated with the development and progression of a variety of
types of cancer (24,25). The present study confirmed that
miR-15b was downregulated in glioma tissue, which is consistent
with previous observations (15).
In addition, previous studies demonstrated that certain miR-15
family members are downregulated in patients with chronic
lymphocytic leukemia, glioma and prostate cancer (11–13),
suggesting that the miR-15 family represents an important class of
tumor suppressors.
The present study demonstrated that the miR-15b
expression was negatively correlated with the cripto-1 expression
in glioma tissue, suggesting an association between the two
molecules. The results of the biological software analysis
indicated that miR-15b has binding sites on cripto-1, thus miR-15b
may regulate cripto-1 in a targeted manner. miR-15b significantly
inhibited cripto-1 expression in glioma cells following
transfection with the miR-15b mimics, and dual luciferase reporter
assays demonstrated that miR-15b directly targets cripto-1.
Additional tests indicated that miR-15b inhibits the proliferation
and invasion of glioma cells while promoting apoptosis, by
inhibiting the expression of MMP-2 and -9. Co-transfection with
miR-15b and cripto-1 overcame the action of miR-15b, however its
effect in invasion was significantly attenuated compared with the
cells transfected with cripto-1 overexpression vector alone. This
further indicated that miR-15b modulates the growth and invasion of
glioma cells by regulating cripto-1. A previous study demonstrated
that overexpression of miR-15b inhibited proliferation by arresting
cell cycle progression and inducing apoptosis, possibly by directly
targeting cyclin D1 in glioma cells (26). Xia et al (27) demonstrated that overexpression of
miR-15b resulted in cell cycle arrest at
G0/G1 phase, and suppression of miR-15b
expression resulted in a reduction of cell populations in
G0/G1 and a corresponding increase of cell
populations in the S phase. Zheng et al (28) demonstrated that miR-15b and miR-152
reduced glioma cell invasion and angiogenesis via neuropilin 2 and
MMP-3. Furthermore, Wu et al (29) demonstrated that downregulation of
microRNA-15b by the hepatitis B virus X enhanced hepatocellular
carcinoma proliferation via fucosyltransferase 2-induced Globo H
expression. These results indicate that miR-15b affects the growth
of tumor cells by regulating the expression of a series of
genes.
In conclusion, the miR-15b expression is negatively
associated with the cripto-1 expression in glioma cells. miR-15b
may subsequently impair growth and invasion of glioma cells through
targeted regulation of cripto-1. This discovery may provide novel
targets for the prevention and treatment of gliomas.
Acknowledgements
The current study was supported by the China Natural
Science Foundation (grant nos. 81201976 and 81000963), Jiangsu
Province's Natural Science Foundation (grant nos. BK2012670 and
BK20141256), Jiangsu Province's Health Department (grant no.
z201318) and Yancheng Medical Science Development Foundation (grant
nos. YK2013003 and YK2013019).
References
|
1
|
Germano I, Swiss V and Casaccia P: Primary
brain tumors, neural stem cell, and brain tumor cancer cells: Where
is the link? Neuropharmacology. 58:903–910. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bianco C, Strizzi L, Normanno N, Khan N
and Salomon DS: Cripto-1: An oncofetal gene with many faces. Curr
Top Dev Biol. 67:85–133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Strizzi L, Bianco C, Normanno N and
Salomon D: Cripto-1: A multifunctional modulator during
embryogenesis and oncogenesis. Oncogene. 24:5731–5741. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Luca A, Lamura L, Strizzi L, Roma C,
D'Antonio A, Margaryan N, Pirozzi G, Hsu MY, Botti G, Mari E, et
al: Expression and functional role of CRIPTO-1 in cutaneous
melanoma. Br J Cancer. 105:1030–1038. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yoon HJ, Hong JS, Shin WJ, Lee YJ, Hong
KO, Lee JI, Hong SP and Hong SD: The role of Cripto-1 in the
tumorigenesis and progression of oral squamous cell carcinoma. Oral
Oncol. 47:1023–1031. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Normanno N, De Luca A, Bianco C, Maiello
MR, Carriero MV, Rehman A, Wechselberger C, Arra C, Strizzi L,
Sanicola M, et al: Cripto-1 overexpression leads to enhanced
invasiveness and resistance to anoikis in human MCF-7 breast cancer
cells. J Cell Physiol. 198:31–39. 2004. View Article : Google Scholar
|
|
7
|
Wu Z, Li G, Wu L, Weng D, Li X and Yao K:
Cripto-1 overexpression is involved in the tumorigenesis of
nasopharyngeal carcinoma. BMC Cancer. 9:3152009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pilgaard L, Mortensen JH, Henriksen M,
Olesen P, Sørensen P, Laursen R, Vyberg M, Agger R, Zachar V, Moos
T, et al: Cripto-1 expression in glioblastoma multiforme. Brain
Pathol. 24:360–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tysnes BB, Satran HA, Mork SJ, Margaryan
NV, Eide GE, Petersen K, Strizzi L and Hendrix MJ: Age-dependent
association between protein expression of the embryonic stem cell
marker cripto-1 and survival of glioblastoma patients. Transl
Oncol. 6:732–741. 2013. View Article : Google Scholar
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bonci D, Coppola V, Musumeci M, Addario A,
Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C,
et al: The miR-15a-miR-16-1 cluster controls prostate cancer by
targeting multiple oncogenic activities. Nat Med. 14:1271–1277.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baraniskin A, Kuhnhenn J, Schlegel U,
Maghnouj A, Zöllner H, Schmiegel W, Hahn S and Schroers R:
Identification of microRNAs in the cerebrospinal fluid as biomarker
for the diagnosis of glioma. Neuro-oncol. 14:29–33. 2012.
View Article : Google Scholar :
|
|
14
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun G, Yan S, Shi L, Wan Z, Jiang N, Li M
and Guo J: Decreased expression of miR-15b in human gliomas is
associated with poor prognosis. Cancer Biother Radiopharm.
30:169–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Giese A, Bjerkvig R, Berens ME and
Westphal M: Cost of migration: Invasion of malignant gliomas and
implications for treatment. J Clin Oncol. 21:1624–1636. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Van Meir EG, Hadjipanayis CG, Norden AD,
Shu HK, Wen PY and Olson JJ: Exciting new advances in
neuro-oncology: The avenue to a cure for malignant glioma. CA
Cancer J Clin. 60:166–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhong XY, Zhang LH, Jia SQ, Shi T, Niu ZJ,
Du H, Zhang GG, Hu Y, Lu AP, Li JY, et al: Positive association of
up-regulated Cripto-1 and down-regulated E-cadherin with tumour
progression and poor prognosis in gastric cancer. Histopathology.
52:560–568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gong YP, Yarrow PM, Carmalt HL, Kwun SY,
Kennedy CW, Lin BP, Xing PX and Gillett DJ: Overexpression of
Cripto and its prognostic significance in breast cancer: A study
with long-term survival. Eur J Surg Oncol. 33:438–443. 2007.
View Article : Google Scholar
|
|
22
|
Adkins HB, Bianco C, Schiffer SG, Rayhorn
P, Zafari M, Cheung AE, Orozco O, Olson D, De Luca A, Chen LL, et
al: Antibody blockade of the Cripto CFC domain suppresses tumor
cell growth in vivo. J Clin Invest. 112:575–587. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strizzi L, Bianco C, Normanno N, Seno M,
Wechselberger C, Wallace-Jones B, Khan NI, Hirota M, Sun Y,
Sanicola M, et al: Epithelial mesenchymal transition is a
characteristic of hyperplasias and tumors in mammary gland from
MMTV-Cripto-1 transgenic mice. J Cell Physiol. 201:266–276. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar :
|
|
25
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun G, Shi L, Yan S, Wan Z, Jiang N, Fu L,
Li M and Guo J: MiR-15b targets cyclin D1 to regulate proliferation
and apoptosis in glioma cells. BioMed Res Int. 2014:6878262014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang
M, Li D, Zhao Y, Ge R, Li G, et al: MicroRNA-15b regulates cell
cycle progression by targeting cyclins in glioma cells. Biochem
Biophys Res Commun. 380:205–210. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng X, Chopp M, Lu Y, Buller B and Jiang
F: MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis
via NRP-2 and MMP-3. Cancer Lett. 329:146–154. 2013. View Article : Google Scholar :
|
|
29
|
Wu CS, Yen CJ, Chou RH, Chen JN, Huang WC,
Wu CY and Yu YL: Downregulation of microRNA-15b by hepatitis B
virus X enhances hepatocellular carcinoma proliferation via
fucosyltransferase 2-induced Globo H expression. Int J Cancer.
134:1638–1647. 2014. View Article : Google Scholar
|